

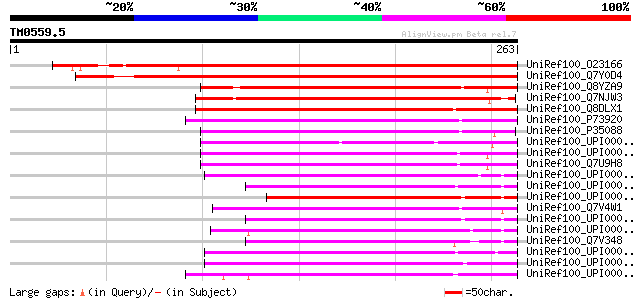
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0559.5
(263 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O23166 Thiol-disulfide interchange like protein [Arabi... 327 2e-88
UniRef100_Q7Y0D4 Putative thioredoxin-like protein [Oryza sativa] 310 3e-83
UniRef100_Q8YZA9 Thioredoxin-like protein [Anabaena sp.] 150 4e-35
UniRef100_Q7NJW3 Thiol:disulfide interchange protein [Gloeobacte... 140 3e-32
UniRef100_Q8DLX1 Thiol:disulfide interchange protein [Synechococ... 135 1e-30
UniRef100_P73920 Thiol:disulfide interchange protein txlA homolo... 131 2e-29
UniRef100_P35088 Thiol:disulfide interchange protein txlA [Synec... 121 2e-26
UniRef100_UPI000032FA1C UPI000032FA1C UniRef100 entry 116 7e-25
UniRef100_UPI0000275B16 UPI0000275B16 UniRef100 entry 114 3e-24
UniRef100_Q7U9H8 Thioredoxin-like protein TxlA [Synechococcus sp.] 114 3e-24
UniRef100_UPI00002E2A2A UPI00002E2A2A UniRef100 entry 110 4e-23
UniRef100_UPI000034587E UPI000034587E UniRef100 entry 109 6e-23
UniRef100_UPI000025FEF4 UPI000025FEF4 UniRef100 entry 108 1e-22
UniRef100_Q7V4W1 Thioredoxin-like protein TxlA precursor [Prochl... 108 1e-22
UniRef100_UPI000028425C UPI000028425C UniRef100 entry 107 3e-22
UniRef100_UPI000029DD0B UPI000029DD0B UniRef100 entry 106 7e-22
UniRef100_Q7V348 Thioredoxin-like protein TxlA precursor [Prochl... 105 1e-21
UniRef100_UPI000032FAD8 UPI000032FAD8 UniRef100 entry 105 2e-21
UniRef100_UPI0000288AE3 UPI0000288AE3 UniRef100 entry 105 2e-21
UniRef100_UPI00003418E7 UPI00003418E7 UniRef100 entry 104 2e-21
>UniRef100_O23166 Thiol-disulfide interchange like protein [Arabidopsis thaliana]
Length = 261
Score = 327 bits (838), Expect = 2e-88
Identities = 166/252 (65%), Positives = 202/252 (79%), Gaps = 17/252 (6%)
Query: 23 FTVNPRHLH--FNTR----KFHTLACQTNPNLDEKDASATSQEIKIVVEPSTVNGESETC 76
F ++PR+L F T+ +F + C+ NP ++S T QE K+V++ + S+
Sbjct: 16 FNLSPRNLQSFFVTQTGAPRFRAVRCKPNP-----ESSETKQE-KLVIDNGETSSASKEV 69
Query: 77 KPTSSTADAS-----GLPQLPTKDINRKVAIASTLAALGLFLATRLDFGVSLKDLTANAL 131
+ +SS AD+S G P+ P KDINR+VA + +AAL LF++TRLDFG+SLKDLTA+AL
Sbjct: 70 ESSSSVADSSSSSSSGFPESPNKDINRRVAAVTVIAALSLFVSTRLDFGISLKDLTASAL 129
Query: 132 PYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDE 191
PYE+ALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKD+VNFVMLNVDNT WEQELDE
