

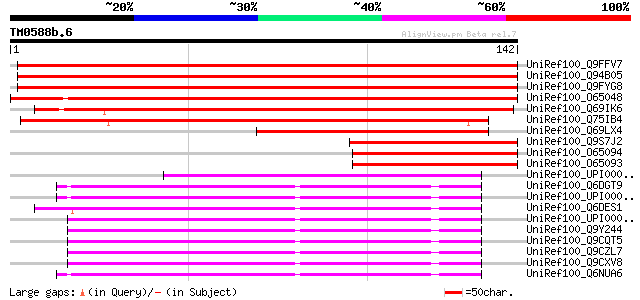
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0588b.6
(142 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FFV7 Gb|AAB95243.1 [Arabidopsis thaliana] 214 2e-55
UniRef100_Q94B05 Hypothetical protein F1N21.15 [Arabidopsis thal... 214 4e-55
UniRef100_Q9FYG8 F1N21.7 [Arabidopsis thaliana] 212 1e-54
UniRef100_O65048 Hypothetical protein Sb08 [Picea mariana] 197 5e-50
UniRef100_Q69IK6 Proteasome maturation factor-like [Oryza sativa] 157 6e-38
UniRef100_Q75IB4 Putative proteasome maturation factor [Oryza sa... 123 8e-28
UniRef100_Q69LX4 Proteasome maturation factor-like protein [Oryz... 89 2e-17
UniRef100_Q9S7J2 Hypothetical protein Sb08 [Picea abies] 88 5e-17
UniRef100_O65094 Hypothetical protein Sb08 [Picea mariana] 84 5e-16
UniRef100_O65093 Hypothetical protein Sb08 [Picea mariana] 83 1e-15
UniRef100_UPI000031698B UPI000031698B UniRef100 entry 72 3e-12
UniRef100_Q6DGT9 Zgc:92775 [Brachydanio rerio] 67 6e-11
UniRef100_UPI0000360ABD UPI0000360ABD UniRef100 entry 66 1e-10
UniRef100_Q6DES1 MGC89819 protein [Xenopus tropicalis] 65 2e-10
UniRef100_UPI00001CB723 UPI00001CB723 UniRef100 entry 64 5e-10
UniRef100_Q9Y244 HSPC036 protein [Homo sapiens] 62 4e-09
UniRef100_Q9CQT5 Mus musculus 14, 17 days embryo head cDNA, RIKE... 61 5e-09
UniRef100_Q9CZL7 Mus musculus 11 days embryo whole body cDNA, RI... 61 5e-09
UniRef100_Q9CXV8 Mus musculus 13 days embryo head cDNA, RIKEN fu... 61 6e-09
UniRef100_Q6NUA6 MGC81104 protein [Xenopus laevis] 60 1e-08
>UniRef100_Q9FFV7 Gb|AAB95243.1 [Arabidopsis thaliana]
Length = 141
Score = 214 bits (546), Expect = 2e-55
Identities = 102/140 (72%), Positives = 120/140 (84%)
Query: 3 EAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
E+ K IAH+IGG++NDALRFGL GVKSDI+ SHPL++A ES + E MKR+ + + YGA
Sbjct: 2 ESQKKIAHEIGGMKNDALRFGLHGVKSDILRSHPLETAYESGKQSQEEMKRRVITHTYGA 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
A PLKMDLDRQILSRFQRP G IPSSMLGLE +TG+LD+FGFEDYL+DPRESET++P+D
Sbjct: 62 ALPLKMDLDRQILSRFQRPPGPIPSSMLGLEVYTGALDNFGFEDYLNDPRESETLKPVDF 121
Query: 123 HHGMEVRLGLSKGPVCPSFM 142
HHGMEVRLGLSKGP PSFM
Sbjct: 122 HHGMEVRLGLSKGPASPSFM 141
>UniRef100_Q94B05 Hypothetical protein F1N21.15 [Arabidopsis thaliana]
Length = 141
Score = 214 bits (544), Expect = 4e-55
Identities = 98/140 (70%), Positives = 119/140 (85%)
Query: 3 EAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
E+ K IAH+IGG++NDALRFGL GVKS+I+GSHPL+S+ ES K E +KR + + YG
Sbjct: 2 ESEKKIAHEIGGVKNDALRFGLHGVKSNIIGSHPLESSYESEKKSKEALKRTVIAHAYGT 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
A PLKMD+DRQILSRFQRP G IPSSMLGLE +TG++DDFGFEDYL+DPR+SET +P+D
Sbjct: 62 ALPLKMDMDRQILSRFQRPPGPIPSSMLGLEVYTGAVDDFGFEDYLNDPRDSETFKPVDF 121
Query: 123 HHGMEVRLGLSKGPVCPSFM 142
HHGMEVRLG+SKGP+ PSFM
Sbjct: 122 HHGMEVRLGISKGPIYPSFM 141
>UniRef100_Q9FYG8 F1N21.7 [Arabidopsis thaliana]
Length = 303
Score = 212 bits (540), Expect = 1e-54
Identities = 97/140 (69%), Positives = 119/140 (84%)
Query: 3 EAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
E+ K IAH+IGG++NDALRFGL GVKS+I+GSHPL+S+ ES K E +KR + + YG
Sbjct: 2 ESEKKIAHEIGGVKNDALRFGLHGVKSNIIGSHPLESSYESEKKSKEALKRTVIAHAYGT 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
A PLKMD+DRQILSRFQRP G IPSSMLGLE +TG++DDFGFEDYL+DPR+SET +P+D
