
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33186.1 750 aa
(750 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
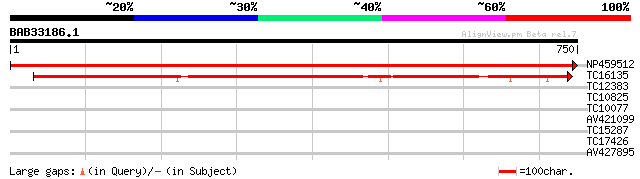
NP459512 PSI P700 apoprotein A1 [Lotus japonicus] 1561 0.0
TC16135 UP|PSAB_LOTJA (P58385) Photosystem I P700 chlorophyll A ... 628 e-180
TC12383 30 1.8
TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Ar... 28 4.1
TC10077 28 4.1
AV421099 28 5.4
TC15287 similar to UP|Q9FMY5 (Q9FMY5) U2 snRNP auxiliary factor,... 28 7.0
TC17426 similar to UP|Q9SX30 (Q9SX30) F24J5.9, partial (29%) 27 9.2
AV427895 27 9.2
>NP459512 PSI P700 apoprotein A1 [Lotus japonicus]
Length = 2253
Score = 1561 bits (4041), Expect = 0.0
Identities = 750/750 (100%), Positives = 750/750 (100%)
Frame = +1
Query: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 60
MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT
Sbjct: 1 MIIRSPEPEVKILVDRDPIKTSFEEWAKPGHFSRTIAKGPDTTTWIWNLHADAHDFDSHT 180
Query: 61 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 120
SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG
Sbjct: 181 SDLEDISRKIFSAHFGQLSIIFLWLSGMYFHGARFSNYEAWLSDPTHIRPSAQVVWPIVG 360
Query: 121 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 180
QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK
Sbjct: 361 QEILNGDVGGGFRGIQITSGFFQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHK 540
Query: 181 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 240
AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF
Sbjct: 541 AAPKLAWFQDVESMLNHHLAGLLGLGSLSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEF 720
Query: 241 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 300
ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI
Sbjct: 721 ILNRDLLAQLYPSFAEGATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAI 900
Query: 301 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 360
LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL
Sbjct: 901 LFLIAGHMYRTNWGIGHGIKDILEAHKGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSL 1080
Query: 361 TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 420
TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY
Sbjct: 1081TIIVAHHMYSMPPYPYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRY 1260
Query: 421 NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 480
NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF
Sbjct: 1261NDLLDRVLRHRDAIISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIF 1440
Query: 481 AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 540
AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT
Sbjct: 1441AQWIQNTHALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFT 1620
Query: 541 IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 600
IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA
Sbjct: 1621IHVTVLILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNA 1800
Query: 601 ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 660
ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY
Sbjct: 1801ISVVIFHFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSY 1980
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 720
GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV
Sbjct: 1981GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIV 2160
Query: 721 QGRAVGVTHYLLGGIATTWAFFLARIIAVG 750
QGRAVGVTHYLLGGIATTWAFFLARIIAVG
Sbjct: 2161QGRAVGVTHYLLGGIATTWAFFLARIIAVG 2250
>TC16135 UP|PSAB_LOTJA (P58385) Photosystem I P700 chlorophyll A apoprotein
A2 (PsaB) (PSI-B), complete
Length = 2647
Score = 628 bits (1619), Expect = e-180
Identities = 344/746 (46%), Positives = 449/746 (60%), Gaps = 33/746 (4%)
Frame = +1
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL+IIFLW SG F
Sbjct: 22 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLAIIFLWTSGNLF 198
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 199 HVAWQGNFEAWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 378
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L +A+ L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 379 IGLRTNGDLYSGALFLLFLSAISLLAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 558
Query: 207 SLSWAGHQVHVSLP--------INQFLNAGVDPKEIPLPHEFILNRDLLAQ--LYPSFAE 256
SL+W GH VHV++P N FL+ LPH L Q LY +
Sbjct: 559 SLAWTGHLVHVAIPGSRGEYVRWNNFLSI--------LPHPQGLGPLFTGQWNLYAQNPD 714
Query: 257 GATPFFTLNWAKYADFLTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIG 316
++ F + LT GG P T LWLTD+AHHHLAIAILFL+AGHMYRTN+GIG
Sbjct: 715 SSSHLFGTSQGAGTAILTLLGGFHPQTQSLWLTDMAHHHLAIAILFLLAGHMYRTNFGIG 894
