

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.11
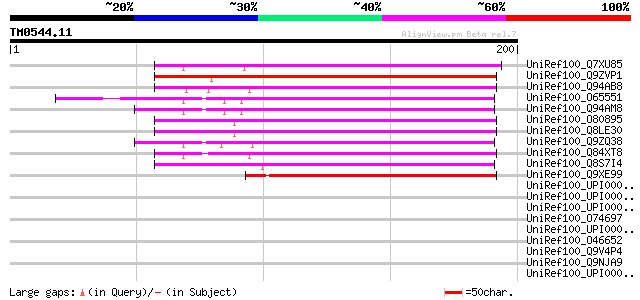
(200 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XU85 OSJNBa0029H02.22 protein [Oryza sativa] 120 3e-26
UniRef100_Q9ZVP1 T7A14.6 protein [Arabidopsis thaliana] 115 7e-25
UniRef100_Q94AB8 At1g05070/T7A14_6 [Arabidopsis thaliana] 112 7e-24
UniRef100_O65551 Hypothetical protein F6I18.100 [Arabidopsis tha... 111 1e-23
UniRef100_Q94AM8 Hypothetical protein At4g30996 [Arabidopsis tha... 110 2e-23
UniRef100_O80895 Expressed protein [Arabidopsis thaliana] 108 6e-23
UniRef100_Q8LE30 Hypothetical protein [Arabidopsis thaliana] 108 6e-23
UniRef100_Q9ZQ38 Expressed protein [Arabidopsis thaliana] 105 9e-22
UniRef100_Q84XT8 Hypothetical protein [Phytophthora sojae] 103 2e-21
UniRef100_Q8S7I4 Hypothetical protein OSJNBa0010I09.2 [Oryza sat... 96 4e-19
UniRef100_Q9XE99 Hypothetical protein T19B17.8 [Arabidopsis thal... 91 1e-17
UniRef100_UPI000036832F UPI000036832F UniRef100 entry 38 0.13
UniRef100_UPI000036832E UPI000036832E UniRef100 entry 38 0.13
UniRef100_UPI000036832D UPI000036832D UniRef100 entry 38 0.13
UniRef100_O74697 Hypothetical protein [Phaeosphaeria nodorum] 37 0.39
UniRef100_UPI00004364C6 UPI00004364C6 UniRef100 entry 35 0.87
UniRef100_O46652 Fertilin alpha-II [Papio anubis] 35 1.1
UniRef100_Q9V4P4 CG30494-PA [Drosophila melanogaster] 35 1.1
UniRef100_Q9NJA9 Paramyosin [Anisakis simplex] 35 1.1
UniRef100_UPI00001D09DB UPI00001D09DB UniRef100 entry 35 1.5
>UniRef100_Q7XU85 OSJNBa0029H02.22 protein [Oryza sativa]
Length = 196
Score = 120 bits (300), Expect = 3e-26
Identities = 64/160 (40%), Positives = 88/160 (55%), Gaps = 23/160 (14%)
Query: 58 GPLSRWRLKE--------------SASTESPCHHPCDCYCSSAESPPD---------TFD 94
GP WR K+ +A+ SP PC C C S T
Sbjct: 31 GPAFYWRYKKGFASSSSSSSVSASAAAVVSPSCPPCSCDCPPPLSLQSIAPGLVNFSTSG 90
Query: 95 CGKHDPVMNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCN 154
CGK+DP +++EM K + +L+EEL LQ+IV E + ++ A+ ++ YQ E EKCN
Sbjct: 91 CGKNDPELSKEMEKQFVDLLNEELKLQQIVAEEHSHHMNATLVEAKRQATQYQREAEKCN 150
Query: 155 VAMETCEEARERAEAELIEEHRVTALWENRAREYGWKDKR 194
A ETCEEARER+EA + +E ++TALWE RAR+ GW+D R
Sbjct: 151 AATETCEEARERSEAAISKEKKLTALWEQRARQLGWQDSR 190
>UniRef100_Q9ZVP1 T7A14.6 protein [Arabidopsis thaliana]
Length = 176
Score = 115 bits (288), Expect = 7e-25
Identities = 56/136 (41%), Positives = 82/136 (60%), Gaps = 1/136 (0%)
Query: 58 GPLSRWRLKESASTESPCHHP-CDCYCSSAESPPDTFDCGKHDPVMNEEMNKDLLTMLSE 116
