
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
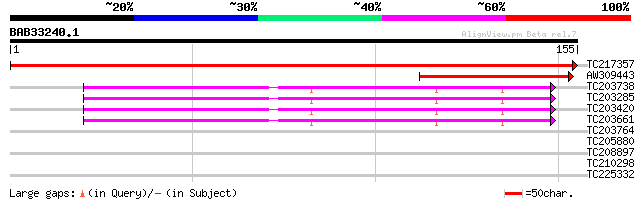
Query= BAB33240.1 155 aa
(155 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217357 ribosomal protein S7 301 1e-82
AW309443 similar to SP|P06361|RR7_T Chloroplast 30S ribosomal pr... 72 8e-14
TC203738 homologue to UP|RS5_CICAR (O65731) 40S ribosomal protei... 64 4e-11
TC203285 homologue to UP|RS5_CICAR (O65731) 40S ribosomal protei... 63 6e-11
TC203420 homologue to UP|RS5_CICAR (O65731) 40S ribosomal protei... 62 1e-10
TC203661 similar to GB|AAO63400.1|28950953|BT005336 At3g26700 {A... 62 1e-10
TC203764 homologue to UP|RS5_CICAR (O65731) 40S ribosomal protei... 33 0.055
TC205880 similar to UP|Q9FQL9 (Q9FQL9) Cytochrome P450, partial ... 28 1.8
TC208897 similar to UP|Q9SXG8 (Q9SXG8) Dof zinc finger protein, ... 27 3.9
TC210298 weakly similar to UP|Q9FJH4 (Q9FJH4) Similarity to ring... 26 6.7
TC225332 similar to UP|O65848 (O65848) Annexin, complete 26 8.8
>TC217357 ribosomal protein S7
Length = 894
Score = 301 bits (770), Expect = 1e-82
Identities = 153/155 (98%), Positives = 155/155 (99%)
Frame = +1
Query: 1 MSRRGTAEKKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLS 60
MSRRGTAE+KTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLS
Sbjct: 427 MSRRGTAEEKTAKSDPIYRNRLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLS 606
Query: 61 VLRQAIRGVTPDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKL 120
VLRQAIRGVTPDIAVKARRVGGSTHQVP+EIGSTQGKALAIRWLLGASRKRPGRNMAFKL
Sbjct: 607 VLRQAIRGVTPDIAVKARRVGGSTHQVPVEIGSTQGKALAIRWLLGASRKRPGRNMAFKL 786
Query: 121 SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR 155
SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR
Sbjct: 787 SSELVDAAKGSGDAIRKKEETHRMAEANRAFAHFR 891
>AW309443 similar to SP|P06361|RR7_T Chloroplast 30S ribosomal protein S7.
[Black nightshade] {Solanum nigrum}, partial (27%)
Length = 405
Score = 72.4 bits (176), Expect = 8e-14
Identities = 36/42 (85%), Positives = 37/42 (87%)
Frame = -3
Query: 113 GRNMAFKLSSELVDAAKGSGDAIRKKEETHRMAEANRAFAHF 154
G NM FKLSSELVDAAKGSGDAI KKEET+RM E NRAFAHF
Sbjct: 382 G*NMVFKLSSELVDAAKGSGDAICKKEETYRMVEVNRAFAHF 257
>TC203738 homologue to UP|RS5_CICAR (O65731) 40S ribosomal protein S5
(Fragment), partial (88%)
Length = 1846
Score = 63.5 bits (153), Expect = 4e-11
Identities = 50/136 (36%), Positives = 75/136 (54%), Gaps = 7/136 (5%)
Frame = +3
Query: 21 RLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVTPDIAVKARRV 80
RL N L+ +GKK LA +II AM+ I T+ NP+ V+ +A+ P A R+
Sbjct: 123 RLTNSLMMHGRNNGKKLLAVRIIKHAMEIIHLLTDQNPIQVIVEAVVNSGP--REDATRI 296
Query: 81 G--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSGD-- 133
G G + ++I + AI L +R+ RN +A L+ EL++AAKGS +
Sbjct: 297 GSAGVVRRQAVDISPLRRVNQAIYLLTTGAREAAFRNIKTIAECLADELINAAKGSSNSY 476
Query: 134 AIRKKEETHRMAEANR 149
AI+KK+E R+A+ANR
Sbjct: 477 AIKKKDEIERVAKANR 524
>TC203285 homologue to UP|RS5_CICAR (O65731) 40S ribosomal protein S5
(Fragment), complete
Length = 1224
Score = 62.8 bits (151), Expect = 6e-11
Identities = 50/136 (36%), Positives = 74/136 (53%), Gaps = 7/136 (5%)
Frame = +1
Query: 21 RLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVTPDIAVKARRV 80
RL N L+ +GKK LA +II AM+ I T+ NP+ V+ A+ P A R+
Sbjct: 313 RLTNSLMMHGRNNGKKLLAVRIIKHAMEIIHLLTDQNPIQVIVDAVINSGP--REDATRI 486
Query: 81 G--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSGD-- 133
G G + ++I + AI L +R+ RN +A L+ EL++AAKGS +
Sbjct: 487 GSAGVVRRQAVDISPLRRVNQAIYLLTTGAREAAFRNIKTIAECLADELINAAKGSSNSY 666
Query: 134 AIRKKEETHRMAEANR 149
AI+KK+E R+A+ANR
Sbjct: 667 AIKKKDEIERVAKANR 714
>TC203420 homologue to UP|RS5_CICAR (O65731) 40S ribosomal protein S5
(Fragment), complete
Length = 1013
