
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
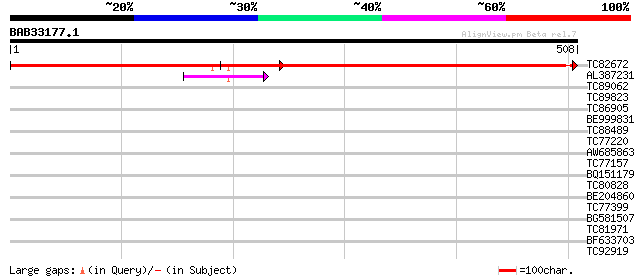
Query= BAB33177.1 508 aa
(508 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82672 maturase [Medicago truncatula] 509 e-145
AL387231 42 7e-04
TC89062 weakly similar to GP|5669672|gb|AAD46419.1| receptor-lik... 32 0.44
TC89823 31 1.3
TC86905 similar to GP|11908126|gb|AAG41492.1 unknown protein {Ar... 30 1.7
BE999831 30 2.2
TC88489 similar to GP|14334978|gb|AAK59666.1 unknown protein {Ar... 30 2.9
TC77220 similar to GP|22597162|gb|AAN03468.1 bZIP transcription ... 30 2.9
AW685863 similar to GP|10798819|dbj putative damage-specific DNA... 29 3.7
TC77157 homologue to GP|13877717|gb|AAK43936.1 OsNAC6 protein-li... 29 3.7
BQ151179 29 3.7
TC80828 29 4.9
BE204860 29 4.9
TC77399 similar to GP|16930765|gb|AAL32042.1 photosystem II reac... 29 4.9
BG581507 similar to GP|7290473|gb| CG15570 gene product {Drosoph... 29 4.9
TC81971 similar to PIR|B96836|B96836 unknown protein T21F11.23 [... 28 6.4
BF633703 similar to GP|19310633|gb| unknown protein {Arabidopsis... 28 8.3
TC92919 homologue to GP|343533|gb|AAA32115.1|| apocytochrome b {... 28 8.3
>TC82672 maturase [Medicago truncatula]
Length = 1559
Score = 509 bits (1311), Expect = e-145
Identities = 253/319 (79%), Positives = 272/319 (84%)
Frame = +3
Query: 190 IFIPQKSIYTFSKKNPRFFFFLYNFYVCEYESIFFFLRTKSFHLRLKSFRVFFERIFFYA 249
+ +P+ FSK NPR FLYNFYVCEYESIF FLR KS HLRLKSF VFFERIFFYA
Sbjct: 606 LLLPKTRFLLFSKSNPRLSLFLYNFYVCEYESIFLFLRNKSSHLRLKSFNVFFERIFFYA 785
Query: 250 KKGHLVEVFVKDFFSTLTFFKDPFIHYVRYQGKSILASKNLPILMNKWKYYFIHLWQCYF 309
K+ HLVEVF KDF TLTFFKDP IHYVRYQGK ILASKN P LMNKWK+YFIHLWQ +F
Sbjct: 786 KREHLVEVFAKDFSYTLTFFKDPLIHYVRYQGKCILASKNSPFLMNKWKHYFIHLWQGFF 965
Query: 310 DVWSQPRTIRINQLSEYSFHFLGYFLKVGLKHSVVRGQMLQNGFLIKIIIKKLDIIVPII 369
VWSQPRT+ INQLSE+SF LGYFL V + SVVR QMLQN FLI+I KKLD+IVPII
Sbjct: 966 YVWSQPRTMNINQLSEHSFQLLGYFLNVRVNRSVVRSQMLQNTFLIEIFNKKLDLIVPII 1145
Query: 370 PIIRLLAKSKFCNVLGNPLSKPSWADLSDFEIIARFLRICRNLSHYYNGSSKKKSLYRIK 429
P+IR LAK+KFCNVLG+P+SKP WAD SDF+II RFLRICRNLSHYYNGSSKKKSLYRIK
Sbjct: 1146PLIRSLAKAKFCNVLGHPISKPVWADSSDFDIIDRFLRICRNLSHYYNGSSKKKSLYRIK 1325
Query: 430 YILRLSCIKTLACKHKSTVRAFSKRLGSEELLEEFFTEEEEILSLIFPTTSSTLRRLHRN 489
