
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33186.1 750 aa
(750 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
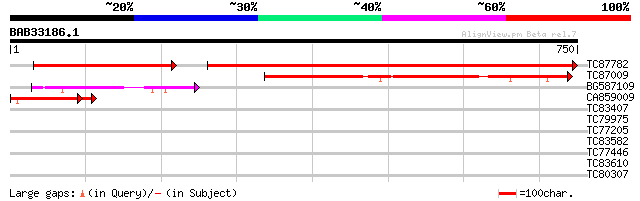
Score E
Sequences producing significant alignments: (bits) Value
TC87782 homologue to SP|P05310|PSAA_PEA Photosystem I P700 chlor... 994 0.0
TC87009 homologue to SP|P58385|PSAB_LOTJA Photosystem I P700 chl... 378 e-105
BG587109 homologue to GP|22091671|em photosystem1 subunit A {Pin... 222 5e-58
CA859009 homologue to PIR|S00703|S00 photosystem I protein A1 - ... 195 6e-55
TC83407 similar to GP|17385678|dbj|BAB78631. putative co-repress... 31 2.0
TC79975 similar to GP|16417944|gb|AAL18924.1 acetyl Co-A acetylt... 30 2.6
TC77205 similar to GP|19347753|gb|AAL86301.1 putative RING-H2 zi... 30 2.6
TC83582 similar to GP|22128707|gb|AAM92819.1 putative ABC transp... 30 4.5
TC77446 homologue to GP|303750|dbj|BAA02116.1| GTP-binding prote... 29 5.8
TC83610 similar to SP|Q9LNQ4|ALA4_ARATH Potential phospholipid-t... 28 9.9
TC80307 similar to GP|20160990|dbj|BAB89924. putative protein ki... 28 9.9
>TC87782 homologue to SP|P05310|PSAA_PEA Photosystem I P700 chlorophyll A
apoprotein A1 (PsaA) (PSI-A). [Garden pea] {Pisum
sativum}, partial (64%)
Length = 2152
Score = 994 bits (2571), Expect = 0.0
Identities = 474/489 (96%), Positives = 482/489 (97%)
Frame = +2
Query: 262 FTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKD 321
FTLNW+KYADFLTFR GLDP+TGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGI+D
Sbjct: 5 FTLNWSKYADFLTFRXGLDPLTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIRD 184
Query: 322 ILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDY 381
ILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMY+MPPYPYLATDY
Sbjct: 185 ILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYAMPPYPYLATDY 364
Query: 382 GTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWV 441
GTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDR+LRHRDAIISHLNWV
Sbjct: 365 GTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRILRHRDAIISHLNWV 544
Query: 442 CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPGITAPGATT 501
CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQP+FAQWIQNTHALAPG TAPGAT
Sbjct: 545 CIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPVFAQWIQNTHALAPGTTAPGATA 724
Query: 502 GTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLI 561
TSLTWGGGDLVAVG KVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLI
Sbjct: 725 STSLTWGGGDLVAVGGKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLI 904
Query: 562 PDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSIS 621
PDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYN+ISVVIFHFSWKMQSDVWGSI+
Sbjct: 905 PDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNSISVVIFHFSWKMQSDVWGSIN 1084
Query: 622 DQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFS 681
DQGVVTHITGGNFAQSSITINGWLRDFLWAQA VIQSYGSSLSAYGLFFLGAHFVWAFS
Sbjct: 1085DQGVVTHITGGNFAQSSITINGWLRDFLWAQAXXVIQSYGSSLSAYGLFFLGAHFVWAFS 1264
