

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0588b.3
(432 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
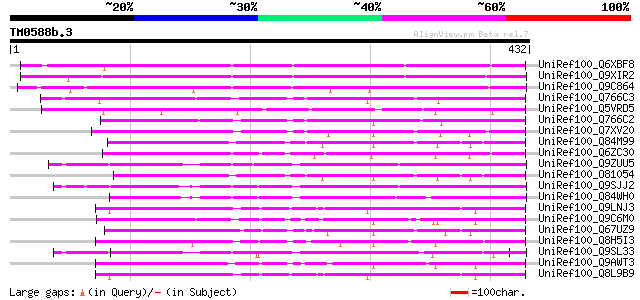
UniRef100_Q6XBF8 CDR1 [Arabidopsis thaliana] 318 1e-85
UniRef100_Q9XIR2 F13O11.13 protein [Arabidopsis thaliana] 310 5e-83
UniRef100_Q9C864 Chloroplast nucleoid DNA binding protein, putat... 278 2e-73
UniRef100_Q766C3 Aspartic proteinase nepenthesin I [Nepenthes gr... 240 5e-62
UniRef100_Q5VRD5 Putative CDR1 [Oryza sativa] 232 1e-59
UniRef100_Q766C2 Aspartic proteinase nepenthesin II [Nepenthes g... 230 7e-59
UniRef100_Q7XV20 OSJNBa0064H22.10 protein [Oryza sativa] 225 2e-57
UniRef100_Q84M99 At2g03200 [Arabidopsis thaliana] 220 5e-56
UniRef100_Q6ZC30 Putative nucleoid DNA-binding protein [Oryza sa... 220 7e-56
UniRef100_Q9ZUU5 Putative chloroplast nucleoid DNA binding prote... 216 8e-55
UniRef100_O81054 Putative chloroplast nucleoid DNA binding prote... 214 4e-54
UniRef100_Q9SJJ2 Putative chloroplast nucleoid DNA binding prote... 211 3e-53
UniRef100_Q84WH0 Putative chloroplast nucleoid DNA binding prote... 206 1e-51
UniRef100_Q9LNJ3 F6F3.10 protein [Arabidopsis thaliana] 206 1e-51
UniRef100_Q9C6M0 Hypothetical protein F2J7.6 [Arabidopsis thaliana] 205 2e-51
UniRef100_Q67UZ9 Putative aspartic proteinase nepenthesin I [Ory... 205 2e-51
UniRef100_Q8H5I3 Putative nucleoid DNA-binding protein cnd41 [Or... 204 3e-51
UniRef100_Q9SL33 Putative chloroplast nucleoid DNA binding prote... 204 3e-51
UniRef100_Q9AWT3 Putative aspartic proteinase nepenthesin I [Ory... 201 3e-50
UniRef100_Q8L9B9 Chloroplast nucleoid DNA binding protein, putat... 201 3e-50
>UniRef100_Q6XBF8 CDR1 [Arabidopsis thaliana]
Length = 437
Score = 318 bits (816), Expect = 1e-85
Identities = 176/430 (40%), Positives = 257/430 (58%), Gaps = 17/430 (3%)
Query: 10 LCFLVVLYLLSCQTPIEAQDAGFSVQLTRQNSPHSPFYKPDNLHRHKLPS-FHQVPKKAF 68
LC L L+L + + GF+ L ++SP SPFY P +L + H+ + F
Sbjct: 12 LCLLSSLFLSNANAKPKL---GFTADLIHRDSPKSPFYNPMETSSQRLRNAIHRSVNRVF 68
Query: 69 APNGPFSTR-----VTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQK 123
+T +TSN+G+YLM +++G+PP I + DTGSDL+W QC+PC CY Q
Sbjct: 69 HFTEKDNTPQPQIDLTSNSGEYLMNVSIGTPPFPIMAIADTGSDLLWTQCAPCDDCYTQV 128
Query: 124 SPMFEPLSSKTFNPIPCDSEQCGSLFSH-SCSP-QKLCAYSYSYADSSVTKGVLARETIT 181
P+F+P +S T+ + C S QC +L + SCS C+YS SY D+S TKG +A +T+T
Sbjct: 129 DPLFDPKTSSTYKDVSCSSSQCTALENQASCSTNDNTCSYSLSYGDNSYTKGNIAVDTLT 188
Query: 182 FSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQC 241
S ++ + + +II GCGH+N+G FN+ G++GLGGGP+SL+ Q+G +FS C
Sbjct: 189 LGS-SDTRPMQLKNIIIGCGHNNAGTFNKKGSGIVGLGGGPVSLIKQLGDSIDG-KFSYC 246
Query: 242 LVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQ-TPYLVTLEGISVGDTFVSFNS 300
LVP + + I+FG + VSG GVV+TPL+++ Q T Y +TL+ ISVG + ++
Sbjct: 247 LVPLTSKKDQTSKINFGTNAIVSGSGVVSTPLIAKASQETFYYLTLKSISVGSKQIQYSG 306
Query: 301 SE-KLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNL 359
S+ + S+GN++IDSGT T LP EFY L E+ S DP G LCY + +L
Sbjct: 307 SDSESSEGNIIIDSGTTLTLLPTEFYSEL-EDAVASSIDAEKKQDPQSGLSLCYSATGDL 365
Query: 360 EGPILTAHFEGADVQLMPIQTFIPPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDR 419
+ P++T HF+GADV+L F+ + + CFA G+ I+GN AQ N L+G+D
Sbjct: 366 KVPVITMHFDGADVKLDSSNAFVQVSEDLVCFAFRGSPSFS-IYGNVAQMNFLVGYDTVS 424
Query: 420 KTISFKPTDC 429
KT+SFKPTDC
Sbjct: 425 KTVSFKPTDC 434
>UniRef100_Q9XIR2 F13O11.13 protein [Arabidopsis thaliana]
Length = 431
Score = 310 bits (794), Expect = 5e-83
Identities = 175/431 (40%), Positives = 247/431 (56%), Gaps = 15/431 (3%)
Query: 10 LCFLVVLYLLSCQTPIEAQDAGFSVQLTRQNSPHSPFY--------KPDNLHRHKLPSFH 61
L F +L LL GF++ L ++SP SPFY + N R S
Sbjct: 4 LIFATLLSLLLLSNVNAYPKDGFTIDLIHRDSPKSPFYNSAETSSQRMRNAIRRSARSTL 63
Query: 62 QVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYK 121
Q +PN P S +TSN G+YLM +++G+PPV I + DTGSDL+W QC+PC CY+
Sbjct: 64 QFSNDDASPNSPQSF-ITSNRGEYLMNISIGTPPVPILAIADTGSDLIWTQCNPCEDCYQ 122
Query: 122 QKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCS-PQKLCAYSYSYADSSVTKGVLARETI 180
Q SP+F+P S T+ + C S QC +L SCS + C+Y+ +Y D+S TKG +A +T+
Sbjct: 123 QTSPLFDPKESSTYRKVSCSSSQCRALEDASCSTDENTCSYTITYGDNSYTKGDVAVDTV 182
Query: 181 TFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQ 240
T S + + + ++I GCGH N+G F+ G+IGLGGG SLVSQ+ +FS
Sbjct: 183 TMGS-SGRRPVSLRNMIIGCGHENTGTFDPAGSGIIGLGGGSTSLVSQLRKSING-KFSY 240
