

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0587.7
(319 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
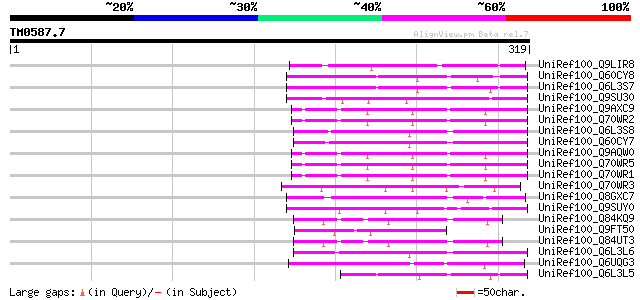
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana] 79 1e-13
UniRef100_Q60CY8 Putative F-box protein [Solanum demissum] 70 1e-10
UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum] 69 2e-10
UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis tha... 58 3e-07
UniRef100_Q9AXC9 S locus F-box (SLF)-S2-like protein [Antirrhinu... 57 9e-07
UniRef100_Q70WR2 S locus F-box (SLF)-S1 protein [Antirrhinum his... 57 9e-07
UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum] 55 3e-06
UniRef100_Q60CY7 Putative F-box protein [Solanum demissum] 55 3e-06
UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein)... 55 3e-06
UniRef100_Q70WR5 S locus F-box (SLF)-S4 protein [Antirrhinum his... 55 3e-06
UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum his... 55 3e-06
UniRef100_Q70WR3 S locus F-box (SLF)-S1E protein [Antirrhinum hi... 54 6e-06
UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabid... 51 4e-05
UniRef100_Q9SUY0 Hypothetical protein AT4g22390 [Arabidopsis tha... 51 4e-05
UniRef100_Q84KQ9 F-box [Prunus mume] 51 4e-05
UniRef100_Q9FT50 Hypothetical protein T25B15_90 [Arabidopsis tha... 51 5e-05
UniRef100_Q84UT3 F-box [Prunus mume] 51 5e-05
UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum] 50 8e-05
UniRef100_Q6UQG3 S3 self-incompatibility locus-linked putative F... 49 2e-04
UniRef100_Q6L3L5 Putative F-box domain containing protein [Solan... 49 2e-04
>UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana]
Length = 364
Score = 79.3 bits (194), Expect = 1e-13
Identities = 51/147 (34%), Positives = 79/147 (53%), Gaps = 8/147 (5%)
Query: 173 SFGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFP--VSLSHISQL 230
++GFGYD D YKVVA+ + H+ + K+Y WR+ SFP V ++ S+
Sbjct: 158 TYGFGYDESEDDYKVVALLQ---QRHQVKIETKIYSTRQKLWRSNTSFPSGVVVADKSRS 214
Query: 231 GVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCI 290
G+YI+GTLNW + + ++ I+S D+ +E+ + LP G L L+ CL +
Sbjct: 215 GIYINGTLNWAATSSSSSWTIISYDMSRDEFKE--LPGPVCCGRGCFTMTLGDLRGCLSM 272
Query: 291 LENNEGDNRFLVWQMKKFGVHNSWIQL 317
+ +G N VW MK+FG SW +L
Sbjct: 273 VCYCKGANAD-VWVMKEFGEVYSWSKL 298
>UniRef100_Q60CY8 Putative F-box protein [Solanum demissum]
Length = 261
Score = 69.7 bits (169), Expect = 1e-10
Identities = 50/156 (32%), Positives = 72/156 (46%), Gaps = 15/156 (9%)
Query: 171 YSSFGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQL 230
Y +GFGYD D YKVV + + VV +Y + WR I F + ++
Sbjct: 39 YLKYGFGYDETRDDYKVVVIQCIYEDSGSCDSVVNIYSLKADSWRTINKFQGNFL-VNSP 97
Query: 231 GVYISGTLNWEVKTDNYTF---IIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKD- 286
