

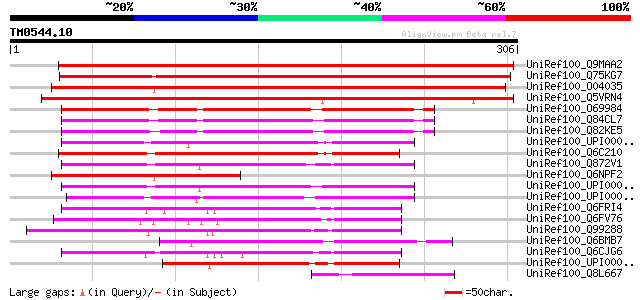
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.10
(306 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9MAA2 T12H1.14 protein [Arabidopsis thaliana] 412 e-114
UniRef100_Q75KG7 Hypothetical protein OJ1489_G03.9 [Oryza sativa] 407 e-112
UniRef100_O04035 F7G19.18 protein [Arabidopsis thaliana] 368 e-100
UniRef100_Q5VRN4 Hypothetical protein OSJNBa0004I20.1 [Oryza sat... 307 2e-82
UniRef100_O69984 Hypothetical protein SCO5808 [Streptomyces coel... 193 5e-48
UniRef100_Q84CL7 Hypothetical protein [Streptomyces murayamaensis] 192 9e-48
UniRef100_Q82KE5 Hypothetical protein [Streptomyces avermitilis] 184 3e-45
UniRef100_UPI000023D766 UPI000023D766 UniRef100 entry 166 9e-40
UniRef100_Q6C210 Yarrowia lipolytica chromosome F of strain CLIB... 165 2e-39
UniRef100_Q872V1 Hypothetical protein B23G1.030 [Neurospora crassa] 164 4e-39
UniRef100_Q6NPF2 At1g08940 [Arabidopsis thaliana] 159 1e-37
UniRef100_UPI000021962A UPI000021962A UniRef100 entry 158 2e-37
UniRef100_UPI0000235FC2 UPI0000235FC2 UniRef100 entry 156 6e-37
UniRef100_Q6FRI4 Similar to tr|Q99288 Saccharomyces cerevisiae Y... 150 5e-35
UniRef100_Q6FV76 Similar to tr|Q99288 Saccharomyces cerevisiae Y... 142 1e-32
UniRef100_Q99288 S.cerevisiae chromosome IV reading frame ORF YD... 141 2e-32
UniRef100_Q6BMB7 Similar to CA5949|IPF3498 Candida albicans IPF3... 126 6e-28
UniRef100_Q6CJG6 Similar to sgd|S0002458 Saccharomyces cerevisia... 122 9e-27
UniRef100_UPI000042C7BD UPI000042C7BD UniRef100 entry 120 5e-26
UniRef100_Q8L667 Hypothetical protein OSJNBb0091B12.12 [Oryza sa... 68 3e-10
>UniRef100_Q9MAA2 T12H1.14 protein [Arabidopsis thaliana]
Length = 316
Score = 412 bits (1059), Expect = e-114
Identities = 198/276 (71%), Positives = 232/276 (83%), Gaps = 1/276 (0%)
Query: 30 VLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISST-SSTD 88
+LPKRIILVRHGES+GNLD +AYTTTPDHKI LT G+ QA+ AGAR+ +ISS SS +
Sbjct: 7 LLPKRIILVRHGESEGNLDTAAYTTTPDHKIQLTDSGLLQAQEAGARLHALISSNPSSPE 66
Query: 89 WRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQ 148
WRVYFYVSPY RTRSTLREIGRSFS+ R++GVREECRIREQDFGNFQV+ER+ A K+ R+
Sbjct: 67 WRVYFYVSPYDRTRSTLREIGRSFSRRRVIGVREECRIREQDFGNFQVKERMRATKKVRE 126
Query: 149 RFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVF 208
RFGRFF+RFP+GESAADVFDRVSSFLESLWRDIDMNRL+ +PS++LN +IVSHGL SRVF
Sbjct: 127 RFGRFFYRFPEGESAADVFDRVSSFLESLWRDIDMNRLHINPSHELNFVIVSHGLTSRVF 186