Query: 241 CLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFNS 300
CLVPF +++ + I+FG VSG+GVV+T +V ++ T Y + LE ISVG + F S
Sbjct: 241 CLVPFTSETGLTSKINFGTNGIVSGDGVVSTSMVKKDPATYYFLNLEAISVGSKKIQFTS 300
Query: 301 S-EKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNL 359
+ +GN++IDSGT T LP FY L E + + DPD LCYR ++
Sbjct: 301 TIFGTGEGNIVIDSGTTLTLLPSNFYYEL-ESVVASTIKAERVQDPDGILSLCYRDSSSF 359
Query: 360 EGPILTAHFEGADVQLMPIQTFIPPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDR 419
+ P +T HF+G DV+L + TF+ + V CFA A + IFGN AQ N L+G+D
Sbjct: 360 KVPDITVHFKGGDVKLGNLNTFVAVSEDVSCFAFAAN-EQLTIFGNLAQMNFLVGYDTVS 418
Query: 420 KTISFKPTDCT 430
T+SFK TDC+
Sbjct: 419 GTVSFKKTDCS 429
>UniRef100_Q9C864 Chloroplast nucleoid DNA binding protein, putative [Arabidopsis
thaliana]
Length = 445
Score = 278 bits (712), Expect = 2e-73
Identities = 159/444 (35%), Positives = 238/444 (52%), Gaps = 27/444 (6%)
Query: 7 FFHLCFLVVLYLLSCQTPIEAQDAGFSVQLTRQNSPHSPFYKP-----DNLHRHKLPSFH 61
F + L + + + + A +V+L ++SPHSP Y P D L+ L S
Sbjct: 6 FLYCSLLAISFFFASNS--SANRENLTVELIHRDSPHSPLYNPHHTVSDRLNAAFLRSIS 63
Query: 62 QVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYK 121
+ + F + + SN G+Y M +++G+PP ++ + DTGSDL W QC PC CYK
Sbjct: 64 R--SRRFTTKTDLQSGLISNGGEYFMSISIGTPPSKVFAIADTGSDLTWVQCKPCQQCYK 121
Query: 122 QKSPMFEPLSSKTFNPIPCDSEQCGSLFSH--SCSPQK-LCAYSYSYADSSVTKGVLARE 178
Q SP+F+ S T+ CDS+ C +L H C K +C Y YSY D+S TKG +A E
Sbjct: 122 QNSPLFDKKKSSTYKTESCDSKTCQALSEHEEGCDESKDICKYRYSYGDNSFTKGDVATE 181
Query: 179 TITFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRF 238
TI+ S ++G + +FGCG++N G F E G+IGLGGGPLSLVSQ+G+ G ++F
Sbjct: 182 TISIDS-SSGSSVSFPGTVFGCGYNNGGTFEETGSGIIGLGGGPLSLVSQLGSSIG-KKF 239
Query: 239 SQCLVPFHADSRTSGTISFGDASDVSG----EGVVTTPLVSEEGQTPYLVTLEGISVGDT 294
S CL A + + I+ G S S +TTPL+ ++ +T Y +TLE ++VG T
Sbjct: 240 SYCLSHTAATTNGTSVINLGTNSIPSNPSKDSATLTTPLIQKDPETYYFLTLEAVTVGKT 299
Query: 295 FVSF-------NSSEKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDL 347
+ + N GN++IDSGT T L FYD ++ + +DP
Sbjct: 300 KLPYTGGGYGLNGKSSKRTGNIIIDSGTTLTLLDSGFYDDFGTAVEESVTGAKRVSDPQG 359
Query: 348 GTQLCYRS-ETNLEGPILTAHFEGADVQLMPIQTFIPPKDGVFCFAMAGTADGDYIFGNF 406
C++S + + P +T HF ADV+L PI F+ + C +M T + I+GN
Sbjct: 360 LLTHCFKSGDKEIGLPAITMHFTNADVKLSPINAFVKLNEDTVCLSMIPTTE-VAIYGNM 418
Query: 407 AQSNILIGFDLDRKTISFKPTDCT 430
Q + L+G+DL+ KT+SF+ DC+
Sbjct: 419 VQMDFLVGYDLETKTVSFQRMDCS 442
>UniRef100_Q766C3 Aspartic proteinase nepenthesin I [Nepenthes gracilis]
Length = 437
Score = 240 bits (613), Expect = 5e-62
Identities = 145/415 (34%), Positives = 215/415 (50%), Gaps = 26/415 (6%)
Query: 26 EAQDAGFSVQLTRQNSPHSPFYKPDNLHRHKLPSFHQVPKKAFAPNGP--FSTRVTSNNG 83
EA+ GF + L +S + K L R ++ + NGP T V + +G
Sbjct: 35 EAKVTGFQIMLEHVDSGKN-LTKFQLLERAIERGSRRLQRLEAMLNGPSGVETSVYAGDG 93
Query: 84 DYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSE 143
+YLM L++G+P ++DTGSDL+W QC PC C+ Q +P+F P S +F+ +PC S+
Sbjct: 94 EYLMNLSIGTPAQPFSAIMDTGSDLIWTQCQPCTQCFNQSTPIFNPQGSSSFSTLPCSSQ 153
Query: 144 QCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHS 203
C +L S +CS C Y+Y Y D S T+G + ET+TF S + + +I FGCG +
Sbjct: 154 LCQALSSPTCS-NNFCQYTYGYGDGSETQGSMGTETLTFGS------VSIPNITFGCGEN 206
Query: 204 NSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGD-ASD 262
N G N G++G+G GPLSL SQ+ +FS C+ P S T + G A+
Sbjct: 207 NQGFGQGNGAGLVGMGRGPLSLPSQLDV----TKFSYCMTPI--GSSTPSNLLLGSLANS 260
Query: 263 VSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFV-----SFNSSEKLSKGNMMIDSGTPA 317
V+ TT + S + T Y +TL G+SVG T + +F + G ++IDSGT
Sbjct: 261 VTAGSPNTTLIQSSQIPTFYYITLNGLSVGSTRLPIDPSAFALNSNNGTGGIIIDSGTTL 320
Query: 318 TYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRS---ETNLEGPILTAHFEGADVQ 374
TY Y + +E Q + LPV N G LC+++ +NL+ P HF+G D++
Sbjct: 321 TYFVNNAYQSVRQEFISQIN-LPVVNGSSSGFDLCFQTPSDPSNLQIPTFVMHFDGGDLE 379
Query: 375 LMPIQTFIPPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDC 429
L FI P +G+ C AM ++ G IFGN Q N+L+ +D +SF C
Sbjct: 380 LPSENYFISPSNGLICLAMGSSSQGMSIFGNIQQQNMLVVYDTGNSVVSFASAQC 434
>UniRef100_Q5VRD5 Putative CDR1 [Oryza sativa]
Length = 454
Score = 232 bits (592), Expect = 1e-59
Identities = 153/432 (35%), Positives = 225/432 (51%), Gaps = 39/432 (9%)
Query: 27 AQDAGFSVQLTRQNSPHSPFYKPD-NLHRHKLPSFHQVPKKAFAPNGPFST--------- 76
