
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BAB33183.1 158 aa
(158 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
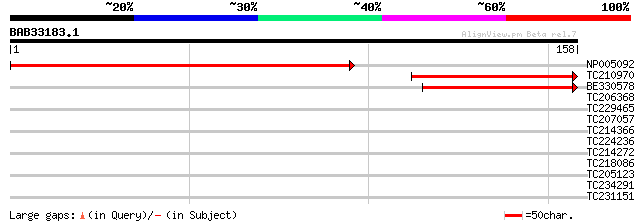
Score E
Sequences producing significant alignments: (bits) Value
NP005092 orf158/159 182 4e-47
TC210970 homologue to UP|NUGC_TOBAC (P12201) NAD(P)H-quinone oxi... 101 1e-22
BE330578 similar to SP|P12201|NUGC_ NAD(P)H-quinone oxidoreducta... 82 1e-16
TC206368 similar to UP|Q8L7U2 (Q8L7U2) AT4g30890/F6I18_200, part... 30 0.37
TC229465 weakly similar to UP|Q8LA09 (Q8LA09) Lectin-like protei... 27 3.2
TC207057 weakly similar to UP|O24515 (O24515) Unconventional myo... 27 4.1
TC214366 homologue to UP|Q7XA96 (Q7XA96) Photosystem I psaH prot... 26 7.0
TC224236 weakly similar to UP|Q941P1 (Q941P1) Sinapoylglucose:ch... 26 7.0
TC214272 similar to UP|Q6NMQ7 (Q6NMQ7) At1g74670, partial (68%) 26 7.0
TC218086 26 9.2
TC205123 weakly similar to GB|AAM19921.1|20453365|AY097405 AT3g2... 26 9.2
TC234291 26 9.2
TC231151 similar to UP|Q7PC87 (Q7PC87) PDR6 ABC transporter, par... 26 9.2
>NP005092 orf158/159
Length = 293
Score = 182 bits (463), Expect = 4e-47
Identities = 86/96 (89%), Positives = 90/96 (93%)
Frame = +1
Query: 1 MQGRLSAWLVKHGLVHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
MQGRLS+WLVKHGL HRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDV+
Sbjct: 1 MQGRLSSWLVKHGLSHRSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVA 180
Query: 61 PGGLLASVYHLTRIECGIYQPEEVCIKIFVPRNNPR 96
PGGLLASVYHLTR+E I QPEEVC+KIFV R NPR
Sbjct: 181PGGLLASVYHLTRLEYDIGQPEEVCMKIFVARGNPR 288
>TC210970 homologue to UP|NUGC_TOBAC (P12201) NAD(P)H-quinone oxidoreductase
chain J, chloroplast (NAD(P)H dehydrogenase, chain J)
(NADH-plastoquinone oxidoreductase subunit J) , partial
(29%)
Length = 694
Score = 101 bits (252), Expect = 1e-22
Identities = 44/46 (95%), Positives = 45/46 (97%)
Frame = -2
Query: 113 ESYDMLGISYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
ESYDMLGISYDNHPRL+RILMPESWIGWPLRKDYI PNFYEIQDAH
Sbjct: 684 ESYDMLGISYDNHPRLRRILMPESWIGWPLRKDYIAPNFYEIQDAH 547
>BE330578 similar to SP|P12201|NUGC_ NAD(P)H-quinone oxidoreductase chain J
chloroplast (EC 1.6.5.-) (NAD(P)H dehydrogenase chain
J), partial (27%)
Length = 136
Score = 82.0 bits (201), Expect = 1e-16
Identities = 37/43 (86%), Positives = 38/43 (88%)
Frame = +2
Query: 116 DMLGISYDNHPRLKRILMPESWIGWPLRKDYITPNFYEIQDAH 158
DMLGISYDNH RL RILMP SWIGWP RKDYI+PN YEIQDAH
Sbjct: 8 DMLGISYDNHSRLIRILMP*SWIGWPSRKDYISPNCYEIQDAH 136
>TC206368 similar to UP|Q8L7U2 (Q8L7U2) AT4g30890/F6I18_200, partial (42%)
Length = 711
Score = 30.4 bits (67), Expect = 0.37
Identities = 11/29 (37%), Positives = 14/29 (47%)
Frame = +2
Query: 82 EEVCIKIFVPRNNPRIPSIFWVWKSADFQ 110
+E+C P NN P FW+WK Q
Sbjct: 8 KEICSNSDTPENNDIAPDAFWLWKPGKHQ 94
>TC229465 weakly similar to UP|Q8LA09 (Q8LA09) Lectin-like protein, partial
(10%)
Length = 798
Score = 27.3 bits (59), Expect = 3.2
Identities = 10/19 (52%), Positives = 11/19 (57%)
Frame = -1
Query: 133 MPESWIGWPLRKDYITPNF 151
MP+ WI WP I PNF
Sbjct: 165 MPKQWINWP*LTKLIQPNF 109
>TC207057 weakly similar to UP|O24515 (O24515) Unconventional myosin, partial
(8%)
Length = 1017
