
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
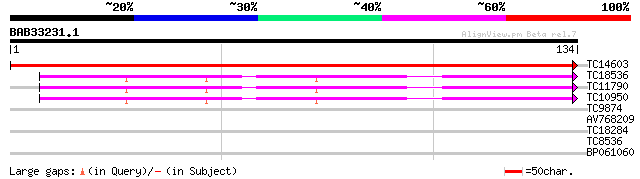
Query= BAB33231.1 134 aa
(134 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14603 UP|RR8_LOTJA (Q9BBQ1) Chloroplast 30S ribosomal protein ... 267 4e-73
TC18536 homologue to UP|AAK62367 (AAK62367) 40S ribosomal protei... 43 2e-05
TC11790 homologue to UP|AAK62367 (AAK62367) 40S ribosomal protei... 42 3e-05
TC10950 homologue to UP|AAK62367 (AAK62367) 40S ribosomal protei... 42 3e-05
TC9874 similar to UP|Q8VZJ1 (Q8VZJ1) AT5g09790/F17I14_20, partia... 27 1.2
AV768209 26 2.0
TC18284 homologue to UP|TBA3_ARATH (P20363) Tubulin alpha-3/alph... 25 4.5
TC8536 weakly similar to UP|AAR92224 (AAR92224) Cystatin, partia... 24 10.0
BP061060 24 10.0
>TC14603 UP|RR8_LOTJA (Q9BBQ1) Chloroplast 30S ribosomal protein S8,
complete
Length = 609
Score = 267 bits (683), Expect = 4e-73
Identities = 134/134 (100%), Positives = 134/134 (100%)
Frame = +3
Query: 1 MGKDTIADIITSIRNADMNRKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLV 60
MGKDTIADIITSIRNADMNRKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLV
Sbjct: 84 MGKDTIADIITSIRNADMNRKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLV 263
Query: 61 LTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREA 120
LTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREA
Sbjct: 264 LTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREA 443
Query: 121 RLEKIGGEVLCYIW 134
RLEKIGGEVLCYIW
Sbjct: 444 RLEKIGGEVLCYIW 485
>TC18536 homologue to UP|AAK62367 (AAK62367) 40S ribosomal protein S15A,
complete
Length = 556
Score = 42.7 bits (99), Expect = 2e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +2
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 62 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLLVMQKHGYIGEFEYVDDHRAGK---IVVEL 232
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 233 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 388
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 389 RKNVGGKVLGFFY 427
>TC11790 homologue to UP|AAK62367 (AAK62367) 40S ribosomal protein S15A,
complete
Length = 474
Score = 42.4 bits (98), Expect = 3e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +1
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 109 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLIVMQKHGYIGEFEYVDDHRAGK---IVVEL 279
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 280 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 435
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 436 RKNVGGKVLGFFY 474
>TC10950 homologue to UP|AAK62367 (AAK62367) 40S ribosomal protein S15A,
complete
Length = 569
Score = 42.4 bits (98), Expect = 3e-05
Identities = 39/133 (29%), Positives = 63/133 (47%), Gaps = 6/133 (4%)
Frame = +2
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ G+I E V HR +V+ L
Sbjct: 80 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLMVMQHHGYIGEFEYVDDHRAGK---IVVEL 250
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 251 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 406
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 407 RKNVGGKVLGFFY 445
>TC9874 similar to UP|Q8VZJ1 (Q8VZJ1) AT5g09790/F17I14_20, partial (22%)
Length = 546
Score = 26.9 bits (58), Expect = 1.2
Identities = 17/51 (33%), Positives = 24/51 (46%), Gaps = 1/51 (1%)
Frame = +3
Query: 44 FIENVRKH-RESDKSFLVLTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNY 93
FI + H +E K ++RY N G + FL R G R+YY+Y
Sbjct: 57 FINGIHNHTQEGKKKQNCKSVRY--NVDGQSRVFLVATRDIAKGERLYYDY 203
>AV768209
Length = 422
Score = 26.2 bits (56), Expect = 2.0
Identities = 20/75 (26%), Positives = 37/75 (48%)
Frame = -3
Query: 58 FLVLTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTD 117
F+ +TL +K S +T ++L ++STP + + +LGG+ + + T R +
Sbjct: 261 FISVTL----GSKESQETPMHLIKVSTPSKDTQCSDAVLMNLLGGVSLTVFYTGRS**S- 97
Query: 118 REARLEKIGGEVLCY 132
ARL+ V+ Y
Sbjct: 96 -SARLQPWSSAVIVY 55
>TC18284 homologue to UP|TBA3_ARATH (P20363) Tubulin alpha-3/alpha-5
chain, partial (44%)
Length = 739
Score = 25.0 bits (53), Expect = 4.5
Identities = 12/18 (66%), Positives = 12/18 (66%)
Frame = +1
Query: 59 LVLTLRYRRNTKGSYKTF 76
LV LRYR N KGSY F
Sbjct: 19 LVXRLRYRINIKGSYFLF 72
>TC8536 weakly similar to UP|AAR92224 (AAR92224) Cystatin, partial (69%)
Length = 543
Score = 23.9 bits (50), Expect = 10.0
Identities = 15/44 (34%), Positives = 24/44 (54%)
Frame = -1
Query: 85 PGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREARLEKIGGE 128
P L I +++I +L + +L RGI TD+ + L + GGE
Sbjct: 234 P*LLIILGHREIGDLLNVRVVEVLD--RGIATDKSSLLPESGGE 109
>BP061060
Length = 311
Score = 23.9 bits (50), Expect = 10.0
Identities = 16/63 (25%), Positives = 31/63 (48%)
Frame = -1
Query: 70 KGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREARLEKIGGEV 129
K + +T ++L ++STP + + +LGG+ + + T R + ARL+ V
Sbjct: 212 KKATETPMHLIKVSTPSKDTQCSDAVLMNLLGGVSLTVFYTGRS**S--SARLQPWSSAV 39
Query: 130 LCY 132
+ Y
Sbjct: 38 IVY 30
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.141 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,756,671
Number of Sequences: 28460
Number of extensions: 18041
Number of successful extensions: 84
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 84
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 84
length of query: 134
length of database: 4,897,600
effective HSP length: 81
effective length of query: 53
effective length of database: 2,592,340
effective search space: 137394020
effective search space used: 137394020
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 50 (23.9 bits)
Description of BAB33231.1