G +++G L W + D TF I+SLDL E + +L LP S YG+ L V++
Sbjct: 98 GKFVNGKLYWALSADVDTFNMCNIISLDLADETWRRLELPDS--YGKGSYPLALGVVESH 155
Query: 287 ----CLCILENNEGDNRFLVWQMKKFGVHNSWIQLY 318
CL +E D VW K GV SW +++
Sbjct: 156 LSVFCLNCIEGTNSD----VWIRKDCGVEVSWTKIF 187
>UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum]
Length = 372
Score = 68.6 bits (166), Expect = 2e-10
Identities = 51/153 (33%), Positives = 73/153 (47%), Gaps = 9/153 (5%)
Query: 171 YSSFGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQL 230
Y +GFGYD D YKVV + + VV +Y + WR I F + ++
Sbjct: 150 YLKYGFGYDETRDDYKVVVIQCIYEDSGSCDSVVNIYSLKADSWRTINKFQGNFL-VNSP 208
Query: 231 GVYISGTLNWEVKTDNYTF---IIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDC 287
G +++G + W + D TF I+SLDL E + +L LP S YG+ L V+
Sbjct: 209 GKFVNGKIYWALSADVDTFNMCNIISLDLADETWRRLELPDS--YGKGSYPLALGVVGSH 266
Query: 288 LCILENN--EGDNRFLVWQMKKFGVHNSWIQLY 318
L +L N EG N VW K GV SW +++
Sbjct: 267 LSVLCLNCIEGTNSD-VWIRKDCGVEVSWTKIF 298
>UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis thaliana]
Length = 408
Score = 58.2 bits (139), Expect = 3e-07
Identities = 46/168 (27%), Positives = 78/168 (46%), Gaps = 23/168 (13%)
Query: 171 YSSFGFGYDCMNDTYKVVAVAVSCFREHKEERV-------VKVYDMGGSFWRNIQS---- 219
Y +G GYD ++D YKVV + F+ E+ + VKV+ + + W+ I+S
Sbjct: 137 YVFYGLGYDSVSDDYKVVRMVQ--FKIDSEDELGCSFPYEVKVFSLKKNSWKRIESVASS 194
Query: 220 ------FPVSLSHISQLGVYISGTLNWEV--KTDNYTF-IIVSLDLGTEEYSQLSLPYSP 270
F L + GV +L+W + + F +IV DL EE+ + P +
Sbjct: 195 IQLLFYFYYHLLYRRGYGVLAGNSLHWVLPRRPGLIAFNLIVRFDLALEEFEIVRFPEAV 254
Query: 271 LYGEDFEFPFLAVLKDCLCILENNEGDNRFLVWQMKKFGVHNSWIQLY 318
G + VL CLC++ N + + VW MK++ V +SW +++
Sbjct: 255 ANGNVDIQMDIGVLDGCLCLMCNYD-QSYVDVWMMKEYNVRDSWTKVF 301
>UniRef100_Q9AXC9 S locus F-box (SLF)-S2-like protein [Antirrhinum hispanicum]
Length = 376
Score = 56.6 bits (135), Expect = 9e-07
Identities = 43/153 (28%), Positives = 73/153 (47%), Gaps = 12/153 (7%)
Query: 174 FGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQ--SFPVS-LSHISQL 230
+GFG C ND YKVV + +H + VY + W++I+ S P+ + H
Sbjct: 159 YGFGNTC-NDCYKVVLIESVGPEDHHIN--IYVYYSDTNSWKHIEDDSTPIKYICHFPCN 215
Query: 231 GVYISGTLNWEVKTDN--YTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCL 288
++ G +W + + Y I++ D+ TE + +++ P+ L F L L +CL
Sbjct: 216 ELFFKGAFHWNANSTDIFYADFILTFDIITEVFKEMAYPHC-LAQFSNSFLSLMSLNECL 274
Query: 289 CIL---ENNEGDNRFLVWQMKKFGVHNSWIQLY 318
++ E E F +W MK++GV SW + Y
Sbjct: 275 AMVRYKEWMEEPELFDIWVMKQYGVRESWTKQY 307
>UniRef100_Q70WR2 S locus F-box (SLF)-S1 protein [Antirrhinum hispanicum]
Length = 376
Score = 56.6 bits (135), Expect = 9e-07
Identities = 43/153 (28%), Positives = 73/153 (47%), Gaps = 12/153 (7%)
Query: 174 FGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQ--SFPVS-LSHISQL 230