A GFSV+ ++SP SPF+ P H L + + +A A G S+
Sbjct: 29 ASGGGFSVEFIHRDSPRSPFHDPAFTAHGRALAAARRSVARAAAIAGSASSSASGGGAAD 88
Query: 77 ----RVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSP---MFEP 129
+V S + +YLM + LGSPP + + DTGSDLVW +C + + F+P
Sbjct: 89 DVVSKVVSRSFEYLMTVNLGSPPRSMLAIADTGSDLVWVKCKKGNNDTSSAAAPTTQFDP 148
Query: 130 LSSKTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNG- 188
S T+ + C ++ C +L +C CAY Y+Y D S T GVL+ ET TF +G
Sbjct: 149 SRSSTYGRVSCQTDACEALGRATCDDGSNCAYLYAYGDGSNTTGVLSTETFTFDDGGSGR 208
Query: 189 --DELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQM-GALYGSRRFSQCLVPF 245
++ VG + FGC + +G+F + G++GLGGG +SLV+Q+ GA RRFS CLVP
Sbjct: 209 SPRQVRVGGVKFGCSTATAGSFPAD--GLVGLGGGAVSLVTQLGGATSLGRRFSYCLVPH 266
Query: 246 HADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFNSSEKLS 305
++ S ++FG +DV+ G +TPLV+ + T Y V L+ + VG+ V+ +S ++
Sbjct: 267 SVNA--SSALNFGALADVTEPGAASTPLVAGDVDTYYTVVLDSVKVGNKTVASAASSRI- 323
Query: 306 KGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCY-----RSETNLE 360
++DSGT T+L +V+EL + +L PV + PD QLCY E
Sbjct: 324 ----IVDSGTTLTFLDPSLLGPIVDELSRRITLPPVQS-PDGLLQLCYNVAGREVEAGES 378
Query: 361 GPILTAHF-EGADVQLMPIQTFIPPKDGVFCFAMAGTADGD--YIFGNFAQSNILIGFDL 417
P LT F GA V L P F+ ++G C A+ T + I GN AQ NI +G+DL
Sbjct: 379 IPDLTLEFGGGAAVALKPENAFVAVQEGTLCLAIVATTEQQPVSILGNLAQQNIHVGYDL 438
Query: 418 DRKTISFKPTDC 429
D T++F DC
Sbjct: 439 DAGTVTFAGADC 450
>UniRef100_Q766C2 Aspartic proteinase nepenthesin II [Nepenthes gracilis]
Length = 438
Score = 230 bits (586), Expect = 7e-59
Identities = 132/363 (36%), Positives = 198/363 (54%), Gaps = 23/363 (6%)
Query: 76 TRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTF 135
T V + +G+YLM + +G+P ++DTGSDL+W QC PC C+ Q +P+F P S +F
Sbjct: 87 TPVYAGDGEYLMNVAIGTPDSSFSAIMDTGSDLIWTQCEPCTQCFSQPTPIFNPQDSSSF 146
Query: 136 NPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGD 195
+ +PC+S+ C L S +C+ + C Y+Y Y D S T+G +A ET TF + + V +
Sbjct: 147 STLPCESQYCQDLPSETCNNNE-CQYTYGYGDGSTTQGYMATETFTFETSS------VPN 199
Query: 196 IIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTI 255
I FGCG N G N G+IG+G GPLSL SQ+G +FS C+ + S + T+
Sbjct: 200 IAFGCGEDNQGFGQGNGAGLIGMGWGPLSLPSQLGV----GQFSYCMTSY--GSSSPSTL 253
Query: 256 SFGDASDVSGEGVVTTPLV-SEEGQTPYLVTLEGISVGDTFVSFNSS----EKLSKGNMM 310
+ G A+ EG +T L+ S T Y +TL+GI+VG + SS + G M+
Sbjct: 254 ALGSAASGVPEGSPSTTLIHSSLNPTYYYITLQGITVGGDNLGIPSSTFQLQDDGTGGMI 313
Query: 311 IDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETN---LEGPILTAH 367
IDSGT TYLPQ+ Y+ + + Q +L VD + G C++ ++ ++ P ++
Sbjct: 314 IDSGTTLTYLPQDAYNAVAQAFTDQINLPTVD-ESSSGLSTCFQQPSDGSTVQVPEISMQ 372
Query: 368 FEGADVQLMPIQTFIPPKDGVFCFAMAGTAD-GDYIFGNFAQSNILIGFDLDRKTISFKP 426
F+G + L I P +GV C AM ++ G IFGN Q + +DL +SF P
Sbjct: 373 FDGGVLNLGEQNILISPAEGVICLAMGSSSQLGISIFGNIQQQETQVLYDLQNLAVSFVP 432
Query: 427 TDC 429
T C
Sbjct: 433 TQC 435
>UniRef100_Q7XV20 OSJNBa0064H22.10 protein [Oryza sativa]
Length = 444
Score = 225 bits (573), Expect = 2e-57
Identities = 134/377 (35%), Positives = 194/377 (50%), Gaps = 30/377 (7%)
Query: 69 APNGPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFE 128
A G V + NG++LM +++G+P + +VDTGSDLVW QC PC C+KQ +P+F+
Sbjct: 79 AGGGDLQVPVHAGNGEFLMDVSIGTPALAYSAIVDTGSDLVWTQCKPCVDCFKQSTPVFD 138
Query: 129 PLSSKTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNG 188
P SS T+ +PC S C L + C+ C Y+Y+Y DSS T+GVLA ET T +
Sbjct: 139 PSSSSTYATVPCSSASCSDLPTSKCTSASKCGYTYTYGDSSSTQGVLATETFTLAKSK-- 196
Query: 189 DELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHAD 248
+ ++FGCG +N G G++GLG GPLSLVSQ+G +FS CL D
Sbjct: 197 ----LPGVVFGCGDTNEGDGFSQGAGLVGLGRGPLSLVSQLGL----DKFSYCLTSL--D 246
Query: 249 SRTSGTISFGDASDVS-----GEGVVTTPLVSEEGQTP-YLVTLEGISVGDTFVSFNSS- 301
+ + G + +S V TTPL+ Q Y V+L+ I+VG T +S SS
Sbjct: 247 DTNNSPLLLGSLAGISEASAAASSVQTTPLIKNPSQPSFYYVSLKAITVGSTRISLPSSA 306
Query: 302 ---EKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSET- 357
+ G +++DSGT TYL + Y L + Q + LP + +G LC+R+
Sbjct: 307 FAVQDDGTGGVIVDSGTSITYLEVQGYRALKKAFAAQMA-LPAADGSGVGLDLCFRAPAK 365
Query: 358 ---NLEGPILTAHFEGADVQLMPIQTF--IPPKDGVFCFAMAGTADGDYIFGNFAQSNIL 412
+E P L HF+G +P + + + G C + G+ G I GNF Q N
Sbjct: 366 GVDQVEVPRLVFHFDGGADLDLPAENYMVLDGGSGALCLTVMGSR-GLSIIGNFQQQNFQ 424
Query: 413 IGFDLDRKTISFKPTDC 429
+D+ T+SF P C