YILRLSCIKTLACKHKSTVRAF KR GSEELLEEFFTEEEEILSLIFP SSTL RL+RN
Sbjct: 1326YILRLSCIKTLACKHKSTVRAFLKRSGSEELLEEFFTEEEEILSLIFPRDSSTLHRLNRN 1505
Query: 490 RIWYLDILFSNDNDLINHD 508
RIWYLDILFS NDL+N +
Sbjct: 1506RIWYLDILFS--NDLVNDE 1556
Score = 304 bits (778), Expect = 6e-83
Identities = 163/263 (61%), Positives = 186/263 (69%), Gaps = 17/263 (6%)
Frame = +1
Query: 1 MEEYQLYLELDRSRQQDFLYPLVFHEYIYGLAYSHDLNRSIFVENFGYDNKYSLLIVKRL 60
M+EYQ+YLE RSRQQDFLYPL+F EYIYGLAYSH+ NRS+F+EN G D+KYSLLIVKRL
Sbjct: 37 MKEYQVYLERARSRQQDFLYPLIFREYIYGLAYSHNFNRSVFLENVGSDSKYSLLIVKRL 216
Query: 61 ITRMYQQNHLIISANDSNKNRFLRYNKNFYSQIISEGFAIIVEILFSLQLSSSLEEAEII 120
ITRMYQQNHLIISANDSNKN F YNKN YSQIISEGFAI+VEI F L+LSSSLEEAEII
Sbjct: 217 ITRMYQQNHLIISANDSNKNPFWGYNKNIYSQIISEGFAIVVEIPFFLELSSSLEEAEII 396
Query: 121 KSYKNLRSIHSIFPFFEDKVTYLNYISDIRVPYPIHLEILVQILRYWVKDAPLFHLLRLF 180
KSYKNLRSIHSIFPF EDK TY NY+SDIR+PYPIHLEILVQILRYWVKDAP F + +
Sbjct: 397 KSYKNLRSIHSIFPFLEDKFTYFNYVSDIRIPYPIHLEILVQILRYWVKDAPFFSFITIV 576
Query: 181 ---------LYDYCNWNSIFIPQ--------KSIYTFSKKNPRFFFFLYNFYVCEYESIF 223
Y Y + F + I+ + N FF++ N + + +
Sbjct: 577 SL*F**LE*FYYYQKLDFYFFQKVIRDYPCSSIIFMYVNMNLSSFFYVINPLIYD*NLLT 756
Query: 224 FFLRTKSFHLRLKSFRVFFERIF 246
FFL F + F RIF
Sbjct: 757 FFLNEFFFMQKENIL*KFLLRIF 825
>AL387231
Length = 524
Score = 41.6 bits (96), Expect = 7e-04
Identities = 27/93 (29%), Positives = 41/93 (44%), Gaps = 16/93 (17%)
Frame = +1
Query: 156 HLEILVQILRYWVKDAPLFHLLRLFLYDYCNWNSIFIPQ----------------KSIYT 199
HL I IL +W+ + RLFL Y N +F+ + SI
Sbjct: 97 HLTINFSILVFWMVNEANVTANRLFLIHYVNLFRLFVVKHMFF*RQLYIFSCFTYSSIVQ 276
Query: 200 FSKKNPRFFFFLYNFYVCEYESIFFFLRTKSFH 232
F K F F Y F++ +Y +++FFL+ K F+
Sbjct: 277 FGKNMVFSFLFFYAFFL*QYINMYFFLKKKKFY 375
>TC89062 weakly similar to GP|5669672|gb|AAD46419.1| receptor-like kinase
{Hordeum vulgare}, partial (26%)
Length = 1309
Score = 32.3 bits (72), Expect = 0.44
Identities = 16/41 (39%), Positives = 24/41 (58%)
Frame = +2
Query: 188 NSIFIPQKSIYTFSKKNPRFFFFLYNFYVCEYESIFFFLRT 228
NSI Q ++YT +KK P FFF + + Y ++F+ L T
Sbjct: 2 NSIIYNQTNLYTMNKKQP-FFFLFSSPLIYSYITLFYLLLT 121
>TC89823
Length = 1017
Score = 30.8 bits (68), Expect = 1.3
Identities = 19/69 (27%), Positives = 33/69 (47%), Gaps = 6/69 (8%)
Frame = -3