GP W L E+ + S P C C CS+ + DC KHDP +NE+ K+ +L+E
Sbjct: 25 GPPLYWHLTEALAAVSASSCPSCPCECSTYSAVTIPKDCAKHDPEVNEDTEKNYAELLTE 84
Query: 117 ELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNVAMETCEEARERAEAELIEEHR 176
EL L+ + E ++ D ++ A+ +S YQ E +KCN METCEEARE+AE L E+ +
Sbjct: 85 ELKLREAESLEKHKRADMGLLEAKKVTSSYQKEADKCNSGMETCEEAREKAELALAEQKK 144
Query: 177 VTALWENRAREYGWKD 192
+T+ WE RAR+ GW++
Sbjct: 145 LTSRWEERARQKGWRE 160
>UniRef100_Q94AB8 At1g05070/T7A14_6 [Arabidopsis thaliana]
Length = 184
Score = 112 bits (279), Expect = 7e-24
Identities = 59/144 (40%), Positives = 85/144 (58%), Gaps = 9/144 (6%)
Query: 58 GPLSRWRLKES--ASTESPCHH-PCDCYCSSAESPPDTF------DCGKHDPVMNEEMNK 108
GP W L E+ A + S C PC+C SA + P DC KHDP +NE+ K
Sbjct: 25 GPPLYWHLTEALAAVSASSCPSCPCECSTYSAVTIPKELSNASFADCAKHDPEVNEDTEK 84
Query: 109 DLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNVAMETCEEARERAE 168
+ +L+EEL L+ + E ++ D ++ A+ +S YQ E +KCN METCEEARE+AE
Sbjct: 85 NYAELLTEELKLREAESLEKHKRADMGLLEAKKVTSSYQKEADKCNSGMETCEEAREKAE 144
Query: 169 AELIEEHRVTALWENRAREYGWKD 192
L E+ ++T+ WE RAR+ GW++
Sbjct: 145 LALAEQKKLTSRWEERARQKGWRE 168
>UniRef100_O65551 Hypothetical protein F6I18.100 [Arabidopsis thaliana]
Length = 2895
Score = 111 bits (277), Expect = 1e-23
Identities = 69/186 (37%), Positives = 92/186 (49%), Gaps = 20/186 (10%)
Query: 19 ISEHTASSQSPRSSNGSVSVDVHITCESPSDIVQVIVEFGPLSRWRLKE----SASTESP 74
IS +S PR S + C +V +V GP W+ + S S
Sbjct: 2715 ISAFCSSEAMPRRSGDCMR------CLVIFAVVSALVVCGPALYWKFNKGFVGSTRANSL 2768
Query: 75 CHHPCDCYC----SSAESPP-----DTFDCGKHDPVMNEEMNKDLLTMLSEELNLQRIVT 125
C PC C C S + P DCG DP + +EM K + +L+EEL LQ V
Sbjct: 2769 CP-PCVCDCPPPLSLLQIAPGLANLSITDCGSDDPELKQEMEKQFVDLLTEELKLQEAVA 2827
Query: 126 NETLEQTDRLVMNARNASSHYQMETEKCNVAMETCEEARERAEAELIEEHRVTALWENRA 185
+E + + A+ +S YQ E EKCN A E CE ARERAEA LI+E ++T+LWE RA
Sbjct: 2828 DEHSRHMNVTLAEAKRVASQYQKEAEKCNAATEICESARERAEALLIKERKITSLWEKRA 2887
Query: 186 REYGWK 191
R+ GW+
Sbjct: 2888 RQSGWE 2893
>UniRef100_Q94AM8 Hypothetical protein At4g30996 [Arabidopsis thaliana]
Length = 172
Score = 110 bits (276), Expect = 2e-23
Identities = 62/155 (40%), Positives = 83/155 (53%), Gaps = 14/155 (9%)
Query: 50 IVQVIVEFGPLSRWRLKE----SASTESPCHHPCDCYC----SSAESPP-----DTFDCG 96
+V +V GP W+ + S S C PC C C S + P DCG
Sbjct: 17 VVSALVVCGPALYWKFNKGFVGSTRANSLCP-PCVCDCPPPLSLLQIAPGLANLSITDCG 75
Query: 97 KHDPVMNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNVA 156
DP + +EM K + +L+EEL LQ V +E + + A+ +S YQ E EKCN A