Query: 209 LMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEWGLSPDMIADQKW 268
LMKWFKW+VEQF+ LNN GN E RVM+LG GG+YSLA+HHT+ E+ WGLSP+MIADQKW
Sbjct: 187 LMKWFKWSVEQFEGLNNPGNSEIRVMELGQGGDYSLAIHHTEEELATWGLSPEMIADQKW 246
Query: 269 RAHATKGAWHDQGPLSLDTFFDHLHDSDNEDNDDQT 304
RA+A KG W + FFDH+ DSD E + T
Sbjct: 247 RANAHKGEWKEDCKWYFGDFFDHMADSDKECETEAT 282
>UniRef100_Q75KG7 Hypothetical protein OJ1489_G03.9 [Oryza sativa]
Length = 295
Score = 407 bits (1046), Expect = e-112
Identities = 193/272 (70%), Positives = 226/272 (82%), Gaps = 2/272 (0%)
Query: 31 LPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWR 90
LPKRIILVRHGESQGNLD SAYTTTPD++I LTP G+ QAR AG I V+SS ++ W+
Sbjct: 13 LPKRIILVRHGESQGNLDMSAYTTTPDYRIPLTPLGVDQARAAGKGILDVVSSAAN--WK 70
Query: 91 VYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQRF 150
VYFYVSPY RTR+TLREIG +F +HR++G REECR+REQDFGNFQV+ER+ A+KETR RF
Sbjct: 71 VYFYVSPYERTRATLREIGAAFPRHRVIGAREECRVREQDFGNFQVEERMRAVKETRDRF 130
Query: 151 GRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFLM 210
GRFFFRFP+GESAADVFDRV+SFLESLWRDIDM RL D S + NL+IVSHGL SRVFLM
Sbjct: 131 GRFFFRFPEGESAADVFDRVASFLESLWRDIDMGRLEQDASCETNLVIVSHGLTSRVFLM 190
Query: 211 KWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEWGLSPDMIADQKWRA 270
KWFKWTV+QF+ LNNF N EFRVMQLG GEYSL +HHT E+ WGLSP+MIADQ+WRA
Sbjct: 191 KWFKWTVDQFERLNNFDNCEFRVMQLGPAGEYSLLIHHTKEELQRWGLSPEMIADQQWRA 250
Query: 271 HATKGAWHDQGPLSLDTFFDHLHDSDNEDNDD 302
A + +W D+ L TFFDH ++ DN+D+DD
Sbjct: 251 SANRRSWADECSSFLATFFDHWNEDDNDDDDD 282
>UniRef100_O04035 F7G19.18 protein [Arabidopsis thaliana]
Length = 281
Score = 368 bits (944), Expect = e-100
Identities = 177/279 (63%), Positives = 223/279 (79%), Gaps = 5/279 (1%)
Query: 26 HKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTS 85
++ +LPKRIIL+RHGES GN+D AY TTPDHKI LT +G QAR AG ++R +IS+ S
Sbjct: 3 NEKKMLPKRIILMRHGESAGNIDAGAYATTPDHKIPLTEEGRAQAREAGKKMRALISTQS 62
Query: 86 S----TDWRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERIN 141
+WRVYFYVSPY RTR+TLRE+G+ FS+ R++GVREECRIREQDFGNFQV+ER+
Sbjct: 63 GGACGENWRVYFYVSPYERTRTTLREVGKGFSRKRVIGVREECRIREQDFGNFQVEERMR 122
Query: 142 ALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSH 201
+KETR+RFGRFF+RFP+GESAADV+DRVSSFLES+WRD+DMNR DPS++LNL+IVSH
Sbjct: 123 VVKETRERFGRFFYRFPEGESAADVYDRVSSFLESMWRDVDMNRHQVDPSSELNLVIVSH 182
Query: 202 GLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEWGLSPD 261