Score = 26.9 bits (58), Expect = 4.1
Identities = 10/29 (34%), Positives = 19/29 (65%)
Frame = +3
Query: 129 KRILMPESWIGWPLRKDYITPNFYEIQDA 157
++ L PE+ P+R+D++ P Y+ +DA
Sbjct: 159 RKSLAPENATVQPVRRDFVLPRGYDSEDA 245
>TC214366 homologue to UP|Q7XA96 (Q7XA96) Photosystem I psaH protein,
partial (42%)
Length = 763
Score = 26.2 bits (56), Expect = 7.0
Identities = 10/35 (28%), Positives = 21/35 (59%)
Frame = +2
Query: 26 IETLQIKPEDWHSIAVILYVYGYNYLRSQCAYDVS 60
I+ LQ+ + IA+ +Y+Y + Y++ C ++S
Sbjct: 53 IQELQVPFHAFP*IAIYIYIYTHTYIQEFCIKEIS 157
>TC224236 weakly similar to UP|Q941P1 (Q941P1) Sinapoylglucose:choline
sinapoyltransferase, partial (23%)
Length = 507
Score = 26.2 bits (56), Expect = 7.0
Identities = 14/58 (24%), Positives = 29/58 (49%), Gaps = 6/58 (10%)
Frame = +2
Query: 22 DYQG-IETLQIKPEDWHSIAVILYV-----YGYNYLRSQCAYDVSPGGLLASVYHLTR 73
+Y G + L +KP+ W ++ I++V G++Y +++ A S L+ + R
Sbjct: 278 EYNGSLPNLTLKPQSWTKVSSIIFVDLPAGTGFSYPKTERAVQQSSSKLVRHAHQFIR 451
>TC214272 similar to UP|Q6NMQ7 (Q6NMQ7) At1g74670, partial (68%)
Length = 784
Score = 26.2 bits (56), Expect = 7.0
Identities = 12/36 (33%), Positives = 19/36 (52%)
Frame = +2
Query: 36 WHSIAVILYVYGYNYLRSQCAYDVSPGGLLASVYHL 71
WHS+ + YV+ + + + GLL S+YHL
Sbjct: 647 WHSMILCFYVFWC--ITPWALFAFNESGLLISIYHL 748
>TC218086
Length = 1385
Score = 25.8 bits (55), Expect = 9.2
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = +1
Query: 114 SYDMLGISYDNHPRLKRILMPESWIGWPLR 143
SY M G++ HP L R+ +P+ PLR
Sbjct: 496 SYGMGGVAVHVHPLLSRLALPQYNAHLPLR 585
>TC205123 weakly similar to GB|AAM19921.1|20453365|AY097405 AT3g23600/MDB19_9
{Arabidopsis thaliana;} , partial (97%)
Length = 1010
Score = 25.8 bits (55), Expect = 9.2
Identities = 20/58 (34%), Positives = 28/58 (47%), Gaps = 8/58 (13%)
Frame = -1
Query: 48 YNYLRSQC-AYDV------SPGGLLASVYHLTRIECGIYQPEEVCI-KIFVPRNNPRI 97
Y+ R++C YDV SPGG L ++C I QP V + +IF P+I
Sbjct: 797 YSLNRARCLTYDV*QASPASPGGPLLPPLRFLHLQCYISQPNRVKLWEIF*QNWQPQI 624
>TC234291
Length = 542
Score = 25.8 bits (55), Expect = 9.2
Identities = 12/26 (46%), Positives = 14/26 (53%)
Frame = +3
Query: 56 AYDVSPGGLLASVYHLTRIECGIYQP 81
A+ VSP L VY+L EC I P
Sbjct: 216 AFQVSPSSSLDLVYYLRYTECKIISP 293
>TC231151 similar to UP|Q7PC87 (Q7PC87) PDR6 ABC transporter, partial (11%)
Length = 744
Score = 25.8 bits (55), Expect = 9.2
Identities = 10/38 (26%), Positives = 19/38 (49%)
Frame = +3
Query: 17 RSLGFDYQGIETLQIKPEDWHSIAVILYVYGYNYLRSQ 54
++LGFDY + + W + + ++ YG +L Q
Sbjct: 348 QNLGFDYDFLPVVAAAHVGWVILFMFVFAYGIKFLNFQ 461
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.143 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,334,864
Number of Sequences: 63676
Number of extensions: 146351
Number of successful extensions: 646
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 646
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 646
length of query: 158
length of database: 12,639,632
effective HSP length: 89
effective length of query: 69
effective length of database: 6,972,468
effective search space: 481100292
effective search space used: 481100292
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 55 (25.8 bits)
Description of BAB33183.1