Sbjct: 76 SDDPELKQEMEKQFVDLLTEELKLQEAVADEHSRHMNVTLAEAKRVASQYQKEAEKCNAA 135
Query: 157 METCEEARERAEAELIEEHRVTALWENRAREYGWK 191
E CE ARERAEA LI+E ++T+LWE RAR+ GW+
Sbjct: 136 TEICESARERAEALLIKERKITSLWEKRARQSGWE 170
>UniRef100_O80895 Expressed protein [Arabidopsis thaliana]
Length = 183
Score = 108 bits (271), Expect = 6e-23
Identities = 55/143 (38%), Positives = 81/143 (56%), Gaps = 8/143 (5%)
Query: 58 GPLSRWRLKESASTESPCHHPCDCYCSSAE--------SPPDTFDCGKHDPVMNEEMNKD 109
GP W L E+ + + C C CSS S DC K DP +NE+ K+
Sbjct: 25 GPPLYWHLTEALAVSATSCSACVCDCSSLPLLTIPTGLSNGSFTDCAKRDPEVNEDTEKN 84
Query: 110 LLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNVAMETCEEARERAEA 169
+L+EEL + + E ++ D ++ A+ +S YQ E +KCN METCEEARE+AE
Sbjct: 85 YAELLTEELKQREAASMEKHKRVDTGLLEAKKITSSYQKEADKCNSGMETCEEAREKAEK 144
Query: 170 ELIEEHRVTALWENRAREYGWKD 192
L+E+ ++T++WE RAR+ G+KD
Sbjct: 145 ALVEQKKLTSMWEQRARQKGYKD 167
>UniRef100_Q8LE30 Hypothetical protein [Arabidopsis thaliana]
Length = 183
Score = 108 bits (271), Expect = 6e-23
Identities = 55/143 (38%), Positives = 81/143 (56%), Gaps = 8/143 (5%)
Query: 58 GPLSRWRLKESASTESPCHHPCDCYCSSAE--------SPPDTFDCGKHDPVMNEEMNKD 109
GP W L E+ + + C C CSS S DC K DP +NE+ K+
Sbjct: 25 GPPLYWHLTEALAVSATSCSACVCDCSSLPLLTIPTGLSNGSFTDCAKRDPEVNEDTEKN 84
Query: 110 LLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNVAMETCEEARERAEA 169
+L+EEL + + E ++ D ++ A+ +S YQ E +KCN METCEEARE+AE
Sbjct: 85 YAELLTEELKQREAASMEKHKRVDTGLLEAKKITSSYQKEADKCNSGMETCEEAREKAEK 144
Query: 170 ELIEEHRVTALWENRAREYGWKD 192
L+E+ ++T++WE RAR+ G+KD
Sbjct: 145 ALVEQKKLTSMWEQRARQKGYKD 167
>UniRef100_Q9ZQ38 Expressed protein [Arabidopsis thaliana]
Length = 173
Score = 105 bits (261), Expect = 9e-22
Identities = 60/156 (38%), Positives = 84/156 (53%), Gaps = 15/156 (9%)
Query: 50 IVQVIVEFGPLSRWRLKE-----SASTESPCHHPCDCY----CSSAESPPDTFD-----C 95
+V ++ GP W+L + + ST S C PC C S + P + C
Sbjct: 17 VVSALLVCGPALYWKLNKGFVGSARSTNSICP-PCVCDFPPPLSLLQIAPGLANLSITGC 75
Query: 96 GKHDPVMNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNV 155
G DP + EEM K + +L+EEL LQ V +E + + A+ +S YQ E EKCN
Sbjct: 76 GSDDPELKEEMEKPFVDLLTEELKLQEAVADEHSRHMNVTLAEAKRVASQYQKEAEKCNA 135
Query: 156 AMETCEEARERAEAELIEEHRVTALWENRAREYGWK 191
A E CE ARERA+A L++E ++T LWE RAR+ GW+
Sbjct: 136 ATEICESARERAQALLLKERKITFLWERRARQLGWE 171
>UniRef100_Q84XT8 Hypothetical protein [Phytophthora sojae]
Length = 189
Score = 103 bits (258), Expect = 2e-21