Sbjct: 425 FVYDVGHDTLSFAPVQC 441
>UniRef100_Q84M99 At2g03200 [Arabidopsis thaliana]
Length = 461
Score = 220 bits (561), Expect = 5e-56
Identities = 137/368 (37%), Positives = 201/368 (54%), Gaps = 33/368 (8%)
Query: 82 NGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCD 141
+G++LM+L++G+P V +VDTGSDL+W QC PC C+ Q +P+F+P S +++ + C
Sbjct: 104 SGEFLMELSIGNPAVKYSAIVDTGSDLIWTQCKPCTECFDQPTPIFDPEKSSSYSKVGCS 163
Query: 142 SEQCGSLFSHSCSPQK-LCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGC 200
S C +L +C+ K C Y Y+Y D S T+G+LA ET TF DE + I FGC
Sbjct: 164 SGLCNALPRSNCNEDKDACEYLYTYGDYSSTRGLLATETFTFE-----DENSISGIGFGC 218
Query: 201 GHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGD- 259
G N G G++GLG GPLSL+SQ+ +FS CL DS S ++ G
Sbjct: 219 GVENEGDGFSQGSGLVGLGRGPLSLISQL----KETKFSYCLTSIE-DSEASSSLFIGSL 273
Query: 260 --------ASDVSGEGVVTTPLVSEEGQTP-YLVTLEGISVGDTFVSFNSS----EKLSK 306
+ + GE T L+ Q Y + L+GI+VG +S S +
Sbjct: 274 ASGIVNKTGASLDGEVTKTMSLLRNPDQPSFYYLELQGITVGAKRLSVEKSTFELAEDGT 333
Query: 307 GNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYR---SETNLEGPI 363
G M+IDSGT TYL + + L EE + S LPVD+ G LC++ + N+ P
Sbjct: 334 GGMIIDSGTTITYLEETAFKVLKEEFTSRMS-LPVDDSGSTGLDLCFKLPDAAKNIAVPK 392
Query: 364 LTAHFEGADVQLMPIQTFI--PPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKT 421
+ HF+GAD++L P + ++ GV C AM G+++G IFGN Q N + DL+++T
Sbjct: 393 MIFHFKGADLEL-PGENYMVADSSTGVLCLAM-GSSNGMSIFGNVQQQNFNVLHDLEKET 450
Query: 422 ISFKPTDC 429
+SF PT+C
Sbjct: 451 VSFVPTEC 458
>UniRef100_Q6ZC30 Putative nucleoid DNA-binding protein [Oryza sativa]
Length = 448
Score = 220 bits (560), Expect = 7e-56
Identities = 135/369 (36%), Positives = 195/369 (52%), Gaps = 25/369 (6%)
Query: 78 VTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNP 137
V ++ G+YLM L +G+PP+ +VDTGSDL+W QC+PC C Q +P F P S T+
Sbjct: 85 VAASQGEYLMDLAIGTPPLRYTAMVDTGSDLIWTQCAPCVLCADQPTPYFRPARSATYRL 144
Query: 138 IPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDII 197
+PC S C +L +C + +C Y Y Y D + T GVLA ET TF + N +++V D+
Sbjct: 145 VPCRSPLCAALPYPACFQRSVCVYQYYYGDEASTAGVLASETFTFGA-ANSSKVMVSDVA 203
Query: 198 FGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTS----- 252
FGCG+ NSG N G++GLG GPLSLVSQ+ G RFS CL F + +
Sbjct: 204 FGCGNINSGQL-ANSSGMVGLGRGPLSLVSQL----GPSRFSYCLTSFLSPEPSRLNFGV 258
Query: 253 -GTISFGDASDVSGEGVVTTPLVSEEG-QTPYLVTLEGISVGDTFVSFN----SSEKLSK 306
T++ +AS SG V +TPLV + Y ++L+GIS+G + + +
Sbjct: 259 FATLNGTNASS-SGSPVQSTPLVVNAALPSLYFMSLKGISLGQKRLPIDPLVFAINDDGT 317
Query: 307 GNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCY----RSETNLEGP 362
G + IDSGT T+L Q+ YD + EL LP ND ++G + C+ + P
Sbjct: 318 GGVFIDSGTSLTWLQQDAYDAVRRELVSVLRPLPPTNDTEIGLETCFPWPPPPSVAVTVP 377
Query: 363 ILTAHFEGADVQLMPIQTF--IPPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRK 420
+ HF+G +P + + I G C AM + D I GN+ Q N+ I +D+
Sbjct: 378 DMELHFDGGANMTVPPENYMLIDGATGFLCLAMIRSGDAT-IIGNYQQQNMHILYDIANS 436
Query: 421 TISFKPTDC 429
+SF P C
Sbjct: 437 LLSFVPAPC 445
>UniRef100_Q9ZUU5 Putative chloroplast nucleoid DNA binding protein [Arabidopsis
thaliana]
Length = 392
Score = 216 bits (551), Expect = 8e-55
Identities = 144/403 (35%), Positives = 207/403 (50%), Gaps = 31/403 (7%)
Query: 33 SVQLTRQNSPHSPFYKPDNLHRHKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLG 92
S+ T +SPH + D + R S ++ K P++ + N YLMKL +G
Sbjct: 12 SLFTTTASSPHG--FTIDLIQRRSNSSSSRLSKNQLQGASPYADTLFDYN-IYLMKLQVG 68
Query: 93 SPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHS 152
+PP +I +DTGSDL+W QC PC CY Q +P+F+P +S TF C+
Sbjct: 69 TPPFEIEAEIDTGSDLIWTQCMPCTNCYSQYAPIFDPSNSSTFKEKRCNGNS-------- 120
Query: 153 CSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNEND 212
C Y YAD++ +KG LA ET+T S T+G+ V+ + GCGH NS F
Sbjct: 121 ------CHYKIIYADTTYSKGTLATETVTIHS-TSGEPFVMPETTIGCGH-NSSWFKPTF 172
Query: 213 MGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVV-TT 271
G++GL GP SL++QMG Y L+ + S+ + I+FG + V+G+GVV TT
Sbjct: 173 SGMVGLSWGPSSLITQMGGEYPG------LMSYCFASQGTSKINFGTNAIVAGDGVVSTT 226
Query: 272 PLVSEEGQTPYLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVE 330
++ Y + L+ +SVGDT V + ++ +GN++IDSGT TY P Y LV
Sbjct: 227 MFLTTAKPGLYYLNLDAVSVGDTHVETMGTTFHALEGNIIIDSGTTLTYFPVS-YCNLVR 285
Query: 331 ELKVQSSLLPVDNDPDLGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQTFIPP-KDGV 388