Sbjct: 130 PYEEALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDKVNFVMLNVDNTKWEQELDE 189
Query: 192 FGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEAR 251
FGVEGIPHFAFLD+EGNEEGNVVGRLPRQ L+ENV+ALA G+ S+P+AR VG+YSS+E+R
Sbjct: 190 FGVEGIPHFAFLDREGNEEGNVVGRLPRQYLVENVNALAAGKQSIPYARAVGQYSSSESR 249
Query: 252 KVHQVVDPRSHG 263
KVHQV DP SHG
Sbjct: 250 KVHQVTDPLSHG 261
>UniRef100_Q7Y0D4 Putative thioredoxin-like protein [Oryza sativa]
Length = 262
Score = 310 bits (793), Expect = 3e-83
Identities = 155/230 (67%), Positives = 179/230 (77%), Gaps = 11/230 (4%)
Query: 35 RKFHTLACQTNPNLDEKDASATSQEIKIVVEPSTVNGESETCKPTSSTADASGLPQLPTK 94
R+ +L+C P+ E EP+ + +E SS + +G P P K
Sbjct: 43 RRLGSLSCIAPPDSAEPQTD----------EPAAKDDSTEDKAEASSASQDAGNPTFPNK 92
Query: 95 DINRKVAIASTLAALGLFLATRLDFG-VSLKDLTANALPYEQALSNGKPTVVEFYADWCE 153
D++R++A+AST+ A+GLF RLDFG VSLKDL ANA PYE+ALSNGKPTVVEFYADWCE
Sbjct: 93 DLSRRIALASTIGAVGLFAYQRLDFGGVSLKDLAANATPYEEALSNGKPTVVEFYADWCE 152
Query: 154 VCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNV 213
VCRELAPDVYK+EQQYKDRVNFVMLNVDNT WEQELDEFGVEGIPHFAFLDKEGNEEGNV
Sbjct: 153 VCRELAPDVYKVEQQYKDRVNFVMLNVDNTKWEQELDEFGVEGIPHFAFLDKEGNEEGNV 212
Query: 214 VGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
VGRLP+Q L+NV ALA GE +VPHARVVG++SSAE+RKVHQV DPRSHG
Sbjct: 213 VGRLPKQYFLDNVVALASGEPTVPHARVVGQFSSAESRKVHQVADPRSHG 262
>UniRef100_Q8YZA9 Thioredoxin-like protein [Anabaena sp.]
Length = 192
Score = 150 bits (378), Expect = 4e-35
Identities = 81/167 (48%), Positives = 108/167 (64%), Gaps = 7/167 (4%)
Query: 100 VAIASTLA-ALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
VAIA T+A LGL T VSL L A + P E A+SNGKP++VEFYA+WC VC+++
Sbjct: 25 VAIALTVALVLGLRTETA---AVSLDKLYAASTPLEVAISNGKPSMVEFYANWCTVCQKM 81
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
APD+ + EQQY D++NFVMLNVDNT W E+ ++ V+GIPHF FL+K G +G P
Sbjct: 82 APDIAEFEQQYADKMNFVMLNVDNTKWLPEMLKYRVDGIPHFVFLNKTGESLAQAIGDQP 141
Query: 219 RQLLLENVDALARGEASVPHARVVGKYS--SAEARKVHQVVDPRSHG 263
++ N++AL G A +PHA+ G+ S S + DPRSHG
Sbjct: 142 HTIMASNLEALVTGSA-LPHAQTSGQVSQVSTPVAPIGSQDDPRSHG 187
>UniRef100_Q7NJW3 Thiol:disulfide interchange protein [Gloeobacter violaceus]
Length = 179
Score = 140 bits (353), Expect = 3e-32