Identities = 58/148 (39%), Positives = 80/148 (53%), Gaps = 15/148 (10%)
Query: 58 GPLSRWRLKE-------SASTESPCHHPCDCYCSSAESPPDTF------DCGKHDPVMNE 104
GP W E S+ST +PC CDC S P DC K DP ++
Sbjct: 28 GPPLYWHFVEGLAAVNHSSSTCAPCI--CDCSSQPILSIPQGLSNTSFGDCAKPDPEVSG 85
Query: 105 EMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNVAMETCEEAR 164
+ K+ +L+EEL L+ E + D ++ A+ +S YQ E +KCN METCEEAR
Sbjct: 86 DTEKNFAELLAEELKLRENQAVENQQHADMALLEAKKIASQYQKEADKCNSGMETCEEAR 145
Query: 165 ERAEAELIEEHRVTALWENRAREYGWKD 192
E+AE L+ + ++TALWE RAR+ GWK+
Sbjct: 146 EKAELTLVTQKKLTALWELRARKKGWKE 173
>UniRef100_Q8S7I4 Hypothetical protein OSJNBa0010I09.2 [Oryza sativa]
Length = 183
Score = 96.3 bits (238), Expect = 4e-19
Identities = 49/139 (35%), Positives = 69/139 (49%), Gaps = 5/139 (3%)
Query: 58 GPLSRWRLKESASTESPCHHPCDCYCSSAESPPDTFDCGKH-----DPVMNEEMNKDLLT 112
GP W E+ + CDC C + DC K EE K
Sbjct: 36 GPQLYWHASEALGRSTGACPACDCDCDARPLLALPEDCSKQFKDVKSRASGEETEKSFTE 95
Query: 113 MLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNVAMETCEEARERAEAELI 172
+L EEL + + +Q D ++ A+ +S YQ E +KC+ M+TCEEARE++ L+
Sbjct: 96 LLIEELKQREEEATQAQQQADVKLLEAKKLASQYQKEADKCSSGMDTCEEAREKSSEALV 155
Query: 173 EEHRVTALWENRAREYGWK 191
E+ ++TALWE RARE GWK
Sbjct: 156 EQRKLTALWEERARELGWK 174
>UniRef100_Q9XE99 Hypothetical protein T19B17.8 [Arabidopsis thaliana]
Length = 125
Score = 91.3 bits (225), Expect = 1e-17
Identities = 44/99 (44%), Positives = 66/99 (66%), Gaps = 1/99 (1%)
Query: 94 DCGKHDPVMNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKC 153
DC +H+ +EE M++EEL L+ E + DRL+++A+ A+S YQ E +KC
Sbjct: 20 DCMRHEEG-SEESESSFTEMVAEELKLREAQAQEDEWRADRLLLDAKKAASQYQKEADKC 78
Query: 154 NVAMETCEEARERAEAELIEEHRVTALWENRAREYGWKD 192
++ METCE ARE+AEA L E+ R++ +WE RAR+ GWK+
Sbjct: 79 SMGMETCELAREKAEAALDEQRRLSYMWELRARQGGWKE 117
>UniRef100_UPI000036832F UPI000036832F UniRef100 entry
Length = 331
Score = 38.1 bits (87), Expect = 0.13
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 11/89 (12%)
Query: 88 SPPDTFDCGKHDPVMNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQ 147
SPP+ C D + +DLL L E E Q+DR+ S YQ
Sbjct: 98 SPPEVDTCINEDVESLRKTVQDLLARLQEA---------EQQHQSDRVAFEV--TLSRYQ 146
Query: 148 METEKCNVAMETCEEARERAEAELIEEHR 176
E E+ NVA++ E+ E EAE+ E R
Sbjct: 147 KEAEQSNVALQREEDRVEEKEAEVRELQR 175
>UniRef100_UPI000036832E UPI000036832E UniRef100 entry
Length = 365
Score = 38.1 bits (87), Expect = 0.13
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 11/89 (12%)