GL SRVFL KWFKWTV +F+ LNNFGN EFRVM+LG+ GEY+ A+HH++ EM +WG+S D
Sbjct: 183 GLTSRVFLTKWFKWTVAEFERLNNFGNCEFRVMELGASGEYTFAIHHSEEEMLDWGMSKD 242
Query: 262 MIADQKWRAHATK-GAWHDQGPLSLDTFFDHLHDSDNED 299
MI DQK R + +D L L+ +FD L +D+E+
Sbjct: 243 MIDDQKDRVDGCRVTTSNDSCSLHLNEYFDLLDVTDDEE 281
>UniRef100_Q5VRN4 Hypothetical protein OSJNBa0004I20.1 [Oryza sativa]
Length = 375
Score = 307 bits (787), Expect = 2e-82
Identities = 151/291 (51%), Positives = 198/291 (67%), Gaps = 6/291 (2%)
Query: 20 IPNQNNHKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRH 79
+P+Q + P P+RI+LVRHGES+GN+D +AYT PD +I LTPQG A G R+RH
Sbjct: 77 LPHQQRYPPPPRPRRIVLVRHGESEGNVDEAAYTRVPDPRIGLTPQGWRDAEDCGRRLRH 136
Query: 80 VISSTSSTDWRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQER 139
++S+ DW+VYFYVSPY RT TLR +GR+F R+ GVREE R+REQDFGNFQ +++
Sbjct: 137 LLSTGGGDDWKVYFYVSPYRRTLETLRGLGRAFEARRIAGVREEPRLREQDFGNFQDRDK 196
Query: 140 INALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLN--HDPSNDLNLI 197
+ KE R+R+GRFF+RFP+GESAADV+DR++ F E+L DID+ R + + D+N++
Sbjct: 197 MRVEKEIRRRYGRFFYRFPNGESAADVYDRITGFRETLRADIDIGRFQPPGERNPDMNVV 256
Query: 198 IVSHGLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEWG 257
+VSHGL RVFLM+W+KWTV QF+ L N NG VMQ G+GG YSL VHH+ E+ E+G
Sbjct: 257 LVSHGLTLRVFLMRWYKWTVSQFEGLANLSNGGALVMQTGAGGRYSLLVHHSVDELREFG 316
Query: 258 LSPDMIADQKWRAHATKGAWH----DQGPLSLDTFFDHLHDSDNEDNDDQT 304
L+ DMI DQKW+ A G + GP F H HD DD T
Sbjct: 317 LTDDMIEDQKWQMTARPGELNYNFITNGPSFFTHFTHHHHDKHKAAIDDGT 367
>UniRef100_O69984 Hypothetical protein SCO5808 [Streptomyces coelicolor]
Length = 219
Score = 193 bits (490), Expect = 5e-48
Identities = 103/225 (45%), Positives = 138/225 (60%), Gaps = 19/225 (8%)
Query: 32 PKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRV 91
P+RI+LVRHGES GN+D + Y PDH ++LT +G QA G +R V S R+
Sbjct: 4 PRRIVLVRHGESIGNVDDTVYEREPDHALALTDRGRAQAEETGEGLREVFGSE-----RI 58
Query: 92 YFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQRFG 151
YVSPY RT TLR L+ +REE R+REQD+GN+Q ++ + K R +G
Sbjct: 59 SVYVSPYRRTHETLRAFRLDPD---LIRIREEPRLREQDWGNWQDRDDVRLQKAYRDAYG 115
Query: 152 RFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFLMK 211
FF+RF GES ADV+DRV FLESL+R + DP + N+++V+HGLA R+F M+
Sbjct: 116 HFFYRFAQGESGADVYDRVGGFLESLFRSFE------DPDHPPNVLLVTHGLAMRLFCMR 169
Query: 212 WFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEW 256
WF WTV +F+ L N GN E R++ LG+ G+YSL DR W
Sbjct: 170 WFHWTVAEFESLANPGNAEMRMLVLGADGKYSL-----DRPFERW 209