Identities = 74/168 (44%), Positives = 107/168 (63%), Gaps = 6/168 (3%)
Query: 97 NRKVAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCR 156
N VA+AS L A+ +F A R + +++ L A A+P ++AL+NGKPT+VEFYADWC C+
Sbjct: 15 NGIVALASVLIAVLVFFAAR-NQAPTMQQLAAEAVPLDEALANGKPTLVEFYADWCASCQ 73
Query: 157 ELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGR 216
+AP V +++++Y ++ NFVMLNVDN W EL ++ V GIPH+AFLD G+ +G
Sbjct: 74 TMAPTVARLKERYGEQANFVMLNVDNPRWLPELQQYKVSGIPHYAFLDAAARPLGSAIGL 133
Query: 217 LPRQLLLENVDALARGEASVPHARVVGKYSS--AEARKVHQVVDPRSH 262
P Q+L +N+ ALA G ++P A +S +KV Q PR H
Sbjct: 134 QPSQILEDNLTALAAGAETLPKAGAPAGQTSKFTPPKKVDQ---PRDH 178
>UniRef100_Q8DLX1 Thiol:disulfide interchange protein [Synechococcus elongatus]
Length = 182
Score = 135 bits (339), Expect = 1e-30
Identities = 65/169 (38%), Positives = 109/169 (64%), Gaps = 3/169 (1%)
Query: 97 NRKVAIASTLAALGLFLATRLDFG-VSLKDLTANALPYEQALSNGKPTVVEFYADWCEVC 155
N +A+ + L + L D ++L + +A P+E+A NGKPT++EFYA+WC C
Sbjct: 14 NFVIAVVAILISFALLAGLGSDNQPLTLAEQAKHATPWEEAQVNGKPTLLEFYANWCSTC 73
Query: 156 RELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVG 215
R +A D+ +++ QY+DRVNFVMLNVDN+ W E+++F V+GIPHF FL+++ + +G
Sbjct: 74 RSMAKDLGELKAQYRDRVNFVMLNVDNSKWLPEIEQFNVDGIPHFVFLNRQLEPQAVAIG 133
Query: 216 RLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVHQ-VVDPRSHG 263
P++++ +N++AL+ + +P+ ++ G+ S + Q DPRSHG
Sbjct: 134 LQPKEVMAKNLEALS-NDLPLPYGQLTGEMSQLPSPLARQRSDDPRSHG 181
>UniRef100_P73920 Thiol:disulfide interchange protein txlA homolog [Synechocystis
sp.]
Length = 180
Score = 131 bits (329), Expect = 2e-29
Identities = 68/174 (39%), Positives = 100/174 (57%), Gaps = 3/174 (1%)
Query: 92 PTKDINRKVAIASTLAALGLFLATRLDF-GVSLKDLTANALPYEQALSNGKPTVVEFYAD 150
P K N +A+ + + ++L + G+SL+ A+P AL NG+PT+VEFYAD
Sbjct: 3 PAKIRNALLAVVAIALSAAVYLGFQTQTQGISLEAQAQRAIPLATALDNGRPTLVEFYAD 62
Query: 151 WCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEE 210
WC C+ +APD+ ++++ Y VNF MLNVDN W E+ + V+GIPHF +LD G
Sbjct: 63 WCTSCQAMAPDLAELKKNYGGSVNFAMLNVDNNKWLPEVLRYRVDGIPHFVYLDDTGTAI 122
Query: 211 GNVVGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVH-QVVDPRSHG 263
+G P ++L +N+ AL E +P+A V G+ S E R + PRSHG
Sbjct: 123 AESIGEQPLRVLEQNITALVAHE-PIPYANVTGQTSVVENRTIEADPTSPRSHG 175
>UniRef100_P35088 Thiol:disulfide interchange protein txlA [Synechococcus sp.]