Query: 88 SPPDTFDCGKHDPVMNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQ 147
SPP+ C D + +DLL L E E Q+DR+ S YQ
Sbjct: 132 SPPEVDTCINEDVESLRKTVQDLLARLQEA---------EQQHQSDRVAFEV--TLSRYQ 180
Query: 148 METEKCNVAMETCEEARERAEAELIEEHR 176
E E+ NVA++ E+ E EAE+ E R
Sbjct: 181 KEAEQSNVALQREEDRVEEKEAEVRELQR 209
>UniRef100_UPI000036832D UPI000036832D UniRef100 entry
Length = 390
Score = 38.1 bits (87), Expect = 0.13
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 11/89 (12%)
Query: 88 SPPDTFDCGKHDPVMNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQ 147
SPP+ C D + +DLL L E E Q+DR+ S YQ
Sbjct: 157 SPPEVDTCINEDVESLRKTVQDLLARLQEA---------EQQHQSDRVAFEV--TLSRYQ 205
Query: 148 METEKCNVAMETCEEARERAEAELIEEHR 176
E E+ NVA++ E+ E EAE+ E R
Sbjct: 206 KEAEQSNVALQREEDRVEEKEAEVRELQR 234
>UniRef100_O74697 Hypothetical protein [Phaeosphaeria nodorum]
Length = 576
Score = 36.6 bits (83), Expect = 0.39
Identities = 37/161 (22%), Positives = 69/161 (41%), Gaps = 11/161 (6%)
Query: 16 HPFISEHTASSQSPRSSNGSVSVDVHITCESPSDIVQVIVEFGPLSRWRLKESASTESPC 75
H F+ + TA+ + +S+D S I QV+ F + RL++S+ + +
Sbjct: 72 HAFLPKPTANRSKIANYGAFISIDSRPVSNSRGTIKQVVAAF----KERLRKSSQSLTAV 127
Query: 76 HHPCDCYCSSAESPPDTFDCGKHDPVMNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRL 135
P +C + PPD++D +P ++ M D +L NL R E + + +
Sbjct: 128 KDP--FFCMNIICPPDSYD-PNIEPAKDDVMFDDSKVVLEVVDNLLRSHYPEAIMEME-- 182
Query: 136 VMNARNASSHYQMETEKCNVAMETCEEARERAEAELIEEHR 176
V +A H + E+ + + A + AE++ E R
Sbjct: 183 VEPPTSAQQHAGSDIEE--IESHVPDPAHLKPPAEMVREPR 221
>UniRef100_UPI00004364C6 UPI00004364C6 UniRef100 entry
Length = 379
Score = 35.4 bits (80), Expect = 0.87
Identities = 27/98 (27%), Positives = 42/98 (42%), Gaps = 7/98 (7%)
Query: 102 MNEEMNKDLLTMLSEELNLQRIVTNETLEQTDRLVMNARNASSHYQMETEKCNVAMETCE 161
M E+ K+ T+ EE ++R+ E + QT+ M + K + E E
Sbjct: 197 MKEKTVKEKETIKEEEEEMERVKVEEKMVQTEEEEMEEVKIKKRRRRRIMKEEIIQEE-E 255
Query: 162 EARERAEAELIEEHRVTALWENRAREYGWKDKRRTRLR 199
E E E+ EE + + +E WK+KRR R R
Sbjct: 256 NIIEEEEEEMSEEE------DKKTQEDKWKNKRRRRGR 287
>UniRef100_O46652 Fertilin alpha-II [Papio anubis]
Length = 825
Score = 35.0 bits (79), Expect = 1.1
Identities = 24/80 (30%), Positives = 38/80 (47%), Gaps = 2/80 (2%)
Query: 64 RLKESASTESPCHHPCDCYCSSAESPPDTFDCGKHDPVMNEEMNKDLLTMLSEELNLQRI 123
R +ES + C++ C+C S +PPD + G V + K ++S E N I
Sbjct: 674 RPEESCHGKGVCNNLRHCHCGSGFAPPDCKNPGNGGSVDSGPAGKPSDEIISREENRNHI 733
Query: 124 VTNETLEQTDRLVMNARNAS 143
V + L++ D V N +N S
Sbjct: 734 VGHGNLQRGD--VSNNKNKS 751