>UniRef100_Q84CL7 Hypothetical protein [Streptomyces murayamaensis]
Length = 219
Score = 192 bits (488), Expect = 9e-48
Identities = 103/225 (45%), Positives = 134/225 (58%), Gaps = 19/225 (8%)
Query: 32 PKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRV 91
P+RI+LVRHGES+GN D + Y PDH + LT G QAR G R+R + RV
Sbjct: 4 PRRIVLVRHGESEGNADDTVYEREPDHALRLTEPGRLQARATGERLREIFGRE-----RV 58
Query: 92 YFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQRFG 151
YVSPY RT T RE G L+ +REE R+REQD+GN+Q ++ + K R +G
Sbjct: 59 SVYVSPYRRTHETFREFGLDPD---LVRIREEPRLREQDWGNWQDRDDVRLQKAYRDAYG 115
Query: 152 RFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFLMK 211
FF+RF GES ADV+DRV SFLESL R + P + N+++V+HGL R+F M+
Sbjct: 116 HFFYRFAQGESGADVYDRVGSFLESLHRSFEA------PDHPPNVLLVTHGLTMRLFCMR 169
Query: 212 WFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEW 256
WF WTV F+ L+N GN E R + LG+ G Y+L DR W
Sbjct: 170 WFHWTVADFESLSNPGNAETRTLVLGTDGRYTL-----DRPFARW 209
>UniRef100_Q82KE5 Hypothetical protein [Streptomyces avermitilis]
Length = 219
Score = 184 bits (467), Expect = 3e-45
Identities = 101/225 (44%), Positives = 134/225 (58%), Gaps = 19/225 (8%)
Query: 32 PKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRV 91
P+RI+LVRHGES GN D + Y PDH + LT G QA G ++R + V
Sbjct: 4 PRRIVLVRHGESAGNADDTVYEREPDHALPLTEVGWRQAEERGKQLRELFGQEG-----V 58
Query: 92 YFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQERINALKETRQRFG 151
YVSPY RT TLR L+ VREE R+REQD+GN+Q ++ + K R +G
Sbjct: 59 SVYVSPYRRTLETLRAFHLDPD---LVRVREEPRLREQDWGNWQDRDDVRLQKAYRDAYG 115
Query: 152 RFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFLMK 211
FF+RF GES ADV+DRV FLESL+R + P + N++IV+HGLA R+F M+
Sbjct: 116 HFFYRFAQGESGADVYDRVGGFLESLFRSFEA------PDHPPNVLIVTHGLAMRLFCMR 169
Query: 212 WFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEW 256
WF W+V +F+ L+N GN E R++ LG+ G+YSL DR W
Sbjct: 170 WFHWSVAEFESLSNPGNAEMRMLVLGNDGKYSL-----DRPFERW 209
>UniRef100_UPI000023D766 UPI000023D766 UniRef100 entry
Length = 494
Score = 166 bits (419), Expect = 9e-40
Identities = 96/223 (43%), Positives = 131/223 (58%), Gaps = 20/223 (8%)
Query: 32 PKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRV 91
P+ IILVRHG+S+GN + + T PDH++ LTP+G +QA AG R+R ++ D +
Sbjct: 4 PRLIILVRHGQSEGNKNREIHQTVPDHRVKLTPEGWSQAHDAGRRLRSLL----RPDDTL 59
Query: 92 YFYVSPYARTRSTLR---------EIGRSFSKHRLLGVREECRIREQDFGNFQ-VQERIN 141