Query: 165 RYWVKDAPLFHLLRLFLYDYC--NWNSIFIPQKSIYTFSKKNPR----FFFFLYNFYVCE 218
R +V D + H L L LY+YC + + S+++F+ R F F L V +
Sbjct: 622 RSYVSDISIHHCLSLQLYEYCILRFRTTHPSLTSLFSFNNTQTRS*ILFHFLLTLKQVLK 443
Query: 219 YESIFFFLR 227
Y +F +++
Sbjct: 442 YSVVFHYVK 416
>TC86905 similar to GP|11908126|gb|AAG41492.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 824
Score = 30.4 bits (67), Expect = 1.7
Identities = 24/92 (26%), Positives = 38/92 (41%), Gaps = 4/92 (4%)
Frame = +1
Query: 168 VKDAPLFHLLRLFLYDYCNWNSIFIPQKSIYTFSKKNPRFFFFLYNFYVCEYESIFFFLR 227
V P+F L F NS F+ +++ T KN + FL+ Y+ + F+FL+
Sbjct: 73 VSTHPVFLFLLFFPSLIFTPNSFFLAPRNLDTTKSKNSSYPDFLFQKYLLGFFLFFYFLK 252
Query: 228 TKSFHL----RLKSFRVFFERIFFYAKKGHLV 255
F K + F +I + G LV
Sbjct: 253 FHFFFTFSVQEKKKINIEFLKINCFLSFGFLV 348
>BE999831
Length = 346
Score = 30.0 bits (66), Expect = 2.2
Identities = 21/71 (29%), Positives = 32/71 (44%), Gaps = 1/71 (1%)
Frame = -3
Query: 209 FFLYNFYVCEYESI-FFFLRTKSFHLRLKSFRVFFERIFFYAKKGHLVEVFVKDFFSTLT 267
+ L F+V +I F SFH+ KS +F ++ KKG F +F ST
Sbjct: 299 YHLSTFFVHHINNINTIFFNNFSFHITTKSIDIFMMVQYWLMKKG----TFSNNFVSTFV 132
Query: 268 FFKDPFIHYVR 278
K+ F+ V+
Sbjct: 131 SSKNKFVVEVK 99
>TC88489 similar to GP|14334978|gb|AAK59666.1 unknown protein {Arabidopsis
thaliana}, partial (47%)
Length = 1187
Score = 29.6 bits (65), Expect = 2.9
Identities = 23/80 (28%), Positives = 33/80 (40%), Gaps = 15/80 (18%)
Frame = -1
Query: 187 WNSIFIPQKSIYTFSKKNPRFFFFLYNFYVCEYESIF--------FFLRTKS-------F 231
W PQ+ I+ NP FFFL+NF + + + FFL K F
Sbjct: 473 WQK*HTPQRQIFFLHLFNPFLFFFLHNFLTTLFTTFW*SFNLFGLFFLFLKKGVP*NCFF 294
Query: 232 HLRLKSFRVFFERIFFYAKK 251
+L K + F ++F KK
Sbjct: 293 NL*SKLNQSLFLKLFEIRKK 234
>TC77220 similar to GP|22597162|gb|AAN03468.1 bZIP transcription factor ATB2
{Glycine max}, partial (65%)
Length = 1341
Score = 29.6 bits (65), Expect = 2.9
Identities = 12/29 (41%), Positives = 18/29 (61%)
Frame = +1
Query: 203 KNPRFFFFLYNFYVCEYESIFFFLRTKSF 231
K+P FFF L +F + Y+ +FF+ SF
Sbjct: 28 KDPPFFFSLLSF*ISMYDLYYFFINLSSF 114
>AW685863 similar to GP|10798819|dbj putative damage-specific DNA binding
protein 2 {Oryza sativa (japonica cultivar-group)},
partial (9%)
Length = 485
Score = 29.3 bits (64), Expect = 3.7
Identities = 18/43 (41%), Positives = 24/43 (54%)
Frame = -1