Query: 682 LMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGGIATTWAF 741
LMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGGIATTWAF
Sbjct: 1265LMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLLGGIATTWAF 1444
Query: 742 FLARIIAVG 750
FLARIIAVG
Sbjct: 1445FLARIIAVG 1471
Score = 165 bits (418), Expect = 5e-41
Identities = 89/194 (45%), Positives = 120/194 (60%), Gaps = 5/194 (2%)
Frame = +3
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 1521 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 1697
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 1698 HVAWQGNFEAWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 1877
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L + + L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 1878 IGLRTNEDLYTGALFLLFLSVISLLAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 2057
Query: 207 SLSWAGHQVHVSLP 220
SL+WAGH VHV++P
Sbjct: 2058 SLAWAGHLVHVAIP 2099
>TC87009 homologue to SP|P58385|PSAB_LOTJA Photosystem I P700 chlorophyll A
apoprotein A2 (PsaB) (PSI-B). {Lotus japonicus}, partial
(56%)
Length = 1445
Score = 378 bits (971), Expect = e-105
Identities = 194/423 (45%), Positives = 260/423 (60%), Gaps = 16/423 (3%)
Frame = +1
Query: 338 LYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGF 397
LY+ + S H QL + L LG +T +VA HMYS+P Y ++A D+ TQ +L+THH +I GF
Sbjct: 1 LYDTINNSIHFQLGLALXSLGVITSLVAQHMYSLPAYAFIAQDFTTQAALYTHHQYIAGF 180
Query: 398 LIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHND 457
++ GA AH AIF +RDY+P +++L R+L H++AIISHL+W +FLGFH+ GLY+HND
Sbjct: 181 IMTGAFAHGAIFFIRDYNPEQNEDNVLARMLEHKEAIISHLSWASLFLGFHTLGLYVHND 360
Query: 458 TMSALGRPQDMFSDTAIQLQPIFAQWIQNTH--------ALAPGITAPGATTGTSLTWGG 509
M A G P+ I ++PIFAQWIQ+ H L +P + G S+ W
Sbjct: 361 VMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTSYGFDVLLSSTNSPALSAGRSI-WLP 522
Query: 510 GDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANLGF 569
G L A+ L + +G DFLVHH A +H T LIL+KG L AR S+L+PDK + G+
Sbjct: 523 GWLNAINENSNSLFLTIGPGDFLVHHAIALGLHTTTLILVKGALDARGSKLMPDKKDFGY 702
Query: 570 RFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQSDVWGSISDQGVVTHI 629
FPCDGPGRGGTC +SAWD +L +FWM N I V F++ WK + G++S
Sbjct: 703 SFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTFYWHWKHITLWQGNVS-------- 858
Query: 630 TGGNFAQSSITINGWLRDFLWAQASQVIQSYG----SSLSAYGLFFLGAHFVWAFSLMFL 685
F +SS + GWLRD+LW +SQ+I Y +SLS + FL H VWA MFL
Sbjct: 859 ---QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMNSLSVWAWMFLFGHLVWATGFMFL 1029
Query: 686 FSGRGYWQELIESIVWAHNKLKVAP----ATQPRALSIVQGRAVGVTHYLLGGIATTWAF 741
S RGYWQELIE++ WAH + +A +P ALSIVQ R VG+ H+ +G I T AF
Sbjct: 1030ISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALSIVQARLVGLVHFSVGYIFTYAAF 1209
Query: 742 FLA 744
+A
Sbjct: 1210LIA 1218
>BG587109 homologue to GP|22091671|em photosystem1 subunit A {Pinus
parviflora}, partial (29%)
Length = 791
Score = 222 bits (565), Expect = 5e-58