Query: 317 HGIKDILEAH--KGPFTGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHHMYSMPPY 374
H IKD+LEAH G G+GHKGLY+ + S H QL + LA LG +T +VA HMYS+P Y
Sbjct: 895 HSIKDLLEAHIPPGGRLGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQHMYSLPAY 1074
Query: 375 PYLATDYGTQLSLFTHHMWIGGFLIVGAAAHAAIFMVRDYDPTTRYNDLLDRVLRHRDAI 434
++A DY TQ +L+THH +I GF++ GA AH AIF +RDY+P +++L R+L H++AI
Sbjct: 1075AFIAQDYTTQAALYTHHQYIAGFIMTGAFAHGAIFFIRDYNPEQNEDNVLARMLDHKEAI 1254
Query: 435 ISHLNWVCIFLGFHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTH------ 488
ISHL+W +FLGFH+ GLY+HND M A G P+ I ++PIFAQWIQ+ H
Sbjct: 1255ISHLSWASLFLGFHTLGLYVHNDVMLAFGTPEKQ-----ILIEPIFAQWIQSAHGKTSYG 1419
Query: 489 --ALAPGITAPGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVL 546
L +P G S+ W G L A+ L + +G DFLVHH A +H T L
Sbjct: 1420FDVLLSSTNSPAFNAGRSI-WLPGWLNAINENSNSLFLTIGPGDFLVHHAIALGLHTTTL 1596
Query: 547 ILLKGVLFARSSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIF 606
IL+KG L AR S+L+PDK + G+ FPCDGPGRGGTC +SAWD +L +FWM N I V F
Sbjct: 1597ILVKGALDARGSKLMPDKKDFGYSFPCDGPGRGGTCDISAWDAFYLAVFWMLNTIGWVTF 1776
Query: 607 HFSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYG----S 662
++ WK + G++S F +SS + GWLRD+LW +SQ+I Y +
Sbjct: 1777YWHWKHITLWQGNVS-----------QFNESSTYLMGWLRDYLWLNSSQLINGYNPFGMN 1923
Query: 663 SLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAP----ATQPRALS 718
SLS + FL H VWA MFL S RGYWQELIE++ WAH + +A +P ALS
Sbjct: 1924SLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHERTPLANLIRWRDKPVALS 2103
Query: 719 IVQGRAVGVTHYLLGGIATTWAFFLA 744
IVQ R VG+ H+ +G I T AF +A
Sbjct: 2104IVQARLVGLAHFSVGYIFTYAAFLIA 2181
>TC12383
Length = 506
Score = 29.6 bits (65), Expect = 1.8
Identities = 13/43 (30%), Positives = 21/43 (48%), Gaps = 3/43 (6%)
Frame = +3
Query: 276 RGGLDPVTGGLWL---TDIAHHHLAIAILFLIAGHMYRTNWGI 315
+ G TGG W+ + HH + ++ L+AG RT G+
Sbjct: 369 KSGFPGKTGGFWICQNVEDFHHESTVVVVVLVAGRRRRTRIGL 497
>TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Arabidopsis
thaliana;}, partial (32%)
Length = 1091
Score = 28.5 bits (62), Expect = 4.1
Identities = 17/44 (38%), Positives = 22/44 (49%)
Frame = +2
Query: 649 LWAQASQVIQSYGSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYW 692
LW++ +VI S SS S FFL HFV L+ G +W
Sbjct: 542 LWSEC-KVISSLTSSCSLSYSFFLSPHFVLEIWLVESLHGSKFW 670
>TC10077
Length = 560
Score = 28.5 bits (62), Expect = 4.1
Identities = 21/68 (30%), Positives = 30/68 (43%)
Frame = +3
Query: 142 FQIWRASGITSELQLYCTAIGGLVFAALMLFAGWFHYHKAAPKLAWFQDVESMLNHHLAG 201
FQ+W SE L+ AIGG+V A ++F H KLA + L + G
Sbjct: 216 FQLWFMDMPLSERVLW--AIGGMVLTAGLVFISKRKSHNQRQKLAAIKRAARKLEGRMPG 389
Query: 202 LLGLGSLS 209
++ LS
Sbjct: 390 MMAEPRLS 413
>AV421099
Length = 428
Score = 28.1 bits (61), Expect = 5.4
Identities = 12/32 (37%), Positives = 19/32 (58%), Gaps = 1/32 (3%)
Frame = +3
Query: 590 VFLGLFWMYNA-ISVVIFHFSWKMQSDVWGSI 620
+F + W N+ I++ + HF+W VWGSI
Sbjct: 327 MFTCIIWAVNSQIALTMCHFTWIQHLFVWGSI 422
>TC15287 similar to UP|Q9FMY5 (Q9FMY5) U2 snRNP auxiliary factor, small
subunit, partial (81%)
Length = 1263
Score = 27.7 bits (60), Expect = 7.0
Identities = 15/38 (39%), Positives = 19/38 (49%)
Frame = -2
Query: 354 LAMLGSLTIIVAHHMYSMPPYPYLATDYGTQLSLFTHH 391
LA+L SL +HH S P +PY Y SL H+
Sbjct: 860 LAVLRSLPSPDSHHASSFPHHPYHCFSYHCSSSLENHN 747
>TC17426 similar to UP|Q9SX30 (Q9SX30) F24J5.9, partial (29%)
Length = 999
Score = 27.3 bits (59), Expect = 9.2
Identities = 16/46 (34%), Positives = 24/46 (51%)
Frame = -2
Query: 661 GSSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKL 706
GSSL +FFL + W +S + G +W+ SIV + NK+
Sbjct: 401 GSSLGKTWIFFLSSSHAWIYSRDPILWGCIFWE---NSIVESKNKV 273
>AV427895
Length = 189
Score = 27.3 bits (59), Expect = 9.2
Identities = 13/43 (30%), Positives = 23/43 (53%)
Frame = -1
Query: 92 GARFSNYEAWLSDPTHIRPSAQVVWPIVGQEILNGDVGGGFRG 134
GARF+++ WL T + S+ + +P +++ GGG G
Sbjct: 123 GARFNSHSPWLIRKTSVTSSSTIPFP---EKLRENTAGGGEGG 4
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.141 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,154,472
Number of Sequences: 28460
Number of extensions: 258633
Number of successful extensions: 1654
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1643
length of query: 750
length of database: 4,897,600
effective HSP length: 97
effective length of query: 653
effective length of database: 2,136,980
effective search space: 1395447940
effective search space used: 1395447940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Description of BAB33186.1