Query: 200 FSKKNPRFFFFLYNFYVCEYESIFFFLRTKSFHLRLKSFRVFF 242
F NP FFFF ++F+ +E FF R F +R+ SF V F
Sbjct: 290 FIFNNPIFFFFEWSFF---FEWFFFIRRRTLFRVRV-SFNVNF 174
>TC77157 homologue to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like protein
{Arabidopsis thaliana}, partial (66%)
Length = 1998
Score = 29.3 bits (64), Expect = 3.7
Identities = 18/50 (36%), Positives = 29/50 (58%)
Frame = -2
Query: 322 QLSEYSFHFLGYFLKVGLKHSVVRGQMLQNGFLIKIIIKKLDIIVPIIPI 371
Q+ E+ ++ L++G K SV G I III+KL I+VP++P+
Sbjct: 1268 QVHEHILQWIS-ILELGAKTSVQSG--------INIIIRKLKILVPLVPL 1146
>BQ151179
Length = 786
Score = 29.3 bits (64), Expect = 3.7
Identities = 32/119 (26%), Positives = 48/119 (39%)
Frame = -3
Query: 173 LFHLLRLFLYDYCNWNSIFIPQKSIYTFSKKNPRFFFFLYNFYVCEYESIFFFLRTKSFH 232
++ L+ LF Y + I + I K FF+ FY+ Y I F L F
Sbjct: 379 IYILVNLFSSSYL*YIYIHTKSQWIN*LK*KK*IIFFYSILFYLTYYNPI*FLLLFSLFP 200
Query: 233 LRLKSFRVFFERIFFYAKKGHLVEVFVKDFFSTLTFFKDPFIHYVRYQGKSILASKNLP 291
L S + +FF H ++ +K FF L F + I+ + Q K + NLP
Sbjct: 199 L---SLSITIY*LFFILYLKHTLKPLIK*FF--LIKFFNTIIYK*QKQKKLYIKKYNLP 38
>TC80828
Length = 721
Score = 28.9 bits (63), Expect = 4.9
Identities = 13/38 (34%), Positives = 21/38 (55%)
Frame = +1
Query: 301 FIHLWQCYFDVWSQPRTIRINQLSEYSFHFLGYFLKVG 338
F+ W CYF+ T + +Q++ + FHF +FL G
Sbjct: 403 FLLAWTCYFN------TSKWSQMNSFLFHFYLFFLLFG 498
>BE204860
Length = 273
Score = 28.9 bits (63), Expect = 4.9
Identities = 16/62 (25%), Positives = 26/62 (41%), Gaps = 3/62 (4%)
Frame = -2
Query: 168 VKDAPLFHLLRLFLYDYCNWNSIF---IPQKSIYTFSKKNPRFFFFLYNFYVCEYESIFF 224
VKD FH ++ C WN ++ IP+ ++ K+ Y Y+ +FF
Sbjct: 248 VKDFVQFHQGTTWVVHICRWNDVY*NGIPRTKLF*IGHKSKVLLLGKYT*YIDPLVDVFF 69
Query: 225 FL 226
L
Sbjct: 68 TL 63
>TC77399 similar to GP|16930765|gb|AAL32042.1 photosystem II reaction center
{Retama raetam}, complete
Length = 718
Score = 28.9 bits (63), Expect = 4.9
Identities = 15/45 (33%), Positives = 24/45 (53%)
Frame = +1
Query: 187 WNSIFIPQKSIYTFSKKNPRFFFFLYNFYVCEYESIFFFLRTKSF 231
W PQK ++ + FF L N+Y+C Y +F+FL + +F
Sbjct: 505 WIVFVKPQKDMHLRNWTVQSFFVVLKNYYLCIY--LFWFLFSSTF 633
>BG581507 similar to GP|7290473|gb| CG15570 gene product {Drosophila
melanogaster}, partial (2%)
Length = 562
Score = 28.9 bits (63), Expect = 4.9