E DP LCY ++T P++T HF GAD+ L +I G
Sbjct: 286 EAVDHYVTAVRTADPTGNDMLCYYTDTIDIFPVITMHFSGGADLVLDKYNMYIETITRGT 345
Query: 389 FCFA-MAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDCT 430
FC A + D IFGN AQ+N L+G+D +SF PT+C+
Sbjct: 346 FCLAIICNNPPQDAIFGNRAQNNFLVGYDSSSLLVSFSPTNCS 388
>UniRef100_O81054 Putative chloroplast nucleoid DNA binding protein [Arabidopsis
thaliana]
Length = 353
Score = 214 bits (545), Expect = 4e-54
Identities = 135/363 (37%), Positives = 196/363 (53%), Gaps = 33/363 (9%)
Query: 87 MKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQCG 146
M+L++G+P V +VDTGSDL+W QC PC C+ Q +P+F+P S +++ + C S C
Sbjct: 1 MELSIGNPAVKYSAIVDTGSDLIWTQCKPCTECFDQPTPIFDPEKSSSYSKVGCSSGLCN 60
Query: 147 SLFSHSCSPQK-LCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNS 205
+L +C+ K C Y Y+Y D S T+G+LA ET TF DE + I FGCG N
Sbjct: 61 ALPRSNCNEDKDACEYLYTYGDYSSTRGLLATETFTFE-----DENSISGIGFGCGVENE 115
Query: 206 GAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGD------ 259
G G++GLG GPLSL+SQ+ +FS CL DS S ++ G
Sbjct: 116 GDGFSQGSGLVGLGRGPLSLISQL----KETKFSYCLTSIE-DSEASSSLFIGSLASGIV 170
Query: 260 ---ASDVSGEGVVTTPLVSEEGQTP-YLVTLEGISVGDTFVSFNSS----EKLSKGNMMI 311
+ + GE T L+ Q Y + L+GI+VG +S S + G M+I
Sbjct: 171 NKTGASLDGEVTKTMSLLRNPDQPSFYYLELQGITVGAKRLSVEKSTFELAEDGTGGMII 230
Query: 312 DSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYR---SETNLEGPILTAHF 368
DSGT TYL + + L EE + S LPVD+ G LC++ + N+ P + HF
Sbjct: 231 DSGTTITYLEETAFKVLKEEFTSRMS-LPVDDSGSTGLDLCFKLPDAAKNIAVPKMIFHF 289
Query: 369 EGADVQLMPIQTFI--PPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKTISFKP 426
+GAD++L P + ++ GV C AM G+++G IFGN Q N + DL+++T+SF P
Sbjct: 290 KGADLEL-PGENYMVADSSTGVLCLAM-GSSNGMSIFGNVQQQNFNVLHDLEKETVSFVP 347
Query: 427 TDC 429
T+C
Sbjct: 348 TEC 350
>UniRef100_Q9SJJ2 Putative chloroplast nucleoid DNA binding protein [Arabidopsis
thaliana]
Length = 396
Score = 211 bits (537), Expect = 3e-53
Identities = 139/399 (34%), Positives = 208/399 (51%), Gaps = 33/399 (8%)
Query: 37 TRQNSPHSPFYKPDNLHRHKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPV 96
T + PH + D +HR S + + P++ V N+ YLMKL +G+PP
Sbjct: 22 TTASPPHG--FTMDLIHRRSNASSRV--SNTQSGSSPYANTVFDNSV-YLMKLQVGTPPF 76
Query: 97 DIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCSPQ 156
+I ++DTGS++ W QC PC CY+Q +P+F+P S TF CD HS
Sbjct: 77 EIQAIIDTGSEITWTQCLPCVHCYEQNAPIFDPSKSSTFKEKRCD--------GHS---- 124
Query: 157 KLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVI 216
C Y Y D + T G LA ETIT S T+G+ V+ + I GCGH+NS F + G++
Sbjct: 125 --CPYEVDYFDHTYTMGTLATETITLHS-TSGEPFVMPETIIGCGHNNSW-FKPSFSGMV 180
Query: 217 GLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSE 276
GL GP SL++QMG Y L+ + + + I+FG + V+G+GVV+T +
Sbjct: 181 GLNWGPSSLITQMGGEYPG------LMSYCFSGQGTSKINFGANAIVAGDGVVSTTMFMT 234
Query: 277 EGQTP-YLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVEELKV 334
+ Y + L+ +SVG+T + + ++ +GN++IDSGT TY P Y LV +
Sbjct: 235 TAKPGFYYLNLDAVSVGNTRIETMGTTFHALEGNIVIDSGTTLTYFPVS-YCNLVRQAVE 293
Query: 335 QSSLLPVDNDPDLGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQTFIPPKD-GVFCFA 392
DP LCY S+T P++T HF G D+ L ++ + GVFC A
Sbjct: 294 HVVTAVRAADPTGNDMLCYNSDTIDIFPVITMHFSGGVDLVLDKYNMYMESNNGGVFCLA 353
Query: 393 -MAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDCT 430
+ + + IFGN AQ+N L+G+D +SF PT+C+
Sbjct: 354 IICNSPTQEAIFGNRAQNNFLVGYDSSSLLVSFSPTNCS 392
>UniRef100_Q84WH0 Putative chloroplast nucleoid DNA binding protein [Arabidopsis
thaliana]
Length = 395
Score = 206 bits (524), Expect = 1e-51
Identities = 135/352 (38%), Positives = 190/352 (53%), Gaps = 29/352 (8%)
Query: 84 DYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSE 143
+YLMKL +G+PP +I ++DTGS+ +W QC PC CY Q +P+F+P S TF I CD+
Sbjct: 64 EYLMKLQIGTPPFEIEAVLDTGSEHIWTQCLPCVHCYNQTAPIFDPSKSSTFKEIRCDTH 123
Query: 144 QCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHS 203
HS C Y Y S TKG L ET+T S T+G V+ + I GCG +
Sbjct: 124 ------DHS------CPYELVYGGKSYTKGTLVTETVTIHS-TSGQPFVMPETIIGCGRN 170
Query: 204 NSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDV 263
NSG F GV+GL GP SL++QMG Y L+ + + + I+FG + V
Sbjct: 171 NSG-FKPGFAGVVGLDRGPKSLITQMGGEYPG------LMSYCFAGKGTSKINFGANAIV 223
Query: 264 SGEGVV-TTPLVSEEGQTPYLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATYLP 321
+G+GVV TT V Y + L+ +SVG+T + + + KGN++IDSG+ TY P
