
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33231.1 134 aa
(134 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
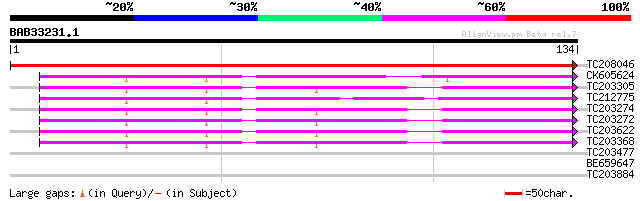
TC208046 homologue to UP|RR8_PHAAN (P59033) Chloroplast 30S ribo... 243 2e-65
CK605624 43 4e-05
TC203305 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete 43 5e-05
TC212775 similar to UP|Q8UW22 (Q8UW22) Ribosomal protein S15 iso... 43 5e-05
TC203274 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete 43 5e-05
TC203272 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete 43 5e-05
TC203622 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete 43 5e-05
TC203368 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete 43 5e-05
TC203477 homologue to UP|RS1A_ARATH (P42798) 40S ribosomal prote... 34 0.023
BE659647 34 0.023
TC203884 homologue to UP|RS1A_ARATH (P42798) 40S ribosomal prote... 34 0.023
>TC208046 homologue to UP|RR8_PHAAN (P59033) Chloroplast 30S ribosomal
protein S8, complete
Length = 816
Score = 243 bits (620), Expect = 2e-65
Identities = 121/134 (90%), Positives = 127/134 (94%)
Frame = +1
Query: 1 MGKDTIADIITSIRNADMNRKRTVQIPFTNITENTVKILLREGFIENVRKHRESDKSFLV 60
MGKDTIADIIT IRNADMN+K VQIPFTNITEN VKILLREGF+ENVRKHRE+DK FLV
Sbjct: 97 MGKDTIADIITYIRNADMNKKGMVQIPFTNITENIVKILLREGFVENVRKHRENDKYFLV 276
Query: 61 LTLRYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREA 120
LTLRYRRN KGSYK+FLNLKRISTPGLRIY NYQ+IPRILGGMGIVILSTSRGIMTDREA
Sbjct: 277 LTLRYRRNRKGSYKSFLNLKRISTPGLRIYSNYQRIPRILGGMGIVILSTSRGIMTDREA 456
Query: 121 RLEKIGGEVLCYIW 134
RLE+IGGEVLCYIW
Sbjct: 457 RLERIGGEVLCYIW 498
>CK605624
Length = 674
Score = 43.1 bits (100), Expect = 4e-05
Identities = 38/133 (28%), Positives = 61/133 (45%), Gaps = 6/133 (4%)
Frame = +3
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + SI NA+ KR V I P + + ++++ + G+I E V HR +V+ L
Sbjct: 117 DALKSINNAEKAGKRQVLIRPSSKVIVKFLQVMQKHGYIGEFEEVDDHRSGK---IVVQL 287
Query: 64 RYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGG--MGIVILSTSRGIMTDREAR 121
R N G N+K + R+L G VIL+TS GIM EAR
Sbjct: 288 NGRLNKTGVISPRYNVKLADLEKWTV--------RLLPSRQFGYVILTTSAGIMDHEEAR 443
Query: 122 LEKIGGEVLCYIW 134
+ + G+++ + +
Sbjct: 444 RKHVSGKIIGFFY 482
>TC203305 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete
Length = 748
Score = 42.7 bits (99), Expect = 5e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +1
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 61 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLLVMQKHGYIGEFEYVDDHRAGK---IVVEL 231
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 232 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 387
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 388 RKNVGGKVLGFFY 426
>TC212775 similar to UP|Q8UW22 (Q8UW22) Ribosomal protein S15 isoform,
partial (98%)
Length = 449
Score = 42.7 bits (99), Expect = 5e-05
Identities = 40/131 (30%), Positives = 63/131 (47%), Gaps = 4/131 (3%)
Frame = +2
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + SI NA+ R V++ P + + ++++LR G+I E V HR +V+ L
Sbjct: 56 DALRSISNAEKRGFRQVRVHPSSKVIVKFLEVMLRHGYIGEFEVVDDHRGGK---IVVNL 226
Query: 64 RYRRNTKGSYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREARLE 123
R N G + I L + N R G +IL+TS GI+ EAR +
Sbjct: 227 IGRINKTGVISPRFD---IQLSDLEKWINNLLPSR---QFGYLILTTSYGIIDHEEARQK 388
Query: 124 KIGGEVLCYIW 134
K GG++L Y++
Sbjct: 389 KTGGKILGYVY 421
>TC203274 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete
Length = 1174
Score = 42.7 bits (99), Expect = 5e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +1