Identities = 34/113 (30%), Positives = 47/113 (41%), Gaps = 20/113 (17%)
Frame = -1
Query: 167 WVKDAPLFHLLRLFLYDYCNWNSI------FIPQKSI----YTFSKK-NPRFFFFLYNFY 215
W K H R F Y + N+N F QKSI +FSK N F FF N
Sbjct: 526 WHKFGFFIHKFRFFFYPFINFNFFG*QVFNFFIQKSINKP*NSFSKHGNFEFIFFSPNSR 347
Query: 216 V-CEYES----IFFFLRTKSFH----LRLKSFRVFFERIFFYAKKGHLVEVFV 259
V C +E+ F FL S + F +F+ + F+ G ++E+ V
Sbjct: 346 VNCRFENGWVLEFDFLSPNSRSGKPIILDSHFSIFYLKFSFFWVMGFVLEMKV 188
>TC81971 similar to PIR|B96836|B96836 unknown protein T21F11.23 [imported] -
Arabidopsis thaliana, partial (13%)
Length = 721
Score = 28.5 bits (62), Expect = 6.4
Identities = 12/36 (33%), Positives = 19/36 (52%)
Frame = +3
Query: 210 FLYNFYVCEYESIFFFLRTKSFHLRLKSFRVFFERI 245
F N C+YE+ F F K L ++ +F+ER+
Sbjct: 600 FCSNILQCKYEATFLFSFVKLLPLNFENVYLFYERV 707
>BF633703 similar to GP|19310633|gb| unknown protein {Arabidopsis thaliana},
partial (12%)
Length = 471
Score = 28.1 bits (61), Expect = 8.3
Identities = 14/38 (36%), Positives = 24/38 (62%)
Frame = -2
Query: 412 LSHYYNGSSKKKSLYRIKYILRLSCIKTLACKHKSTVR 449
++++YN S K + + I Y+ +LSC +T K K+T R
Sbjct: 464 IAYFYNRSKIKPNSFHISYLHKLSC-RTYGNKLKTTDR 354
>TC92919 homologue to GP|343533|gb|AAA32115.1|| apocytochrome b {Trypanosoma
brucei}, partial (3%)
Length = 626
Score = 28.1 bits (61), Expect = 8.3
Identities = 17/65 (26%), Positives = 33/65 (50%)
Frame = +2
Query: 230 SFHLRLKSFRVFFERIFFYAKKGHLVEVFVKDFFSTLTFFKDPFIHYVRYQGKSILASKN 289
S+ ++K +++ + FY KG+ + ++ +FS L+FF IH + L+S
Sbjct: 17 SYWTKIK*YKI----VLFYKFKGYYCQFKIQLYFSLLSFFLY*TIHQIYSISHLFLSSHT 184
Query: 290 LPILM 294
L L+
Sbjct: 185 LSYLL 199
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.330 0.145 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,958,701
Number of Sequences: 36976
Number of extensions: 334020
Number of successful extensions: 3121
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 3050
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3104
length of query: 508
length of database: 9,014,727
effective HSP length: 100
effective length of query: 408
effective length of database: 5,317,127
effective search space: 2169387816
effective search space used: 2169387816
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 60 (27.7 bits)
Description of BAB33177.1