>UniRef100_Q9V4P4 CG30494-PA [Drosophila melanogaster]
Length = 2262
Score = 35.0 bits (79), Expect = 1.1
Identities = 43/165 (26%), Positives = 71/165 (42%), Gaps = 28/165 (16%)
Query: 17 PFISEHTASSQSPRSSNGSVSVDVHITCESPSDIVQVIVEFGPLSRWRLKES--ASTESP 74
P EH+ S+ SV+V + + +D++ VE P S + S ASTE P
Sbjct: 844 PIEGEHSQHSEKETPE----SVEVGLDSPNVADVITTSVEVIPTSTTAVTTSDGASTEMP 899
Query: 75 CHHPCDCYCSSAESPPDTFDCGKHDPVMNEEMNKDLLTMLSEEL-----NLQRIVTNETL 129
P YC +A PP + +++ V ++ D T L EE+ NL+ ++
Sbjct: 900 --EPPKIYCETA--PPAAEEIEQNETVETLSVSAD--TTLPEEIVSQDGNLEASEQTASI 953
Query: 130 EQTDRLVMNARNASSHYQMETEKCNVAMETCEEARERAEAELIEE 174
+ T + +N ME++ C +E + RE E E + E
Sbjct: 954 DVTFQANLN--------NMESKGC---VEVAVDVRESKEDETLME 987
>UniRef100_Q9NJA9 Paramyosin [Anisakis simplex]
Length = 869
Score = 35.0 bits (79), Expect = 1.1
Identities = 29/99 (29%), Positives = 45/99 (45%), Gaps = 3/99 (3%)
Query: 103 NEEMNKDLLTMLSEELNLQRIV--TNETLEQTDRLVMNARNASSHYQMETEKCNVAMETC 160
N E K + + + LQ + T L+QT A+ S E E+C A++
Sbjct: 563 NLEAQKTIKKQSEQIIQLQANLEDTQRQLQQTLDQYALAQRKISALSAELEECKTALDNA 622
Query: 161 EEARERAEAELIEEH-RVTALWENRAREYGWKDKRRTRL 198
AR++AEA+L E H R++ L + K+K T L
Sbjct: 623 IRARKQAEADLEEAHVRISDLTSINSNLTAIKNKLETEL 661
>UniRef100_UPI00001D09DB UPI00001D09DB UniRef100 entry
Length = 1470
Score = 34.7 bits (78), Expect = 1.5
Identities = 40/136 (29%), Positives = 58/136 (42%), Gaps = 19/136 (13%)
Query: 41 HITCESPSDIVQVIVEFGPLSRWRLKESAS--TESPCHHPCDCYCSSAESPPDTFDC--- 95
++ +S SD + V F S ESA TE P H D S E T+
Sbjct: 1032 YLEFDSLSDKSKTRVSFSRYSFQEKPESAPSPTEQPWAHSLDQ--DSLEDDSTTYQRFGK 1089
Query: 96 ---GKHDPVMNEEMNKDLLTMLSEEL--------NLQRIVTNETLEQTDRLVMNARNASS 144
G DP NEE+ +LL L EEL N RI+TNE + D ++ R ++
Sbjct: 1090 VSQGILDPGKNEELTDELLGCLVEELLAIDEKDNNSCRIMTNEA-DTRDLNLVFGRRGNT 1148
Query: 145 HYQMETEKCNVAMETC 160
++ E +V ++ C
Sbjct: 1149 IEGLDRETTDVKLQRC 1164
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.128 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 308,791,790
Number of Sequences: 2790947
Number of extensions: 11099360
Number of successful extensions: 40055
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 39987
Number of HSP's gapped (non-prelim): 126
length of query: 200
length of database: 848,049,833
effective HSP length: 121
effective length of query: 79
effective length of database: 510,345,246
effective search space: 40317274434
effective search space used: 40317274434
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0544.11