Sbjct: 62 ALPLKMDMDRQILSRFQRPPGPIPSSMLGLEVYTGAVDDFGFEDYLNDPRDSETFKPVDF 121
Query: 123 HHGMEVRLGLSKGPVCPSFM 142
HHGMEVRLG+SKGP+ PSF+
Sbjct: 122 HHGMEVRLGISKGPIYPSFI 141
>UniRef100_O65048 Hypothetical protein Sb08 [Picea mariana]
Length = 141
Score = 197 bits (500), Expect = 5e-50
Identities = 97/142 (68%), Positives = 113/142 (79%), Gaps = 1/142 (0%)
Query: 1 MEEAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLY 60
M+ +S+ H+IGGI D LRFGL GVKSDI+ SHP+Q QE+A K MKR + Y
Sbjct: 1 MDPVQQSLPHEIGGI-TDTLRFGLAGVKSDILISHPVQVIQENAQKQQREMKRNILAKTY 59
Query: 61 GAAFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPL 120
G+AFPLKMD+++QILSRFQRP G +PSSMLGLE+ TGSLD+F FEDYL+DPRESET P
Sbjct: 60 GSAFPLKMDIEQQILSRFQRPPGELPSSMLGLESLTGSLDEFSFEDYLNDPRESETFVPA 119
Query: 121 DMHHGMEVRLGLSKGPVCPSFM 142
DMHHGMEVRLGLSKGPVC SFM
Sbjct: 120 DMHHGMEVRLGLSKGPVCRSFM 141
>UniRef100_Q69IK6 Proteasome maturation factor-like [Oryza sativa]
Length = 141
Score = 157 bits (396), Expect = 6e-38
Identities = 81/135 (60%), Positives = 99/135 (73%), Gaps = 2/135 (1%)
Query: 8 IAHQIGGIQNDALRFGLQ-GVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPL 66
+ +IGG +D LRFG+ VK D+ HPLQ++ +K KR +YG+AF +
Sbjct: 7 VKKEIGG-NHDVLRFGVNDSVKGDLAPPHPLQASVHKDAKFWADKKRFGAEAIYGSAFNI 65
Query: 67 KMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGM 126
+ DLD QILS+FQRP GA+PSSMLG EA TGSLDDFGFEDYL+ P++SE+ R DMHHGM
Sbjct: 66 RKDLDAQILSKFQRPPGALPSSMLGYEALTGSLDDFGFEDYLNLPQDSESFRAPDMHHGM 125
Query: 127 EVRLGLSKGPVCPSF 141
EVRLGLSKGPVCPSF
Sbjct: 126 EVRLGLSKGPVCPSF 140
>UniRef100_Q75IB4 Putative proteasome maturation factor [Oryza sativa]
Length = 281
Score = 123 bits (309), Expect = 8e-28
Identities = 66/135 (48%), Positives = 89/135 (65%), Gaps = 4/135 (2%)
Query: 4 AAKSIAHQIGGIQNDALRFGLQG-VKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
A+ SI + G +D L FG+ VK D+ HP+Q+ +K KR +YG+
Sbjct: 2 ASGSIVKKEVGENHDVLLFGVNNSVKGDLAPQHPIQATVHKEAKFWADKKRFGAEAIYGS 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
AF ++ DLD QILS+FQRP GA+PSSMLG EA TGSLDDFGFEDYL+ P++S++ DM
Sbjct: 62 AFNIRKDLDAQILSKFQRPPGALPSSMLGYEALTGSLDDFGFEDYLNLPQDSDSFHAPDM 121
Query: 123 HHGME---VRLGLSK 134
HHGME +++G+ K
Sbjct: 122 HHGMEKNLLKIGICK 136
>UniRef100_Q69LX4 Proteasome maturation factor-like protein [Oryza sativa]
Length = 258
Score = 89.4 bits (220), Expect = 2e-17
Identities = 42/65 (64%), Positives = 51/65 (77%)
Query: 70 LDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVR 129
+D + RFQRP GA+PSSMLG EA TGSLDDFGFEDYL+ P++S++ DMHHGMEV
Sbjct: 29 VDDLSVCRFQRPPGALPSSMLGYEALTGSLDDFGFEDYLNLPQDSDSFHAPDMHHGMEVL 88
Query: 130 LGLSK 134
LG+ K
Sbjct: 89 LGICK 93
>UniRef100_Q9S7J2 Hypothetical protein Sb08 [Picea abies]
Length = 48
Score = 87.8 bits (216), Expect = 5e-17
Identities = 40/47 (85%), Positives = 42/47 (89%)
Query: 96 TGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGLSKGPVCPSFM 142
TGSLD+F FEDYL+DPRESET P DMHHGMEVRLGLSKGPVC SFM
Sbjct: 2 TGSLDEFSFEDYLNDPRESETFVPADMHHGMEVRLGLSKGPVCRSFM 48
>UniRef100_O65094 Hypothetical protein Sb08 [Picea mariana]
Length = 46
Score = 84.3 bits (207), Expect = 5e-16
Identities = 38/46 (82%), Positives = 41/46 (88%)
Query: 97 GSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGLSKGPVCPSFM 142
GSLD+F FEDYL+DPRESET P DMHHGMEVRLGLSKGPVC SF+