Length = 191
Score = 121 bits (303), Expect = 2e-26
Identities = 62/166 (37%), Positives = 93/166 (55%), Gaps = 4/166 (2%)
Query: 100 VAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELA 159
+A A L L + + + SL L A PYE A++N +P ++EFYADWC C+ +A
Sbjct: 17 IAAALVLTILVVLGSRQPSAAASLASLAEQATPYEVAIANDRPMLLEFYADWCTSCQAMA 76
Query: 160 PDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPR 219
+ ++Q Y DR++FVMLN+DN W E+ ++ V+GIP F +L+ +G +G +G LPR
Sbjct: 77 GRIAALKQDYSDRLDFVMLNIDNDKWLPEVLDYNVDGIPQFVYLNGQGQPQGISIGELPR 136
Query: 220 QLLLENVDALARGEASVPHARVVGKYSSAEA---RKVHQVVDPRSH 262
+L N+DAL + +P+ G S A DPRSH
Sbjct: 137 SVLAANLDALVEAQ-PLPYTNARGNLSEFSADLQPSRSSQTDPRSH 181
>UniRef100_UPI000032FA1C UPI000032FA1C UniRef100 entry
Length = 169
Score = 116 bits (290), Expect = 7e-25
Identities = 68/171 (39%), Positives = 93/171 (53%), Gaps = 10/171 (5%)
Query: 100 VAIASTLAALGLFLATRLDFGVSLKDLTAN-ALPYEQALSNGKPTVVEFYADWCEVCREL 158
V + +L +GL R G L D A +LP E AL NG+PT+VEFYADWCE CR +
Sbjct: 2 VLLLVSLLVVGLIWLQRGSDGRDLLDQMARRSLPLETALQNGRPTLVEFYADWCESCRTM 61
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVG-RL 217
AP + +++Q+ + ++ V+LNVDN WE ++ F V GIPH D +G G +G R
Sbjct: 62 APSIETMQEQHPE-IDLVLLNVDNPAWEAQIKRFSVNGIPHLQLFDADGESLGQSIGVRQ 120
Query: 218 PRQLLLENVDALARGEASVPHARVVGKYSSAE-----ARKVHQVVDPRSHG 263
P++ LE + G +P VG SS E R V + PRSHG
Sbjct: 121 PQE--LEALAKALEGSGPLPQLAGVGAISSLEPVEESRRDVSSSIGPRSHG 169
>UniRef100_UPI0000275B16 UPI0000275B16 UniRef100 entry
Length = 184
Score = 114 bits (285), Expect = 3e-24
Identities = 62/168 (36%), Positives = 97/168 (56%), Gaps = 5/168 (2%)
Query: 100 VAIASTLAALGL-FLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
+ +A+ + A+GL + L G L+ L +L + ALSNG+PT++EFYADWC+VCRE+
Sbjct: 18 LVVAAIVLAVGLTMMRGGLQSGSPLEQLARRSLTPDVALSNGRPTMIEFYADWCQVCREM 77
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
AP + K+EQ + +++ VM+NVDN W+ +D + V GIP + EG+ G +G
Sbjct: 78 APAMLKLEQSTQQQLDVVMVNVDNPRWQDLVDRYDVNGIPQLNLFNAEGDAVGRSIGLRR 137
Query: 219 RQLLLENVDALARGEASVPHARVVGKYS---SAEARKVHQVVDPRSHG 263
Q L DAL +G + +P + +G+ S ++ PRSHG
Sbjct: 138 EQELSLLADALIQG-SPLPSLQGLGRVSERTDVAPQQASLTAGPRSHG 184
>UniRef100_Q7U9H8 Thioredoxin-like protein TxlA [Synechococcus sp.]
