
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33186.1 750 aa
(750 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
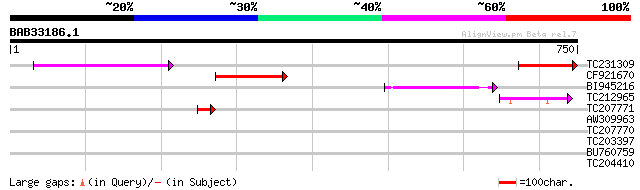
TC231309 homologue to UP|PSAB_LOTJA (P58385) Photosystem I P700 ... 162 5e-40
CF921670 126 3e-29
BI945216 SP|P06407|PSAB Photosystem I P700 chlorophyll A apoprot... 124 2e-28
TC212965 UP|PSAB_ANTMA (Q33332) Photosystem I P700 chlorophyll A... 91 2e-18
TC207771 similar to GB|AAF65219.1|7637533|AF223227 photosystem p... 45 2e-04
AW309963 similar to GP|336870|gb|AA alpha-apoprotein {Euglena gr... 42 0.001
TC207770 homologue to UP|O22445 (O22445) Developmentally regulat... 33 0.46
TC203397 similar to UP|GPX4_CITSI (Q06652) Probable phospholipid... 30 3.0
BU760759 weakly similar to GP|14090380|dbj P0037C04.27 {Oryza sa... 30 3.9
TC204410 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc f... 29 8.8
>TC231309 homologue to UP|PSAB_LOTJA (P58385) Photosystem I P700 chlorophyll
A apoprotein A2 (PsaB) (PSI-B), partial (27%)
Length = 910
Score = 162 bits (410), Expect = 5e-40
Identities = 78/78 (100%), Positives = 78/78 (100%)
Frame = +2
Query: 673 GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLL 732
GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLL
Sbjct: 2 GAHFVWAFSLMFLFSGRGYWQELIESIVWAHNKLKVAPATQPRALSIVQGRAVGVTHYLL 181
Query: 733 GGIATTWAFFLARIIAVG 750
GGIATTWAFFLARIIAVG
Sbjct: 182 GGIATTWAFFLARIIAVG 235
Score = 157 bits (396), Expect = 2e-38
Identities = 86/190 (45%), Positives = 114/190 (59%), Gaps = 5/190 (2%)
Frame = +3
Query: 32 FSRTIAKGPDTTTWIWNLHADAHDFDSHTSDLED-ISRKIFSAHFGQLSIIFLWLSGMYF 90
FS+ +A+ P TT IW A AHDF+SH E+ + + IF++HFGQL IIFLW SG F
Sbjct: 285 FSQGLAQDP-TTRRIWFGIATAHDFESHDDITEERLYQNIFASHFGQLVIIFLWTSGNLF 461
Query: 91 HGARFSNYEAWLSDPTHIRPSAQVVW-PIVGQEILNGDVGGGFRG-IQIT-SGFFQIWRA 147
H A N+EAW+ DP H+RP A +W P GQ + GG G + I SG +Q W
Sbjct: 462 HVAWQGNFEAWVQDPLHVRPIAHAIWDPHFGQPAVEAFTRGGALGPVNIAYSGVYQWWYT 641
Query: 148 SGITSELQLYCTAIGGLVFAALMLFAGWFHYH-KAAPKLAWFQDVESMLNHHLAGLLGLG 206
G+ + LY A+ L + + L AGW H K P ++WF++ ES LNHHL+GL G+
Sbjct: 642 IGLRTNGDLYTGALFLLFLSTISLIAGWLHLQPKWKPSVSWFKNAESRLNHHLSGLFGVS 821
Query: 207 SLSWAGHQVH 216
SL+W GH VH
Sbjct: 822 SLAWTGHLVH 851
>CF921670
Length = 727
Score = 126 bits (317), Expect = 3e-29
Identities = 62/97 (63%), Positives = 71/97 (72%), Gaps = 2/97 (2%)
Frame = -3
Query: 273 LTFRGGLDPVTGGLWLTDIAHHHLAIAILFLIAGHMYRTNWGIGHGIKDILEAHKGP--F 330
LT GG P T LWLTDIAHHHLAIA LFL+AGHMYRTN+GIGH IKD+LEAH P
Sbjct: 293 LTLLGGFHPQTQSLWLTDIAHHHLAIAFLFLVAGHMYRTNFGIGHSIKDLLEAHTPPGGR 114
Query: 331 TGQGHKGLYEILTTSWHAQLSINLAMLGSLTIIVAHH 367
G+GHKGLY+ + S H QL + LA LG +T +VA H
Sbjct: 113 LGRGHKGLYDTINNSIHFQLGLALASLGVITSLVAQH 3
>BI945216 SP|P06407|PSAB Photosystem I P700 chlorophyll A apoprotein A2
(PsaB) (PSI-B). [Common tobacco] {Nicotiana tabacum},
partial (18%)
Length = 419
Score = 124 bits (311), Expect = 2e-28
Identities = 66/149 (44%), Positives = 85/149 (56%)
Frame = +2
Query: 497 PGATTGTSLTWGGGDLVAVGNKVALLPIPLGTADFLVHHIHAFTIHVTVLILLKGVLFAR 556
P G S+ W G L AV L + +G DFLVHH A +H T LIL+KG L AR
Sbjct: 8 PAFNAGRSI-WLPGWLNAVNENSNSLFLTIGPGDFLVHHAIALGLHTTTLILVKGALDAR 184
Query: 557 SSRLIPDKANLGFRFPCDGPGRGGTCQVSAWDHVFLGLFWMYNAISVVIFHFSWKMQSDV 616
S+L+PDK + G+ FPCDGPGRG TC +SAWD +L +FWM N I V F++ WK +
Sbjct: 185 GSKLMPDKKDFGYSFPCDGPGRGRTCDISAWDAFYLAVFWMLNTIGWVTFYWHWKHITLW 364
Query: 617 WGSISDQGVVTHITGGNFAQSSITINGWL 645
G++S F +SS + GWL
Sbjct: 365 QGNVS-----------QFNESSTYLMGWL 418