Identities = 138/257 (53%), Positives = 153/257 (58%), Gaps = 35/257 (13%)
Frame = +2
Query: 29 PGHFSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLEDISR-----------KIFSAHFGQ 77
PG S+ GP + L D+ + HTS++ + R K+FSAHFGQ
Sbjct: 23 PGQTSK*FFYGPGGPP-LAALSRDSCNGSLHTSEV*TMHRVIPVIWRKSLEKVFSAHFGQ 199
Query: 78 LSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFRGIQI 137
LSIIFLWLSGMYFHGARFSNYEA LSDPTHI PSAQVVWPIVGQEILNGDVGGGFRGIQI
Sbjct: 200 LSIIFLWLSGMYFHGARFSNYEACLSDPTHIGPSAQVVWPIVGQEILNGDVGGGFRGIQI 379
Query: 138 TSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLA-W------FQD 190
TSGFFQIWRASGIT+ Y+ P+LA W F
Sbjct: 380 TSGFFQIWRASGITN-------------------------YNFIVPQLAHWSSQP*CFLL 484
Query: 191 VESMLNHHLAGLLG-----------------LGSLSWAGHQVHVSLPINQFLNAGVDPKE 233
V S++ L LG LG + VHVSLPINQFLNAGVDPKE
Sbjct: 485 VGSIITKQLQNWLGSKM*NLC*ITI*QGY*DLGPFLGQDNPVHVSLPINQFLNAGVDPKE 664
Query: 234 IPLPHEFILNRDLLAQL 250
I LPHEF+LNRDLLAQL
Sbjct: 665 ILLPHEFLLNRDLLAQL 715
>CA859009 homologue to PIR|S00703|S00 photosystem I protein A1 - garden pea
chloroplast, partial (16%)
Length = 823
Score = 195 bits (495), Expect(2) = 6e-55
Identities = 93/104 (89%), Positives = 96/104 (91%), Gaps = 8/104 (7%)
Frame = +2
Query: 1 MIIRSPEP--------EVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHAD 52
MIIRSPEP EVKILVDRDPIKTSFE+WAKPGHFSRTIAKGPDTTTWIWNLHAD
Sbjct: 455 MIIRSPEPKVKILVDPEVKILVDRDPIKTSFEQWAKPGHFSRTIAKGPDTTTWIWNLHAD 634
Query: 53 AHDFDSHTSDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFS 96
AHDFDSHTSDLE+ISRK+FSAHFGQLSIIFLWLSGMYFHGARFS
Sbjct: 635 AHDFDSHTSDLEEISRKVFSAHFGQLSIIFLWLSGMYFHGARFS 766
Score = 38.5 bits (88), Expect(2) = 6e-55
Identities = 18/26 (69%), Positives = 19/26 (72%)
Frame = +1
Query: 90 FHGARFSNYEAWLSDPTHIRPSAQVV 115
F F NYEAWL+ THIRPSAQVV
Sbjct: 745 FPWCSFFNYEAWLN*STHIRPSAQVV 822
>TC83407 similar to GP|17385678|dbj|BAB78631. putative co-repressor protein
{Oryza sativa (japonica cultivar-group)}, partial (15%)
Length = 921
Score = 30.8 bits (68), Expect = 2.0
Identities = 16/46 (34%), Positives = 26/46 (55%), Gaps = 3/46 (6%)
Frame = -2
Query: 404 AHAAIFMVRDYDPTTRYNDLLD---RVLRHRDAIISHLNWVCIFLG 446
A + +F VR R + +L R LR+RD ++ + W+C+FLG
Sbjct: 191 AGSTLFTVRKVQFIYRLSYILVLA*RHLRNRDILVGNSRWICLFLG 54
>TC79975 similar to GP|16417944|gb|AAL18924.1 acetyl Co-A acetyltransferase
{Hevea brasiliensis}, partial (73%)
Length = 1019
Score = 30.4 bits (67), Expect = 2.6
Identities = 17/46 (36%), Positives = 23/46 (49%)
Frame = +2
Query: 652 QASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIE 697
Q S IQS+ +SA G HF W + +FSGRG L++
Sbjct: 602 QDSYAIQSFERGISAQN----GGHFAWEIVPVEIFSGRGKPSTLVD 727
>TC77205 similar to GP|19347753|gb|AAL86301.1 putative RING-H2 zinc finger
protein ATL6 {Arabidopsis thaliana}, partial (38%)
Length = 1317
Score = 30.4 bits (67), Expect = 2.6
Identities = 12/33 (36%), Positives = 21/33 (63%)
Frame = +1
Query: 114 VVWPIVGQEILNGDVGGGFRGIQITSGFFQIWR 146
V+ P+V +E++ G + GG+ G+ +G F WR