Length = 184
Score = 114 bits (284), Expect = 3e-24
Identities = 61/168 (36%), Positives = 97/168 (57%), Gaps = 5/168 (2%)
Query: 100 VAIASTLAALGL-FLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
+ +A+ + A+GL + L G L+ L ++ + ALSNG+PT++EFYADWC+VCRE+
Sbjct: 18 LVVAAIVLAVGLTMMRGGLQSGSPLEQLARRSMTPDVALSNGRPTMIEFYADWCQVCREM 77
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
AP + K+EQ + +++ VM+NVDN W+ +D + V GIP + EG+ G +G
Sbjct: 78 APAMLKLEQSMQQQLDVVMVNVDNPRWQDLVDRYDVNGIPQLNLFNAEGDAVGRSIGLRR 137
Query: 219 RQLLLENVDALARGEASVPHARVVGKYS---SAEARKVHQVVDPRSHG 263
Q L DAL +G + +P + +G+ S ++ PRSHG
Sbjct: 138 EQELSLLADALIQG-SPLPSLQGLGRVSERTDVAPQQASLTAGPRSHG 184
>UniRef100_UPI00002E2A2A UPI00002E2A2A UniRef100 entry
Length = 181
Score = 110 bits (275), Expect = 4e-23
Identities = 60/162 (37%), Positives = 91/162 (56%), Gaps = 2/162 (1%)
Query: 102 IASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPD 161
IA + +L LF LK ++ E A N KPT +EFYA+WCEVC+E+AP
Sbjct: 22 IAVIIISLILFRNLFFQSTYLLKSFGELSVDPEIAFKNNKPTFLEFYAEWCEVCKEMAPK 81
Query: 162 VYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQL 221
V +++ Y+ +NFV LNVDN WE + +F V GIP D++GN + ++G+ +
Sbjct: 82 VSDLKESYEKDINFVFLNVDNQKWENYIRKFDVNGIPQVNLFDRKGNLKSTLIGKQDELI 141
Query: 222 LLENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
+ E++D L R S R +S+ + K ++ V+PRSHG
Sbjct: 142 IKESIDNLEREVESQEEIRNAA-FSAIKENKNNE-VNPRSHG 181
>UniRef100_UPI000034587E UPI000034587E UniRef100 entry
Length = 181
Score = 109 bits (273), Expect = 6e-23
Identities = 55/141 (39%), Positives = 83/141 (58%), Gaps = 2/141 (1%)
Query: 123 LKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDN 182
LK ++ E A N KPT +EFYA+WCEVC+E+AP V +++ Y+ +NFV LNVDN
Sbjct: 43 LKSFGELSVDPEIAFKNNKPTFLEFYAEWCEVCKEMAPKVSDLKETYEKDINFVFLNVDN 102
Query: 183 TNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHARVV 242
WE+ + +F V GIP D++GN + +G+ + E++D L R E P +
Sbjct: 103 QKWEKYIRKFDVNGIPQVNLFDRKGNLKSTFIGKQEELTIKESIDQLDR-EVESPEEILN 161
Query: 243 GKYSSAEARKVHQVVDPRSHG 263
+S+ + K ++ V PRSHG
Sbjct: 162 AAFSAIKENKNNE-VSPRSHG 181
>UniRef100_UPI000025FEF4 UPI000025FEF4 UniRef100 entry
Length = 131
Score = 108 bits (271), Expect = 1e-22
Identities = 52/131 (39%), Positives = 80/131 (60%), Gaps = 4/131 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E A +N KPT +EFYA+WCEVC+E+AP V + +Y+ +NFV LNVDN W + +F
Sbjct: 4 EIAFTNNKPTFLEFYAEWCEVCKEMAPQVSAFKNEYEKDINFVFLNVDNQKWANYIQKFA 63
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHARVV-GKYSSAEARK 252
V GIP D +GN +G+ + + E+++ L +GE P+ ++ K+S+ + K
Sbjct: 64 VNGIPQINLFDNKGNLISTFIGKQNEEEVRESINNLGKGEE--PYEEIINAKFSTIQENK 121
Query: 253 VHQVVDPRSHG 263
++ V PRSHG
Sbjct: 122 NNE-VSPRSHG 131
>UniRef100_Q7V4W1 Thioredoxin-like protein TxlA precursor [Prochlorococcus marinus]
Length = 188
Score = 108 bits (271), Expect = 1e-22