>TC212965 UP|PSAB_ANTMA (Q33332) Photosystem I P700 chlorophyll A apoprotein
A2 (PsaB) (PSI-B), partial (15%)
Length = 466
Score = 90.5 bits (223), Expect = 2e-18
Identities = 49/104 (47%), Positives = 62/104 (59%), Gaps = 8/104 (7%)
Frame = +3
Query: 649 LWAQASQVIQSYG----SSLSAYGLFFLGAHFVWAFSLMFLFSGRGYWQELIESIVWAHN 704
LW +SQ+I Y +SLS + FL H VWA MFL S RGYWQELIE++ WAH
Sbjct: 3 LWLNSSQLINGYNPFGMNSLSVWAWMFLFGHLVWATGFMFLISWRGYWQELIETLAWAHE 182
Query: 705 KLKVAP----ATQPRALSIVQGRAVGVTHYLLGGIATTWAFFLA 744
+ +A +P ALSIVQ R VG+ H+ +G I T AF +A
Sbjct: 183 RTPLANLIRWRDKPVALSIVQARLVGLAHFSVGYIFTYAAFLIA 314
>TC207771 similar to GB|AAF65219.1|7637533|AF223227 photosystem protein
{Pisum sativum;} , partial (3%)
Length = 414
Score = 44.7 bits (104), Expect = 2e-04
Identities = 18/24 (75%), Positives = 20/24 (83%)
Frame = -1
Query: 249 QLYPSFAEGATPFFTLNWAKYADF 272
QLY SFA+GA PFFTLNW+KY F
Sbjct: 414 QLYSSFAKGANPFFTLNWSKYTQF 343
>AW309963 similar to GP|336870|gb|AA alpha-apoprotein {Euglena gracilis},
partial (3%)
Length = 464
Score = 42.0 bits (97), Expect = 0.001
Identities = 19/26 (73%), Positives = 22/26 (84%)
Frame = +1
Query: 180 KAAPKLAWFQDVESMLNHHLAGLLGL 205
KAA KL WFQ+V+SMLN+HL LLGL
Sbjct: 385 KAASKLPWFQNVKSMLNYHLTRLLGL 462
>TC207770 homologue to UP|O22445 (O22445) Developmentally regulated GTP
binding protein, partial (46%)
Length = 910
Score = 33.1 bits (74), Expect = 0.46
Identities = 12/19 (63%), Positives = 14/19 (73%)
Frame = -3
Query: 254 FAEGATPFFTLNWAKYADF 272
FA+G PFFTL W+KY F
Sbjct: 908 FAKGVNPFFTLTWSKYTKF 852
>TC203397 similar to UP|GPX4_CITSI (Q06652) Probable phospholipid
hydroperoxide glutathione peroxidase (PHGPx)
(Salt-associated protein) , partial (94%)
Length = 1010
Score = 30.4 bits (67), Expect = 3.0
Identities = 25/106 (23%), Positives = 42/106 (39%), Gaps = 14/106 (13%)
Frame = +2
Query: 608 FSWKMQSDVWGSISDQGVVTHITGGNFAQSSITINGWLRDFLWAQASQVIQSYGSSLSAY 667
F+WK QS + G+ Q +A+ IN ++ D L+ S ++ + +
Sbjct: 53 FNWKRQSSITGTRPRQV-------SQYAEKGRYINIFVPDALYFSHSYILPPLNNYKIIF 211
Query: 668 GLFF--------------LGAHFVWAFSLMFLFSGRGYWQELIESI 699
FF L +HF+ S +FLF R ++ L I
Sbjct: 212 FFFFSSILLQSQPHPLPSLSSHFLQTTSFVFLFPNRSHYGHLKRQI 349
>BU760759 weakly similar to GP|14090380|dbj P0037C04.27 {Oryza sativa
(japonica cultivar-group)}, partial (17%)
Length = 429
Score = 30.0 bits (66), Expect = 3.9
Identities = 19/45 (42%), Positives = 24/45 (53%)
Frame = -1
Query: 208 LSWAGHQVHVSLPINQFLNAGVDPKEIPLPHEFILNRDLLAQLYP 252
LS +G H SLP FL +DP +PLP L+ LL+Q P
Sbjct: 282 LSGSGISQHASLPSLAFLPVSLDPHLLPLP--LSLHLHLLSQNKP 154
>TC204410 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc finger-like
protein), partial (45%)
Length = 471
Score = 28.9 bits (63), Expect = 8.8
Identities = 18/58 (31%), Positives = 24/58 (41%)
Frame = +1
Query: 447 FHSFGLYIHNDTMSALGRPQDMFSDTAIQLQPIFAQWIQNTHALAPGITAPGATTGTS 504
FHS+ +HN T + + F IQ W +NT A P +T TT S
Sbjct: 43 FHSWSYLLHNPTTTPCYKSDFFFLFLQIQ------TWPKNTDAKLPKVTDSAPTTAAS 198
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,086,183
Number of Sequences: 63676
Number of extensions: 664139
Number of successful extensions: 4036
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 3983
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4027
length of query: 750
length of database: 12,639,632
effective HSP length: 104
effective length of query: 646
effective length of database: 6,017,328
effective search space: 3887193888
effective search space used: 3887193888
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Description of BAB33186.1