Sbjct: 1 GSLDEFSFEDYLNDPRESETFVPADMHHGMEVRLGLSKGPVCRSFI 46
>UniRef100_O65093 Hypothetical protein Sb08 [Picea mariana]
Length = 46
Score = 82.8 bits (203), Expect = 1e-15
Identities = 38/46 (82%), Positives = 40/46 (86%)
Query: 97 GSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGLSKGPVCPSFM 142
GSLD+F FEDYL+DPRESET P DMHHGMEVRLGLSKG VC SFM
Sbjct: 1 GSLDEFSFEDYLNDPRESETFVPADMHHGMEVRLGLSKGSVCRSFM 46
>UniRef100_UPI000031698B UPI000031698B UniRef100 entry
Length = 96
Score = 71.6 bits (174), Expect = 3e-12
Identities = 39/89 (43%), Positives = 50/89 (55%)
Query: 44 ASKINEVMKRQCMVNLYGAAFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFG 103
AS + K + M GAA P K+D++RQILS+ +R G I S LGL+ TG +D FG
Sbjct: 1 ASGVEYRRKLEMMAQAQGAAIPAKLDIERQILSQVERLPGLIESKRLGLQIMTGEIDRFG 60
Query: 104 FEDYLSDPRESETIRPLDMHHGMEVRLGL 132
FEDYL+ E D H ME +L L
Sbjct: 61 FEDYLNQASMREEGPRADTHSIMEAKLNL 89
>UniRef100_Q6DGT9 Zgc:92775 [Brachydanio rerio]
Length = 142
Score = 67.4 bits (163), Expect = 6e-11
Identities = 42/119 (35%), Positives = 67/119 (56%), Gaps = 4/119 (3%)
Query: 14 GIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQ 73
G+Q D+LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ +
Sbjct: 27 GVQ-DSLRRGFSSVKNELLPSHPLELSEKNFQLNQDKMNFNTLRNIQGLHAPLKLQMEYR 85
Query: 74 ILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ QR +PSS L L+ GS D GFED L+DP + E + D H E +LGL
Sbjct: 86 AAKQIQR-LPFLPSSNLALDTLRGSDDTIGFEDILNDPVQCEMMG--DPHIMTEYKLGL 141
>UniRef100_UPI0000360ABD UPI0000360ABD UniRef100 entry
Length = 141
Score = 66.2 bits (160), Expect = 1e-10
Identities = 39/119 (32%), Positives = 69/119 (57%), Gaps = 4/119 (3%)
Query: 14 GIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQ 73
G+Q D++R G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ +
Sbjct: 26 GVQ-DSIRSGFSSVKNELMPSHPLELSEKNFQLNRDKMSFSTLRNIQGLHAPLKLQMEYR 84
Query: 74 ILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ QR +PSS L ++ G+ + GFED L+DP +SE + D H +E +LGL
Sbjct: 85 AARQIQR-LPFLPSSNLAVDTLRGNDESVGFEDILNDPAQSEMMG--DPHLMVEYKLGL 140
>UniRef100_Q6DES1 MGC89819 protein [Xenopus tropicalis]
Length = 142
Score = 65.5 bits (158), Expect = 2e-10
Identities = 41/126 (32%), Positives = 72/126 (56%), Gaps = 4/126 (3%)
Query: 8 IAHQIGGIQ-NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPL 66
++ Q GG +D LR G V+S+++ SHPL+ ++++ E + + N+ G PL
Sbjct: 19 LSTQYGGYGVHDNLRTGFTSVRSELMPSHPLELSEKNFQINQEKVNLSTVRNIQGLHAPL 78
Query: 67 KMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGM 126
K+ ++ + + + QR +PSS + L+ G+ + GFED L+DP +SE + + H M
Sbjct: 79 KLQMEFKAVRQVQR-LPFLPSSNIALDTLRGTDESIGFEDILNDPSQSEGMG--EPHLMM 135
Query: 127 EVRLGL 132
E +LGL
Sbjct: 136 EYKLGL 141
>UniRef100_UPI00001CB723 UPI00001CB723 UniRef100 entry
Length = 141
Score = 64.3 bits (155), Expect = 5e-10
Identities = 38/116 (32%), Positives = 67/116 (57%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ QR +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVQR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSELMG--EPHMMVEYKLGL 140
>UniRef100_Q9Y244 HSPC036 protein [Homo sapiens]
Length = 141
Score = 61.6 bits (148), Expect = 4e-09
Identities = 37/116 (31%), Positives = 66/116 (56%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ QR + SS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVQR-LPFLSSSNLSLDVLRGNDETIGFEDILNDPSQSEVMG--EPHLMVEYKLGL 140