Identities = 58/165 (35%), Positives = 93/165 (56%), Gaps = 8/165 (4%)
Query: 106 LAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKI 165
LA + L L+ L+ L +L + AL+NG+PTV+EFYADWC+ CRE+AP + K
Sbjct: 25 LAVVLFLLRGGLNAEAPLEQLARRSLDPDVALTNGRPTVIEFYADWCQACREMAPAMLKT 84
Query: 166 EQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLEN 225
E+ +D+++ V++NVDN W+ +D + V GIP F D +G +G +G ++ L +
Sbjct: 85 ERDREDQLDVVLVNVDNPRWQDLVDRYDVNGIPQLNFFDNKGKLQGLSLGARTQEQLQQL 144
Query: 226 VDALARGEASVPHARVVGKYSSAEARKVH-------QVVDPRSHG 263
DAL + + +P+ VG S+ A + + + P SHG
Sbjct: 145 TDALIQNQ-PLPNFAGVGAISNLNASQERTEKQNDLEAIRPMSHG 188
>UniRef100_UPI000028425C UPI000028425C UniRef100 entry
Length = 196
Score = 107 bits (267), Expect = 3e-22
Identities = 53/142 (37%), Positives = 84/142 (58%), Gaps = 4/142 (2%)
Query: 123 LKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDN 182
LK ++ E A +N KPT +EFYA+WCEVC+E+AP V + +Y+ +NFV LNVDN
Sbjct: 58 LKSFGELSVDPEIAFTNNKPTFLEFYAEWCEVCKEMAPQVSAFKDEYEKDINFVFLNVDN 117
Query: 183 TNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHARVV 242
W + +F V GIP DK+GN +G+ + ++++ L + E P+ ++
Sbjct: 118 QKWSNYIQKFAVNGIPQVNLFDKKGNLISTFIGKQDEMKIRKSINNLEKEEK--PYEEII 175
Query: 243 -GKYSSAEARKVHQVVDPRSHG 263
+YS+ + K ++ V+PRSHG
Sbjct: 176 NAEYSTIQENKNYE-VNPRSHG 196
>UniRef100_UPI000029DD0B UPI000029DD0B UniRef100 entry
Length = 181
Score = 106 bits (264), Expect = 7e-22
Identities = 57/161 (35%), Positives = 89/161 (54%), Gaps = 4/161 (2%)
Query: 105 TLAALGLFLATRLDFGVS--LKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDV 162
++ + LFL F + LK ++ E A +N KPT +EFYA+WCEVC+E+AP V
Sbjct: 23 SIVIISLFLFRNFFFQSTYLLKSFGELSVDPEVAFTNNKPTFLEFYAEWCEVCKEMAPQV 82
Query: 163 YKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLL 222
+ +Y+ +NFV LNVDN W +++F V GIP DK+GN VG+ +
Sbjct: 83 SAFKDEYEKDINFVFLNVDNQKWGNYIEKFAVNGIPQVNLFDKKGNLISTFVGKQDEIKI 142
Query: 223 LENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
+++ + + E S + ++S+ + K H+ V PRSHG
Sbjct: 143 RNSINNIEKEEKSNEEI-INAEFSTIQENKNHE-VSPRSHG 181
>UniRef100_Q7V348 Thioredoxin-like protein TxlA precursor [Prochlorococcus marinus
subsp. pastoris]
Length = 181
Score = 105 bits (262), Expect = 1e-21
Identities = 56/144 (38%), Positives = 84/144 (57%), Gaps = 8/144 (5%)
Query: 123 LKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDN 182
LK +++ E A +N KPT +EFYADWCEVC+E+AP V I+ +Y+ +NFV LNVDN
Sbjct: 43 LKKFGQSSIDPEIAFTNNKPTFLEFYADWCEVCKEMAPKVEDIKNKYEKDINFVFLNVDN 102
Query: 183 TNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDAL---ARGEASVPHA 239
WE+ + F V GIP D + N E +G+ + E++D L RG++ + ++
Sbjct: 103 PKWEKYIRNFNVNGIPQINIFDNDSNLELTFIGKQEEDSIKESLDNLYDKFRGDSQILNS 162
Query: 240 RVVGKYSSAEARKVHQVVDPRSHG 263
++S + K +Q PRSHG
Sbjct: 163 ----EFSIIKDNKNYQ-SSPRSHG 181
>UniRef100_UPI000032FAD8 UPI000032FAD8 UniRef100 entry
Length = 181