+GFG C ND YKVV + +H + VY + W++I+ S P+ + H
Sbjct: 159 YGFGNTC-NDCYKVVLIESVGPEDHHIN--IYVYYSDTNSWKHIEDDSTPIKYICHFPCN 215
Query: 231 GVYISGTLNWEVKTDN--YTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCL 288
++ G +W + + Y I++ D+ TE + +++ P+ L F L L +CL
Sbjct: 216 ELFFKGAFHWNANSTDIFYADFILTFDIITEVFKEMAYPHC-LAQFSNSFLSLMSLNECL 274
Query: 289 CIL---ENNEGDNRFLVWQMKKFGVHNSWIQLY 318
++ E E F +W MK++GV SW + Y
Sbjct: 275 AMVRYKEWMEEPELFDIWVMKQYGVRESWTKQY 307
>UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum]
Length = 327
Score = 55.1 bits (131), Expect = 3e-06
Identities = 44/148 (29%), Positives = 65/148 (43%), Gaps = 8/148 (5%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQLGVYI 234
GFGYD D YKVV + R H VV +Y + W + ++ G ++
Sbjct: 156 GFGYDESRDDYKVVFIDYPIHR-HNHRTVVNIYSLRTKSWTTLHDQLQGFFLLNLHGRFV 214
Query: 235 SGTLNWEVKT--DNYTFI-IVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCIL 291
+G L W + +NY I S DL + +L LP G+D + + V+ L +L
Sbjct: 215 NGKLYWTSSSCINNYKVCNITSFDLADGTWERLELPSC---GKDNSYINVGVVGSDLSLL 271
Query: 292 EN-NEGDNRFLVWQMKKFGVHNSWIQLY 318
G VW MK GV+ SW +L+
Sbjct: 272 YTCQRGAATSDVWIMKHSGVNVSWTKLF 299
>UniRef100_Q60CY7 Putative F-box protein [Solanum demissum]
Length = 383
Score = 55.1 bits (131), Expect = 3e-06
Identities = 44/148 (29%), Positives = 66/148 (43%), Gaps = 9/148 (6%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQLGVYI 234
GFGYD D YKVV + H VV +Y + + W + + ++ G ++
Sbjct: 155 GFGYDESRDDYKVVFIDYPI--RHNHRTVVNIYSLRTNSWTTLHDQLQGIFLLNLHGRFV 212
Query: 235 SGTLNWEVKT--DNYTFI-IVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCIL 291
+G L W T +NY I S DL + L LP G+D + + V+ L +L
Sbjct: 213 NGKLYWTSSTCINNYKVCNITSFDLADGTWGSLELPSC---GKDNSYINVGVVGSDLSLL 269
Query: 292 ENNE-GDNRFLVWQMKKFGVHNSWIQLY 318
+ G VW MK GV+ SW +L+
Sbjct: 270 YTCQLGAATSDVWIMKHSGVNVSWTKLF 297
>UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein) [Antirrhinum
hispanicum]
Length = 376
Score = 55.1 bits (131), Expect = 3e-06
Identities = 42/153 (27%), Positives = 72/153 (46%), Gaps = 12/153 (7%)
Query: 174 FGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQ--SFPVS-LSHISQL 230
+GFG C ND YKVV + +H + VY + W++I+ S P+ + H
Sbjct: 159 YGFGNTC-NDCYKVVLIESVGPEDHHIN--IYVYYSDTNSWKHIEDDSTPIKYICHFPCN 215
Query: 231 GVYISGTLNWEVKTDN--YTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCL 288
++ G +W + + Y I++ D+ TE + +++ P+ L F L L +CL
Sbjct: 216 ELFFKGAFHWNANSTDIFYADFILTFDIITEVFKEMAYPHC-LAQFSNSFLSLMSLNECL 274
Query: 289 CIL---ENNEGDNRFLVWQMKKFGVHNSWIQLY 318
++ E E F +W M ++GV SW + Y
Sbjct: 275 AMVRYKEWMEDPELFDIWVMNQYGVRESWTKQY 307
>UniRef100_Q70WR5 S locus F-box (SLF)-S4 protein [Antirrhinum hispanicum]
Length = 376
Score = 54.7 bits (130), Expect = 3e-06