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 559 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLLVMQKHGYIGEFEYVDDHRAGK---IVVEL 729
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 730 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 885
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 886 RKNVGGKVLGFFY 924
>TC203272 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete
Length = 599
Score = 42.7 bits (99), Expect = 5e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +1
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 49 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLLVMQKHGYIGEFEYVDDHRAGK---IVVEL 219
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 220 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 375
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 376 RKNVGGKVLGFFY 414
>TC203622 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete
Length = 675
Score = 42.7 bits (99), Expect = 5e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +3
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 90 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLLVMQKHGYIGEFEYVDDHRAGK---IVVEL 260
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 261 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 416
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 417 RKNVGGKVLGFFY 455
>TC203368 homologue to UP|Q9SBL9 (Q9SBL9) Wrp15a, complete
Length = 652
Score = 42.7 bits (99), Expect = 5e-05
Identities = 39/133 (29%), Positives = 64/133 (47%), Gaps = 6/133 (4%)
Frame = +2
Query: 8 DIITSIRNADMNRKRTVQI-PFTNITENTVKILLREGFI---ENVRKHRESDKSFLVLTL 63
D + S+ NA+ KR V I P + + + ++ + G+I E V HR +V+ L
Sbjct: 89 DALKSMYNAEKRGKRQVMIRPSSKVIIKFLLVMQKHGYIGEFEYVDDHRAGK---IVVEL 259
Query: 64 RYRRNTKG--SYKTFLNLKRISTPGLRIYYNYQKIPRILGGMGIVILSTSRGIMTDREAR 121
R N G S + + +K I R+ + Q G ++L+TS GIM EAR
Sbjct: 260 NGRLNKCGVISPRFDVGVKEIEGWTARLLPSRQ--------FGYIVLTTSAGIMDHEEAR 415
Query: 122 LEKIGGEVLCYIW 134
+ +GG+VL + +
Sbjct: 416 RKNVGGKVLGFFY 454
>TC203477 homologue to UP|RS1A_ARATH (P42798) 40S ribosomal protein S15a,
partial (77%)
Length = 827
Score = 33.9 bits (76), Expect = 0.023
Identities = 14/31 (45%), Positives = 22/31 (70%)
Frame = +1
Query: 104 GIVILSTSRGIMTDREARLEKIGGEVLCYIW 134
G ++L+TS GIM EAR + +GG+VL + +
Sbjct: 457 GYIVLTTSAGIMDHEEARRKNVGGKVLGFFY 549
>BE659647
Length = 444
Score = 33.9 bits (76), Expect = 0.023
Identities = 14/31 (45%), Positives = 22/31 (70%)
Frame = -1
Query: 104 GIVILSTSRGIMTDREARLEKIGGEVLCYIW 134
G ++L+TS GIM EAR + +GG+VL + +
Sbjct: 231 GYIVLTTSAGIMDHEEARRKNVGGKVLGFFY 139
>TC203884 homologue to UP|RS1A_ARATH (P42798) 40S ribosomal protein S15a,
partial (75%)
Length = 545
Score = 33.9 bits (76), Expect = 0.023
Identities = 14/31 (45%), Positives = 22/31 (70%)
Frame = +1
Query: 104 GIVILSTSRGIMTDREARLEKIGGEVLCYIW 134
G ++L+TS GIM EAR + +GG+VL + +
Sbjct: 217 GYIVLTTSAGIMDHEEARRKNVGGKVLGFFY 309
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.141 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,576,387
Number of Sequences: 63676
Number of extensions: 47937
Number of successful extensions: 200
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 200
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 200
length of query: 134
length of database: 12,639,632
effective HSP length: 87
effective length of query: 47
effective length of database: 7,099,820
effective search space: 333691540
effective search space used: 333691540
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 53 (25.0 bits)
Description of BAB33231.1