Score = 105 bits (261), Expect = 2e-21
Identities = 59/162 (36%), Positives = 87/162 (53%), Gaps = 2/162 (1%)
Query: 102 IASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPD 161
IA + +L LF LK ++ E A N KPT +EFYA+WCEVC+E+AP
Sbjct: 22 IAIVIISLILFRNLLFQSTYLLKSFGELSVDPEIAFKNKKPTFLEFYAEWCEVCKEMAPK 81
Query: 162 VYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQL 221
V ++ +Y+ +NFV LNVDN WE + ++ V GIP DK+GN VG+
Sbjct: 82 VSALKDEYEKDINFVFLNVDNQKWENYIRKYNVNGIPQVNLFDKKGNLRATFVGKQDELS 141
Query: 222 LLENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
+ E++D L + E + K+S + K + ++PRSHG
Sbjct: 142 IRESIDNLHK-EFESKEEILNTKFSEIKENK-NSELNPRSHG 181
>UniRef100_UPI0000288AE3 UPI0000288AE3 UniRef100 entry
Length = 168
Score = 105 bits (261), Expect = 2e-21
Identities = 58/162 (35%), Positives = 83/162 (50%), Gaps = 2/162 (1%)
Query: 102 IASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPD 161
IA + +L LF LK ++ E AL N KPT +EFYA+WCEVC+E+AP
Sbjct: 9 IAVVITSLILFRNIIFQSTYLLKSFGELSVDPEIALKNNKPTFLEFYAEWCEVCKEMAPK 68
Query: 162 VYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQL 221
V ++ +Y+ NFV LNVDN W+ + +F V GIP DK GN +G+
Sbjct: 69 VSLLKDEYEKDFNFVFLNVDNEKWDNYIRKFDVNGIPQVNIFDKTGNLISTFIGKQEEFT 128
Query: 222 LLENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
+ +++D L + S H ++ S V+PRSHG
Sbjct: 129 IRDSMDQLEKETKS--HEEIINNEFSILKENNKNKVNPRSHG 168
>UniRef100_UPI00003418E7 UPI00003418E7 UniRef100 entry
Length = 181
Score = 104 bits (260), Expect = 2e-21
Identities = 61/176 (34%), Positives = 93/176 (52%), Gaps = 6/176 (3%)
Query: 92 PTKDINRKVAIASTLAAL--GLFLATRLDFGVS--LKDLTANALPYEQALSNGKPTVVEF 147
P + N KV + +A + L L L F + LK ++ E A N KPT +EF
Sbjct: 8 PLLNTNLKVILVLLIAVIIIVLVLYRNLFFQSTYLLKSFGELSVEPEIAFKNNKPTFLEF 67
Query: 148 YADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEG 207
YA+WCEVC+E+AP VY ++++Y+ VNFV LNVDN W+ + +F V GIP D++G
Sbjct: 68 YAEWCEVCKEMAPKVYALKEEYQKDVNFVFLNVDNQKWDNYIRKFDVNGIPQVNLFDQKG 127
Query: 208 NEEGNVVGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
N VG+ ++ E++ L A + ++ S + + + PRSHG
Sbjct: 128 NLISTFVGKQKETIIEESLSNLK--SAINSNEEIINAEFSELIKNKNYDISPRSHG 181
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 438,003,731
Number of Sequences: 2790947
Number of extensions: 18241974
Number of successful extensions: 49412
Number of sequences better than 10.0: 2156
Number of HSP's better than 10.0 without gapping: 1347
Number of HSP's successfully gapped in prelim test: 809
Number of HSP's that attempted gapping in prelim test: 47327
Number of HSP's gapped (non-prelim): 2582
length of query: 263
length of database: 848,049,833
effective HSP length: 125
effective length of query: 138
effective length of database: 499,181,458
effective search space: 68887041204
effective search space used: 68887041204
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0559.5