Identities = 42/153 (27%), Positives = 72/153 (46%), Gaps = 12/153 (7%)
Query: 174 FGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQ--SFPVS-LSHISQL 230
+GFG C ND YKVV + +H + VY + W++I+ S P+ + H
Sbjct: 159 YGFGNTC-NDCYKVVLIESVGPEDHHIN--IYVYYSDTNSWKHIEDDSTPIKYICHFPCN 215
Query: 231 GVYISGTLNWEVKTDN--YTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCL 288
++ G +W + + Y I++ D+ TE + +++ P+ L F L L +CL
Sbjct: 216 ELFFKGAFHWNANSTDIFYADFILTFDIITEVFKEMAYPHC-LAQFSNSFLSLMSLNECL 274
Query: 289 CIL---ENNEGDNRFLVWQMKKFGVHNSWIQLY 318
++ E E F +W M ++GV SW + Y
Sbjct: 275 AMVRYKEWMEEPELFDIWVMNQYGVRESWTKQY 307
>UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum hispanicum]
Length = 376
Score = 54.7 bits (130), Expect = 3e-06
Identities = 42/153 (27%), Positives = 72/153 (46%), Gaps = 12/153 (7%)
Query: 174 FGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQ--SFPVS-LSHISQL 230
+GFG C ND YKVV + +H + VY + W++I+ S P+ + H
Sbjct: 159 YGFGNTC-NDCYKVVLIESVGPEDHHIN--IYVYYSDTNSWKHIEDDSTPIKYICHFPCN 215
Query: 231 GVYISGTLNWEVKTDN--YTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCL 288
++ G +W + + Y I++ D+ TE + +++ P+ L F L L +CL
Sbjct: 216 ELFFKGAFHWNANSTDIFYADFILTFDIITEVFKEMAYPHC-LAQFSNSFLSLMSLNECL 274
Query: 289 CIL---ENNEGDNRFLVWQMKKFGVHNSWIQLY 318
++ E E F +W M ++GV SW + Y
Sbjct: 275 AMVRYKEWMEEPELFDIWVMNQYGVRESWTKQY 307
>UniRef100_Q70WR3 S locus F-box (SLF)-S1E protein [Antirrhinum hispanicum]
Length = 384
Score = 53.9 bits (128), Expect = 6e-06
Identities = 44/162 (27%), Positives = 77/162 (47%), Gaps = 18/162 (11%)
Query: 168 RFLYSSFGFGY-DCMNDTYKVVAV----AVSCFREHKEERVVKVYDMGGSFWRNIQSFPV 222
RF + G G+ +C ++ +K+V V +VS + K +V +Y+ WR I+ V
Sbjct: 152 RFYSNIIGQGFGNCDSNFFKIVLVRTIKSVSDYNRDKPYIMVHLYNSNTQSWRLIEGEAV 211
Query: 223 SLSHISQ---LGVYISGTLNWEVKTDN--YTFIIVSLDLGTEEYSQLSLP--YSPLYGED 275
+ +I V+ +G +W Y I++ D+ TE +S+ P + LYG
Sbjct: 212 LVQYIFSSPCTDVFFNGACHWNAGVFGIPYPGSILTFDISTEIFSEFEYPDGFRELYGGC 271
Query: 276 FEFPFLAVLKDCLCILENNEG--DNRFL-VWQMKKFGVHNSW 314
L L +CL ++ N+ D +F+ +W MK +G +SW
Sbjct: 272 L---CLTALSECLSVIRYNDSTKDPQFIEIWVMKVYGNSDSW 310
>UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabidopsis thaliana]
Length = 427
Score = 51.2 bits (121), Expect = 4e-05
Identities = 37/153 (24%), Positives = 72/153 (46%), Gaps = 14/153 (9%)
Query: 171 YSSFGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQL 230
+ ++GFG+D + D YK+V + + ++ VY + WR I + + S
Sbjct: 211 FQTYGFGFDGLTDDYKLVKLVAT----SEDILDASVYSLKADSWRRICNLNYEHNDGSYT 266
Query: 231 -GVYISGTLNWEVKTDNYT-FIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPF----LAVL 284
GV+ +G ++W + ++V+ D+ TEE+ ++ +P ED F + L
Sbjct: 267 SGVHFNGAIHWVFTESRHNQRVVVAFDIQTEEFREMPVPDE---AEDCSHRFSNFVVGSL 323