Sbjct: 913 VIEPVVVREVVEGSL*GGWTGVYYQTGGFLQWR 1011
>TC83582 similar to GP|22128707|gb|AAM92819.1 putative ABC transporter
{Oryza sativa (japonica cultivar-group)}, partial (52%)
Length = 1158
Score = 29.6 bits (65), Expect = 4.5
Identities = 30/123 (24%), Positives = 52/123 (41%), Gaps = 23/123 (18%)
Frame = +2
Query: 45 WIWNLHADAHDFDSHTSDLEDISRKIFSAHFGQLS-------------IIFLWLSG--MY 89
WI +LH + D S D++ R+ + H+G + I+ +LSG Y
Sbjct: 599 WICHLHVNWVDSPSFVEDMKVFQRERLNGHYGVTAFVISNTLSATPFLILITFLSGTICY 778
Query: 90 FH---GARFSNYE----AWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFRGI-QITSGF 141
F FS+Y A + T + + IV ++ +G G +GI + SG+
Sbjct: 779 FMVNLHPGFSHYVFFVLALYASVTVVESLMMAIASIVPNFLMGIIIGAGIQGIFMLVSGY 958
Query: 142 FQI 144
F++
Sbjct: 959 FKL 967
>TC77446 homologue to GP|303750|dbj|BAA02116.1| GTP-binding protein {Pisum
sativum}, complete
Length = 1080
Score = 29.3 bits (64), Expect = 5.8
Identities = 28/103 (27%), Positives = 50/103 (48%), Gaps = 9/103 (8%)
Frame = -2
Query: 441 VCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQ-LQPIF--------AQWIQNTHALA 491
+C F G H+F ++ + + + ++F+ A+ +QP+F + + N +A++
Sbjct: 527 LCCFRG-HNF--FVREIALVSN*KLVNVFTCIAVNFIQPLFNIVKALLVSDIVNNNYAMS 357
Query: 492 PGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVH 534
P I A + T LT +L G LP+ L +ADF VH
Sbjct: 356 PTIVAASDSPETFLTSSVPNLKFNG-----LPVLLNSADFKVH 243
>TC83610 similar to SP|Q9LNQ4|ALA4_ARATH Potential phospholipid-transporting
ATPase 4 (EC 3.6.3.1) (Aminophospholipid flippase 4).,
partial (30%)
Length = 1131
Score = 28.5 bits (62), Expect = 9.9
Identities = 12/32 (37%), Positives = 18/32 (55%), Gaps = 1/32 (3%)
Frame = +3
Query: 590 VFLGLFWMYNA-ISVVIFHFSWKMQSDVWGSI 620
+F + W N IS+ + HF+W +WGSI
Sbjct: 789 MFTCIIWAVNCQISLTMSHFTWIQHLFIWGSI 884
>TC80307 similar to GP|20160990|dbj|BAB89924. putative protein kinase {Oryza
sativa (japonica cultivar-group)}, partial (69%)
Length = 1929
Score = 28.5 bits (62), Expect = 9.9
Identities = 18/57 (31%), Positives = 28/57 (48%), Gaps = 1/57 (1%)
Frame = -3
Query: 512 LVAVGNKVALLPIPLGTADFL-VHHIHAFTIHVTVLILLKGVLFARSSRLIPDKANL 567
L +GN LP PLG + L + +H+ + + L L L R S L+ K++L
Sbjct: 1573 LALIGNSTTSLPCPLGLVEALELRQLHSLGVWLLDLCLGSDELAERLSELLLRKSDL 1403
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.141 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,384,360
Number of Sequences: 36976
Number of extensions: 457952
Number of successful extensions: 2866
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 2126
Number of HSP's successfully gapped in prelim test: 93
Number of HSP's that attempted gapping in prelim test: 699
Number of HSP's gapped (non-prelim): 2274
length of query: 750
length of database: 9,014,727
effective HSP length: 103
effective length of query: 647
effective length of database: 5,206,199
effective search space: 3368410753
effective search space used: 3368410753
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Description of BAB33186.1