Sbjct: 224 AGDGVVSTTVFVKTAKPGFYYLNLDAVSVGNTRIETVGTPFHALKGNIVIDSGSTLTYFP 283
Query: 322 QEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQT 380
E Y LV + Q D+ LCY S+T P++T HF GAD+ L
Sbjct: 284 -ESYCNLVRKAVEQVVTAVRFPRSDI---LCYYSKTIDIFPVITMHFSGGADLVLDKYNM 339
Query: 381 FIPPK-DGVFCFA-MAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDCT 430
++ GVFC A + + + IFGN AQ+N L+G+D +SFKPT+C+
Sbjct: 340 YVASNTGGVFCLAIICNSPIEEAIFGNRAQNNFLVGYDSSSLLVSFKPTNCS 391
>UniRef100_Q9LNJ3 F6F3.10 protein [Arabidopsis thaliana]
Length = 485
Score = 206 bits (523), Expect = 1e-51
Identities = 140/371 (37%), Positives = 189/371 (50%), Gaps = 24/371 (6%)
Query: 72 GPFSTRVTSN----NGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMF 127
G FS+ V S +G+Y +L +G+P +Y ++DTGSD+VW QC+PC CY Q P+F
Sbjct: 125 GGFSSSVVSGLSQGSGEYFTRLGVGTPARYVYMVLDTGSDIVWLQCAPCRRCYSQSDPIF 184
Query: 128 EPLSSKTFNPIPCDSEQCGSLFSHSCSP-QKLCAYSYSYADSSVTKGVLARETITFSSPT 186
+P SKT+ IPC S C L S C+ +K C Y SY D S T G + ET+TF
Sbjct: 185 DPRKSKTYATIPCSSPHCRRLDSAGCNTRRKTCLYQVSYGDGSFTVGDFSTETLTFRRNR 244
Query: 187 NGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFH 246
V + GCGH N G F G++GLG G LS Q G + +++FS CLV
Sbjct: 245 ------VKGVALGCGHDNEGLF-VGAAGLLGLGKGKLSFPGQTGHRF-NQKFSYCLVDRS 296
Query: 247 ADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFV-----SFNSS 301
A S+ S + FG+A+ VS T L + + T Y V L GISVG T V S
Sbjct: 297 ASSKPSSVV-FGNAA-VSRIARFTPLLSNPKLDTFYYVGLLGISVGGTRVPGVTASLFKL 354
Query: 302 EKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETN-LE 360
+++ G ++IDSGT T L + Y + + +V + L D L S N ++
Sbjct: 355 DQIGNGGVIIDSGTSVTRLIRPAYIAMRDAFRVGAKTLKRAPDFSLFDTCFDLSNMNEVK 414
Query: 361 GPILTAHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLD 418
P + HF GADV L P ++ P D G FCFA AGT G I GN Q + +DL
Sbjct: 415 VPTVVLHFRGADVSL-PATNYLIPVDTNGKFCFAFAGTMGGLSIIGNIQQQGFRVVYDLA 473
Query: 419 RKTISFKPTDC 429
+ F P C
Sbjct: 474 SSRVGFAPGGC 484
>UniRef100_Q9C6M0 Hypothetical protein F2J7.6 [Arabidopsis thaliana]
Length = 483
Score = 205 bits (522), Expect = 2e-51
Identities = 131/368 (35%), Positives = 188/368 (50%), Gaps = 31/368 (8%)
Query: 73 PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSS 132
P + T +G+Y ++ +G P ++Y ++DTGSD+ W QC+PC CY Q P+FEP SS
Sbjct: 136 PLISGTTQGSGEYFTRVGIGKPAREVYMVLDTGSDVNWLQCTPCADCYHQTEPIFEPSSS 195
Query: 133 KTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELV 192
++ P+ CD+ QC +L C C Y SY D S T G A ET+T S +
Sbjct: 196 SSYEPLSCDTPQCNALEVSECR-NATCLYEVSYGDGSYTVGDFATETLTIGS------TL 248
Query: 193 VGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTS 252
V ++ GCGHSN G F G++GLGGG L+L SQ+ + FS CLV DS ++
Sbjct: 249 VQNVAVGCGHSNEGLF-VGAAGLLGLGGGLLALPSQL----NTTSFSYCLV--DRDSDSA 301
Query: 253 GTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFNSS----EKLSKGN 308
T+ FG + +S + VV L + + T Y + L GISVG + S ++ G
Sbjct: 302 STVDFG--TSLSPDAVVAPLLRNHQLDTFYYLGLTGISVGGELLQIPQSSFEMDESGSGG 359
Query: 309 MMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQL---CYR--SETNLEGPI 363
++IDSGT T L E Y+ L + V+ +L D + G + CY ++T +E P
Sbjct: 360 IIIDSGTAVTRLQTEIYNSLRDSF-VKGTL---DLEKAAGVAMFDTCYNLSAKTTVEVPT 415
Query: 364 LTAHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKT 421
+ HF G + +P + ++ P D G FC A A TA I GN Q + FDL
Sbjct: 416 VAFHFPGGKMLALPAKNYMIPVDSVGTFCLAFAPTASSLAIIGNVQQQGTRVTFDLANSL 475
Query: 422 ISFKPTDC 429
I F C
Sbjct: 476 IGFSSNKC 483
>UniRef100_Q67UZ9 Putative aspartic proteinase nepenthesin I [Oryza sativa]
Length = 453
Score = 205 bits (522), Expect = 2e-51
Identities = 129/372 (34%), Positives = 188/372 (49%), Gaps = 33/372 (8%)
Query: 80 SNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIP 139
S + +Y++ L +G+PP I L+DTGSDL+W QC C C +Q P+F P S ++ P+
Sbjct: 93 SGDLEYVLDLAVGTPPQPITALLDTGSDLIWTQCDTCTACLRQPDPLFSPRMSSSYEPMR 152
Query: 140 CDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFG 199
C + CG + HSC C Y YSY D + T G A E TF+S + + V + FG
Sbjct: 153 CAGQLCGDILHHSCVRPDTCTYRYSYGDGTTTLGYYATERFTFASSSGETQSV--PLGFG 210
Query: 200 CGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGD 259
CG N G+ N N G++G G PLSLVSQ+ RRFS CL P+ A SR S T+ FG
Sbjct: 211 CGTMNVGSLN-NASGIVGFGRDPLSLVSQLSI----RRFSYCLTPY-ASSRKS-TLQFGS 263
Query: 260 ASDV-----SGEGVVTTPLV-SEEGQTPYLVTLEGISVGDTFVSFNSS----EKLSKGNM 309