Query: 285 KDCLCILENNEGDNRFLVWQMKKFGVHNSWIQL 317
LC++ N+ D +W M ++G SW ++
Sbjct: 324 NGRLCVV-NSCYDVHDDIWVMSEYGEAKSWSRI 355
>UniRef100_Q9SUY0 Hypothetical protein AT4g22390 [Arabidopsis thaliana]
Length = 394
Score = 51.2 bits (121), Expect = 4e-05
Identities = 44/165 (26%), Positives = 76/165 (45%), Gaps = 21/165 (12%)
Query: 171 YSSFGFGYDCMNDTYKVVAVAVSCFREHKEE----RVVKVYDMGGSFWRNI-QSFPVSLS 225
Y +G GYD + D +KVV + +E K++ VKV+ + + W+ + F +
Sbjct: 137 YVFYGLGYDSVGDDFKVVRIVQCKLKEGKKKFPCPVEVKVFSLKKNSWKRVCLMFEFQIL 196
Query: 226 HISQ---------LGVYISGTLNWEVKTDNYTF---IIVSLDLGTEEYSQLSLPYSPLYG 273
IS GV ++ L+W + I+ DL +++ LS P LY
Sbjct: 197 WISYYYHLLPRRGYGVVVNNHLHWILPRRQGVIAFNAIIKYDLASDDIGVLSFP-QELYI 255
Query: 274 EDFEFPFLAVLKDCLCILENNEGDNRFLVWQMKKFGVHNSWIQLY 318
ED + VL C+C++ +E + VW +K++ + SW +LY
Sbjct: 256 ED--NMDIGVLDGCVCLMCYDE-YSHVDVWVLKEYEDYKSWTKLY 297
>UniRef100_Q84KQ9 F-box [Prunus mume]
Length = 428
Score = 51.2 bits (121), Expect = 4e-05
Identities = 42/140 (30%), Positives = 61/140 (43%), Gaps = 19/140 (13%)
Query: 175 GFGYDCMNDTYKVVAVA---VSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQLG 231
GFGYD + YKVV A F +H + V+VY + WR + PV + L
Sbjct: 205 GFGYDPNSKDYKVVRAAQFVSGVFTQHPSK--VEVYSLAADTWREV---PVDIQPHGSLN 259
Query: 232 ----VYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGED-FEFPFLAVLKD 286
+Y G W +I+S D+ E + ++LP S G D +E+ +AV KD
Sbjct: 260 PSYQMYFKGFFYWIAYWTEERNVILSFDMSEEVFHDIALPES---GPDAYEYTSIAVWKD 316
Query: 287 CLCILE---NNEGDNRFLVW 303
L +L NE +W
Sbjct: 317 SLALLTYPVENEAPKTLDLW 336
>UniRef100_Q9FT50 Hypothetical protein T25B15_90 [Arabidopsis thaliana]
Length = 390
Score = 50.8 bits (120), Expect = 5e-05
Identities = 32/98 (32%), Positives = 52/98 (52%), Gaps = 6/98 (6%)
Query: 176 FGYDCMNDTYKVVAVAVSCFREH---KEERVVKVYDMGGSFWRNIQSF--PVSLSHISQL 230
FG+D +D YKV+++ +E + E V V +G S WRN +S P H
Sbjct: 165 FGHDPFHDEYKVLSLFWEVTKEQTVVRSEHQVLVLGVGAS-WRNTKSHHTPHRPFHPYSR 223
Query: 231 GVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPY 268
G+ I G L + +TD +++S DL +EE++ + LP+
Sbjct: 224 GMTIDGVLYYSARTDANRCVLMSFDLSSEEFNLIELPF 261
>UniRef100_Q84UT3 F-box [Prunus mume]
Length = 428
Score = 50.8 bits (120), Expect = 5e-05
Identities = 42/140 (30%), Positives = 61/140 (43%), Gaps = 19/140 (13%)
Query: 175 GFGYDCMNDTYKVVAVA---VSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQLG 231
GFGYD + YKVV A F +H + V+VY + WR + PV + L
Sbjct: 205 GFGYDPNSKDYKVVRAAQFVSGVFTQHPSK--VEVYSLAADTWREV---PVDIQPHGSLN 259
Query: 232 ----VYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGED-FEFPFLAVLKD 286
+Y G W +I+S D+ E + ++LP S G D +E+ +AV KD
Sbjct: 260 PSYQMYFKGFFYWIAYWTEERNVILSFDMSEEVFHDIALPES---GPDAYEYTSIAVWKD 316
Query: 287 CLCILE---NNEGDNRFLVW 303
L +L NE +W
Sbjct: 317 SLVLLTYPVENEAPKTLDLW 336
>UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum]
Length = 384
Score = 50.1 bits (118), Expect = 8e-05
Identities = 42/148 (28%), Positives = 64/148 (42%), Gaps = 9/148 (6%)
Query: 175 GFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSHISQLGVYI 234
GFGYD +D YKVV + H+ VV +Y + + W + + ++ ++
Sbjct: 156 GFGYDESHDDYKVVFINYPSHHNHRS--VVNIYSLRTNSWTTLHDQLQGIFLLNLHCRFV 213
Query: 235 SGTLNWEVKT--DNYTFI-IVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCIL 291
L W T +NY I S DL + L LP G+D + + V+ L +L
Sbjct: 214 KEKLYWTSSTCINNYKVCNITSFDLADGTWESLELPSC---GKDNSYINVGVVGSDLSLL 270
Query: 292 EN-NEGDNRFLVWQMKKFGVHNSWIQLY 318
G VW MK GV+ SW +L+
Sbjct: 271 YTCQRGAANSDVWIMKHSGVNVSWTKLF 298
>UniRef100_Q6UQG3 S3 self-incompatibility locus-linked putative F-box protein S3-
A113 [Petunia integrifolia subsp. inflata]
Length = 376
Score = 48.9 bits (115), Expect = 2e-04
Identities = 35/152 (23%), Positives = 68/152 (44%), Gaps = 10/152 (6%)
Query: 172 SSFGFGYDCMNDTYKVVAVAVSCFREHKEERVVKVYDMGGSFWRNIQSFPVSLSH-ISQL 230
S GFG+D + YKVV ++ +E V +YD WR + V + + +
Sbjct: 153 SGIGFGFDSDANDYKVVRLSEVYKEPCDKEMKVDIYDFSVDSWRELLGQEVPIVYWLPCA 212
Query: 231 GVYISGTLNWEVKTDNYTFIIVSLDLGTEEYSQLSLPYSPLYGEDFEFPFLAVLKDCLCI 290
+ +W D+ +I+ D+ TE++ L +P + +D + L +L C+ +
Sbjct: 213 EILYKRNFHWFAFADD--VVILCFDMNTEKFHNLGMP-DACHFDDGKCYGLVILCKCMTL 269
Query: 291 ------LENNEGDNRFLVWQMKKFGVHNSWIQ 316
+ ++ + +W MK++G SWI+
Sbjct: 270 ICYPDPMPSSPTEKLTDIWIMKEYGEKESWIK 301
>UniRef100_Q6L3L5 Putative F-box domain containing protein [Solanum demissum]
Length = 307
Score = 48.9 bits (115), Expect = 2e-04
Identities = 38/120 (31%), Positives = 58/120 (47%), Gaps = 9/120 (7%)
Query: 204 VKVYDMGGSFWRNIQSFPVSLSHISQLGVYISGTLNWEVKTDNYTFI---IVSLDLGTEE 260
+ +Y + WR I F + ++ G +++G + W + D TF I+SLDL E
Sbjct: 118 INIYSLKADSWRTINKFQGNFL-VNSPGKFVNGKIYWALSADVDTFNMCNIISLDLADET 176
Query: 261 YSQLSLPYSPLYGEDFEFPFLAVLKDCLCILENN--EGDNRFLVWQMKKFGVHNSWIQLY 318
+ +L LP S YG+ L V+ L +L N EG N VW K GV SW +++
Sbjct: 177 WRRLELPDS--YGKGSYPLALGVVGSHLSVLCLNSIEGTNSD-VWIRKDCGVEVSWTKIF 233
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.330 0.141 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 531,407,735
Number of Sequences: 2790947
Number of extensions: 20728440
Number of successful extensions: 48819
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 90
Number of HSP's that attempted gapping in prelim test: 48773
Number of HSP's gapped (non-prelim): 98
length of query: 319
length of database: 848,049,833
effective HSP length: 127
effective length of query: 192
effective length of database: 493,599,564
effective search space: 94771116288
effective search space used: 94771116288
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0587.7