Score = 62.0 bits (149), Expect = 1e-10
Identities = 49/136 (36%), Positives = 74/136 (54%), Gaps = 7/136 (5%)
Frame = +3
Query: 21 RLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVTPDIAVKARRV 80
RL N L+ +GKK +A +II AM+ I T+ NP+ V+ A+ P A R+
Sbjct: 294 RLTNSLMMHGRNNGKKLMAVRIIKHAMEIIHLLTDLNPIQVIVDAVINSGP--REDATRI 467
Query: 81 G--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSGD-- 133
G G + ++I + AI L +R+ RN +A L+ EL++AAKGS +
Sbjct: 468 GSAGVVRRQAVDISPLRRVNQAIYLLTTGAREAAFRNIKTIAECLADELINAAKGSSNSY 647
Query: 134 AIRKKEETHRMAEANR 149
AI+KK+E R+A+ANR
Sbjct: 648 AIKKKDEIERVAKANR 695
>TC203661 similar to GB|AAO63400.1|28950953|BT005336 At3g26700 {Arabidopsis
thaliana;} , partial (88%)
Length = 2451
Score = 62.0 bits (149), Expect = 1e-10
Identities = 49/136 (36%), Positives = 74/136 (54%), Gaps = 7/136 (5%)
Frame = +1
Query: 21 RLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVTPDIAVKARRV 80
RL N L+ +GKK +A +II AM+ I T+ NP+ V+ A+ P A R+
Sbjct: 1735 RLTNSLMMHGRNNGKKLMAVRIIKHAMEIIHLLTDLNPIQVIVDAVINSGP--REDATRI 1908
Query: 81 G--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSGD-- 133
G G + ++I + AI L +R+ RN +A L+ EL++AAKGS +
Sbjct: 1909 GSAGVVRRQAVDISPLRRVNQAIYLLTTGAREAAFRNIKTIAECLADELINAAKGSSNSY 2088
Query: 134 AIRKKEETHRMAEANR 149
AI+KK+E R+A+ANR
Sbjct: 2089 AIKKKDEIERVAKANR 2136
>TC203764 homologue to UP|RS5_CICAR (O65731) 40S ribosomal protein S5
(Fragment), partial (53%)
Length = 588
Score = 33.1 bits (74), Expect = 0.055
Identities = 18/42 (42%), Positives = 25/42 (58%)
Frame = +3
Query: 21 RLVNMLVNRILKHGKKSLAYQIIYRAMKKIQQKTETNPLSVL 62
RL N L+ +GKK LA +II AM+ I T+ NP+ V+
Sbjct: 279 RLTNSLMMHGRNNGKKLLAVRIIKHAMEIIHLLTDQNPIQVI 404
>TC205880 similar to UP|Q9FQL9 (Q9FQL9) Cytochrome P450, partial (50%)
Length = 1154
Score = 28.1 bits (61), Expect = 1.8
Identities = 17/69 (24%), Positives = 30/69 (42%)
Frame = +1
Query: 71 PDIAVKARRVGGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKLSSELVDAAKG 130
P + VK R G + G T+ A+ + W + +RP FK ++E +D G
Sbjct: 112 PTLEVKLERHGVKAFTQDLIAGGTESSAVTVEWAITELLRRP---EIFKKATEELDRVIG 282
Query: 131 SGDAIRKKE 139
+ +K+
Sbjct: 283 RERWVEEKD 309
>TC208897 similar to UP|Q9SXG8 (Q9SXG8) Dof zinc finger protein, partial
(25%)
Length = 1067
Score = 26.9 bits (58), Expect = 3.9
Identities = 15/42 (35%), Positives = 21/42 (49%)
Frame = +1
Query: 81 GGSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRNMAFKLSS 122
GGS VP+ GS + K +++ +S K P N LSS
Sbjct: 232 GGSLRNVPVGGGSRKNKKVSVTASSSSSPKVPDLNPPINLSS 357
>TC210298 weakly similar to UP|Q9FJH4 (Q9FJH4) Similarity to ring finger
protein, partial (11%)
Length = 823
Score = 26.2 bits (56), Expect = 6.7
Identities = 13/35 (37%), Positives = 19/35 (54%)
Frame = -1
Query: 36 KSLAYQIIYRAMKKIQQKTETNPLSVLRQAIRGVT 70
K + +IY + KKI K +T L+ L Q + VT
Sbjct: 640 KPTKHHVIYSSQKKINHKAKTVDLTTLLQIV*EVT 536
>TC225332 similar to UP|O65848 (O65848) Annexin, complete
Length = 1300
Score = 25.8 bits (55), Expect = 8.8
Identities = 12/29 (41%), Positives = 18/29 (61%)
Frame = -1
Query: 86 QVPIEIGSTQGKALAIRWLLGASRKRPGR 114
QVP+ IG+ ++L LGAS+ PG+
Sbjct: 283 QVPVSIGTRLSQSLL*SQTLGASQDPPGK 197
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,675,246
Number of Sequences: 63676
Number of extensions: 42229
Number of successful extensions: 198
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 197
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 198
length of query: 155
length of database: 12,639,632
effective HSP length: 89
effective length of query: 66
effective length of database: 6,972,468
effective search space: 460182888
effective search space used: 460182888
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Description of BAB33240.1