+DV + V TTP++ S + T Y V G++VG + +S G +
Sbjct: 264 LADVGLYDDATGPVQTTPILQSAQNPTFYYVAFTGVTVGARRLRIPASAFALRPDGSGGV 323
Query: 310 MIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNLEG-------- 361
+IDSGT T P +V + Q LP N +C+ + G
Sbjct: 324 IIDSGTALTLFPVAVLAEVVRAFRSQLR-LPFANGSSPDDGVCFAAPAVAAGGGRMARQV 382
Query: 362 --PILTAHFEGADVQLMPIQTFI--PPKDGVFCFAMAGTADGDYIFGNFAQSNILIGFDL 417
P + HF+GAD+ L P + ++ + G C + + D GNF Q ++ + +DL
Sbjct: 383 AVPRMVFHFQGADLDL-PRENYVLEDHRRGHLCVLLGDSGDDGATIGNFVQQDMRVVYDL 441
Query: 418 DRKTISFKPTDC 429
+R+T+SF P +C
Sbjct: 442 ERETLSFAPVEC 453
>UniRef100_Q8H5I3 Putative nucleoid DNA-binding protein cnd41 [Oryza sativa]
Length = 441
Score = 204 bits (520), Expect = 3e-51
Identities = 134/375 (35%), Positives = 195/375 (51%), Gaps = 32/375 (8%)
Query: 72 GPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQC-SPCHGCYKQKSPMFEPL 130
G V ++ YL+ + +G+PP+ + ++DTGSDL+W QC +PC C+ Q +P++ P
Sbjct: 79 GGAEASVHASTATYLVDIAIGTPPLPLTAVLDTGSDLIWTQCDAPCRRCFPQPAPLYAPA 138
Query: 131 SSKTFNPIPCDSEQCGSLFS--HSCSPQKL-CAYSYSYADSSVTKGVLARETITFSSPTN 187
S T+ + C S C +L S CSP CAY +SY D + T GVLA ET T S T
Sbjct: 139 RSATYANVSCRSPMCQALQSPWSRCSPPDTGCAYYFSYGDGTSTDGVLATETFTLGSDT- 197
Query: 188 GDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHA 247
V + FGCG N G+ +N G++G+G GPLSLVSQ+G RFS C PF+A
Sbjct: 198 ----AVRGVAFGCGTENLGS-TDNSSGLVGMGRGPLSLVSQLGV----TRFSYCFTPFNA 248
Query: 248 DSRTSGTISFGDASDVSGEGVVTTPLV------SEEGQTPYLVTLEGISVGDTFVSFNSS 301
T+ + F +S TTP V + + Y ++LEGI+VGDT + + +
Sbjct: 249 ---TAASPLFLGSSARLSSAAKTTPFVPSPSGGARRRSSYYYLSLEGITVGDTLLPIDPA 305
Query: 302 ----EKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCY--RS 355
+ G ++IDSGT T L + + L L LP+ + LG LC+ S
Sbjct: 306 VFRLTPMGDGGVIIDSGTTFTALEERAFVALARAL-ASRVRLPLASGAHLGLSLCFAAAS 364
Query: 356 ETNLEGPILTAHFEGADVQLMPIQTFIPPKD-GVFCFAMAGTADGDYIFGNFAQSNILIG 414
+E P L HF+GAD++L + + GV C M +A G + G+ Q N I
Sbjct: 365 PEAVEVPRLVLHFDGADMELRRESYVVEDRSAGVACLGMV-SARGMSVLGSMQQQNTHIL 423
Query: 415 FDLDRKTISFKPTDC 429
+DL+R +SF+P C
Sbjct: 424 YDLERGILSFEPAKC 438
>UniRef100_Q9SL33 Putative chloroplast nucleoid DNA binding protein [Arabidopsis
thaliana]
Length = 756
Score = 204 bits (520), Expect = 3e-51
Identities = 124/357 (34%), Positives = 188/357 (51%), Gaps = 36/357 (10%)
Query: 85 YLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQ 144
YLMKL +G+PP +I +DTGSD++W QC PC CY Q +P+F+P S TF C+
Sbjct: 421 YLMKLQVGTPPFEIVAEIDTGSDIIWTQCMPCPNCYSQFAPIFDPSKSSTFREQRCNGNS 480
Query: 145 CGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSN 204
C Y YAD + +KG+LA ET+T S T+G+ V+ + GCG N
Sbjct: 481 --------------CHYEIIYADKTYSKGILATETVTIPS-TSGEPFVMAETKIGCGLDN 525
Query: 205 S----GAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDA 260
+ F + G++GL GPLSL+SQM Y L+ + + + I+FG
Sbjct: 526 TNLQYSGFASSSSGIVGLNMGPLSLISQMDLPYPG------LISYCFSGQGTSKINFGTN 579
Query: 261 SDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATY 319
+ V+G+G V + ++ Y + L+ +SV D + + + GN+ IDSGT TY
Sbjct: 580 AIVAGDGTVAADMFIKKDNPFYYLNLDAVSVEDNLIATLGTPFHAEDGNIFIDSGTTLTY 639
Query: 320 LPQEFYDRLVEELKVQSSLLPVDNDPDLGTQ--LCYRSETNLEGPILTAHFE-GADVQLM 376
P + + + E ++ + + V PD+G+ LCY S+T P++T HF GAD+ L
Sbjct: 640 FPMSYCNLVREAVEQVVTAVKV---PDMGSDNLLCYYSDTIDIFPVITMHFSGGADLVLD 696
Query: 377 PIQTFIPP-KDGVFCFAMAGTADGDY--IFGNFAQSNILIGFDLDRKTISFKPTDCT 430
++ G+FC A+ G D +FGN AQ+N L+G+D ISF PT+C+
Sbjct: 697 KYNMYLETITGGIFCLAI-GCNDPSMPAVFGNRAQNNFLVGYDPSSNVISFSPTNCS 752
Score = 193 bits (491), Expect = 7e-48
Identities = 129/388 (33%), Positives = 191/388 (48%), Gaps = 33/388 (8%)
Query: 37 TRQNSPHSPFYKPDNLHRHKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPV 96
T +SPH + D + R S ++ K P++ + N YLMKL +G+PP
Sbjct: 37 TTVSSPHG--FTIDLIQRRSNSSSFRLSKNQLQGASPYADTLFDYN-IYLMKLQVGTPPF 93
Query: 97 DIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCSPQ 156
+I +DTGSDL+W QC PC CY Q P+F+P S TFN C
Sbjct: 94 EIAAEIDTGSDLIWTQCMPCPDCYSQFDPIFDPSKSSTFNEQRCHG-------------- 139
Query: 157 KLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSN----SGAFNEND 212
K C Y Y D++ +KG+LA ET+T S T+G+ V+ + GCG N + F +
Sbjct: 140 KSCHYEIIYEDNTYSKGILATETVTIHS-TSGEPFVMAETTIGCGLHNTDLDNSGFASSS 198
Query: 213 MGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTP 272
G++GL GP SL+SQM Y L+ + + + I+FG + V+G+G V