>UniRef100_Q9CQT5 Mus musculus 14, 17 days embryo head cDNA, RIKEN full-length
enriched library, clone:3200001P07 product:HSPC036
PROTEIN (HYPOTHETICAL 15.8 kDa PROTEIN) (VOLTAGE-GATED K
CHANNEL BETA SUBUNIT 4.1) homolog (RIKEN cDNA
2510048O06) (Mus musculus 18-day embryo whole body cDNA,
RIKEN full-length enriched library, clone:1110004N03
product:HSPC036 PROTEIN (HYPOTHETICAL 15.8 kDa PROTEIN)
(VOLTAGE-GATED K CHANNEL BETA SUBUNIT 4.1) homolog) (Mus
musculus 13 days embryo liver cDNA, RIKEN full- length
enriched library, clone:2510048O06 product:HSPC036
PROTEIN (HYPOTHETICAL 15.8 kDa PROTEIN) (VOLTAGE-GATED K
CHANNEL BETA SUBUNIT 4.1) homolog) [Mus musculus]
Length = 141
Score = 61.2 bits (147), Expect = 5e-09
Identities = 37/116 (31%), Positives = 66/116 (56%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ R +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVHR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSELMG--EPHVMVEHKLGL 140
>UniRef100_Q9CZL7 Mus musculus 11 days embryo whole body cDNA, RIKEN full-length
enriched library, clone:2700059D08 product:HSPC036
PROTEIN (HYPOTHETICAL 15.8 kDa PROTEIN) (VOLTAGE-GATED K
CHANNEL BETA SUBUNIT 4.1) homolog [Mus musculus]
Length = 141
Score = 61.2 bits (147), Expect = 5e-09
Identities = 37/116 (31%), Positives = 66/116 (56%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ R +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVHR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSEQMG--EPHVMVEHKLGL 140
>UniRef100_Q9CXV8 Mus musculus 13 days embryo head cDNA, RIKEN full-length enriched
library, clone:3110001B11 product:HSPC036 PROTEIN
(HYPOTHETICAL 15.8 kDa PROTEIN) (VOLTAGE-GATED K CHANNEL
BETA SUBUNIT 4.1) homolog [Mus musculus]
Length = 141
Score = 60.8 bits (146), Expect = 6e-09
Identities = 37/116 (31%), Positives = 66/116 (56%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSKKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ R +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVHR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSELMG--EPHVMVEHKLGL 140
>UniRef100_Q6NUA6 MGC81104 protein [Xenopus laevis]
Length = 142
Score = 60.1 bits (144), Expect = 1e-08
Identities = 39/119 (32%), Positives = 67/119 (55%), Gaps = 4/119 (3%)
Query: 14 GIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQ 73
GIQ D LR G V+S+++ SHPL+ ++++ E + + N+ G PLK+ ++ +
Sbjct: 27 GIQ-DTLRTGFTSVQSELMPSHPLELSEKNFQINQEKVNFSTVRNIQGLHAPLKLHMEFK 85
Query: 74 ILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ + QR +P S + L+ G+ + FED L+DP +SE + + H ME +LGL
Sbjct: 86 AVKQVQR-LPFLPISNIALDTLRGTDECISFEDILNDPSQSELMG--EPHLMMEYKLGL 141
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 216,610,326
Number of Sequences: 2790947
Number of extensions: 7962678
Number of successful extensions: 15449
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 15405
Number of HSP's gapped (non-prelim): 43
length of query: 142
length of database: 848,049,833
effective HSP length: 118
effective length of query: 24
effective length of database: 518,718,087
effective search space: 12449234088
effective search space used: 12449234088
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0588b.6