Sbjct: 199 SGIVGLNMGPRSLISQMDLPYPG------LISYCFSGQGTSKINFGTNAIVAGDGTVAAD 252
Query: 273 LVSEEGQTPYLVTLEGISVGDTFV-SFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVEE 331
+ ++ Y + L+ +SV D + + + GN++IDSG+ TY P Y LV +
Sbjct: 253 MFIKKDNPFYYLNLDAVSVEDNRIETLGTPFHAEDGNIVIDSGSTVTYFPVS-YCNLVRK 311
Query: 332 LKVQSSLLPVDNDPDLGTQLCYRSETNLEGPILTAHFE-GADVQLMPIQTFIPPKD-GVF 389
Q DP LCY SET P++T HF GAD+ L ++ G+F
Sbjct: 312 AVEQVVTAVRVPDPSGNDMLCYFSETIDIFPVITMHFSGGADLVLDKYNMYMESNSGGLF 371
Query: 390 CFA-MAGTADGDYIFGNFAQSNILIGFD 416
C A + + + IFGN AQ+N L+G+D
Sbjct: 372 CLAIICNSPTQEAIFGNRAQNNFLVGYD 399
>UniRef100_Q9AWT3 Putative aspartic proteinase nepenthesin I [Oryza sativa]
Length = 504
Score = 201 bits (512), Expect = 3e-50
Identities = 128/368 (34%), Positives = 180/368 (48%), Gaps = 27/368 (7%)
Query: 72 GPFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLS 131
GP + V +G+Y ++ +GSP +Y ++DTGSD+ W QC PC CY+Q P+F+P
Sbjct: 154 GPVVSGVGLGSGEYFSRVGVGSPARQLYMVLDTGSDVTWVQCQPCADCYQQSDPVFDPSL 213
Query: 132 SKTFNPIPCDSEQCGSLFSHSC-SPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDE 190
S ++ + CD+ +C L + +C + C Y +Y D S T G A ET+T GD
Sbjct: 214 STSYASVACDNPRCHDLDAAACRNSTGACLYEVAYGDGSYTVGDFATETLTL-----GDS 268
Query: 191 LVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSR 250
V + GCGH N G F G++ LGGGPLS SQ+ A FS CLV DS
Sbjct: 269 APVSSVAIGCGHDNEGLF-VGAAGLLALGGGPLSFPSQISA----TTFSYCLV--DRDSP 321
Query: 251 TSGTISFGDASDVSGEGVVTTPLV-SEEGQTPYLVTLEGISVGDTFVSFNSS----EKLS 305
+S T+ FGDA+D VT PL+ S T Y V L G+SVG +S S +
Sbjct: 322 SSSTLQFGDAADAE----VTAPLIRSPRTSTFYYVGLSGLSVGGQILSIPPSAFAMDSTG 377
Query: 306 KGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCY--RSETNLEGPI 363
G +++DSGT T L Y L + + LP + L CY T++E P
Sbjct: 378 AGGVIVDSGTAVTRLQSSAYAALRDAFVRGTQSLPRTSGVSL-FDTCYDLSDRTSVEVPA 436
Query: 364 LTAHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKT 421
++ F G +P + ++ P D G +C A A T I GN Q + FD + T
Sbjct: 437 VSLRFAGGGELRLPAKNYLIPVDGAGTYCLAFAPTNAAVSIIGNVQQQGTRVSFDTAKST 496
Query: 422 ISFKPTDC 429
+ F C
Sbjct: 497 VGFTTNKC 504
>UniRef100_Q8L9B9 Chloroplast nucleoid DNA binding protein, putative [Arabidopsis
thaliana]
Length = 485
Score = 201 bits (512), Expect = 3e-50
Identities = 137/371 (36%), Positives = 187/371 (49%), Gaps = 24/371 (6%)
Query: 72 GPFSTRVTSN----NGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMF 127
G FS+ V S +G+Y +L +G+P +Y ++DTGSD+VW QC+PC CY Q P+F
Sbjct: 125 GGFSSSVVSGLSQGSGEYFTRLGVGTPARYVYMVLDTGSDIVWLQCAPCRRCYSQSDPIF 184
Query: 128 EPLSSKTFNPIPCDSEQCGSLFSHSCSP-QKLCAYSYSYADSSVTKGVLARETITFSSPT 186
+P SKT+ IPC S C L S C+ +K C Y SY D S T G + ET+TF
Sbjct: 185 DPRKSKTYATIPCSSPHCRRLDSAGCNTRRKTCLYQVSYGDGSFTVGDFSTETLTFRRNR 244
Query: 187 NGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFH 246
V + GCGH N G F G++GLG G LS Q G + +++FS CLV
Sbjct: 245 ------VKGVALGCGHDNEGLF-VGAAGLLGLGKGKLSFPGQTGHRF-NQKFSYCLVDRS 296
Query: 247 ADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFV-----SFNSS 301
A S+ S + FG+A+ VS T L + + T Y V L GISVG T V S
Sbjct: 297 ASSKPSSVV-FGNAA-VSRIARFTPLLSNPKLDTFYYVGLLGISVGGTRVPGVTASLFKL 354
Query: 302 EKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLP-VDNDPDLGTQLCYRSETNLE 360
+++ G ++IDSGT T L + Y + + +V + L N T + ++
Sbjct: 355 DQIGNGGVIIDSGTSVTRLIRPAYIAMRDAFRVGAKTLKRAPNFSLFDTCFDLSNMNEVK 414
Query: 361 GPILTAHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLD 418
P + HF ADV L P ++ P D G FCFA AGT G I GN Q + +DL
Sbjct: 415 VPTVVLHFRRADVSL-PATNYLIPVDTNGKFCFAFAGTMGGLSIIGNIQQQGFRVVYDLA 473
Query: 419 RKTISFKPTDC 429
+ F P C
Sbjct: 474 SSRVGFAPGGC 484
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 780,515,068
Number of Sequences: 2790947
Number of extensions: 35770613
Number of successful extensions: 80256
Number of sequences better than 10.0: 556
Number of HSP's better than 10.0 without gapping: 242
Number of HSP's successfully gapped in prelim test: 314
Number of HSP's that attempted gapping in prelim test: 79122
Number of HSP's gapped (non-prelim): 639
length of query: 432
length of database: 848,049,833
effective HSP length: 130
effective length of query: 302
effective length of database: 485,226,723
effective search space: 146538470346
effective search space used: 146538470346
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0588b.3