F+ SPY RTR T E S + + V EE R+REQDFGNFQ +
Sbjct: 60 QFFTSPYRRTRETTEGILETLTSDETSPSPFRRNNIKVYEEPRLREQDFGNFQPCSAEME 119
Query: 142 ALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSH 201
+ + R +G FF+R P+GESAAD +DRVS F ESLWR + PS ++V+H
Sbjct: 120 RMWQERADYGHFFYRIPNGESAADAYDRVSGFNESLWRQFGEDDF---PS---VCVLVTH 173
Query: 202 GLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSL 244
GL SRVFLMKW+ +TVE F+ L N + EF +M+ +Y L
Sbjct: 174 GLMSRVFLMKWYHFTVEYFEDLRNINHCEFLIMRKQENQKYLL 216
>UniRef100_Q6C210 Yarrowia lipolytica chromosome F of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 289
Score = 165 bits (417), Expect = 2e-39
Identities = 91/208 (43%), Positives = 130/208 (61%), Gaps = 12/208 (5%)
Query: 30 VLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDW 89
+L + IIL+RHGES+GN D S T +H+I LT G QAR AG R+ +++ D
Sbjct: 9 ILTQLIILIRHGESEGNCDKSVNRHTSNHRIKLTANGEEQARDAGKRLADMVNK----DD 64
Query: 90 RVYFYVSPYARTRSTLREIGRSFSKHRL-LGVREECRIREQDFGNFQVQ-ERINALKETR 147
+ FY SPY RTR T + I + + V EE R+REQDFGNFQ +N + R
Sbjct: 65 TLLFYTSPYQRTRQTTQLICEGIEEKNISYKVHEEPRLREQDFGNFQASASEMNKIWNDR 124
Query: 148 QRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRV 207
++G FF+R P+GESAADV+DR + F E+L+R + ++ PS L++V+HG+ RV
Sbjct: 125 SQYGHFFYRIPNGESAADVYDRCAGFNETLFRQFNSDKF---PS---VLVLVAHGIWIRV 178
Query: 208 FLMKWFKWTVEQFDLLNNFGNGEFRVMQ 235
FLMKW++WT E+F+ L N + EF +M+
Sbjct: 179 FLMKWYRWTYEKFESLRNLRHCEFIIMK 206
>UniRef100_Q872V1 Hypothetical protein B23G1.030 [Neurospora crassa]
Length = 570
Score = 164 bits (414), Expect = 4e-39
Identities = 93/223 (41%), Positives = 132/223 (58%), Gaps = 20/223 (8%)
Query: 32 PKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRV 91
P+ IIL+RH +S+GN + + T PDH++ LT +G QA AG R+R ++ + D +
Sbjct: 4 PRLIILIRHAQSEGNKNRDIHQTIPDHRVKLTDEGWQQAYDAGRRLRKLLRA----DDTI 59
Query: 92 YFYVSPYARTRSTLREIGRSFS---------KHRLLGVREECRIREQDFGNFQ-VQERIN 141
F+ SPY RTR T I + + K + V EE R+REQDFGNFQ +
Sbjct: 60 QFFTSPYRRTRETTEGILATLTSDDPEPSPFKRNHIKVYEEPRLREQDFGNFQPCSAEME 119
Query: 142 ALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSH 201
+ + R +G FF+R P+GESAAD +DRVS F ESLWR + N D + ++V+H
Sbjct: 120 RMWQERADYGHFFYRIPNGESAADAYDRVSGFNESLWR-----QFNDDDFASV-CVLVTH 173
Query: 202 GLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSL 244
GL SRVFLMKW+ ++VE F+ L N + EF +M+ G+Y L
Sbjct: 174 GLMSRVFLMKWYHFSVEYFEDLRNVNHCEFLIMRKNDSGKYIL 216
>UniRef100_Q6NPF2 At1g08940 [Arabidopsis thaliana]
Length = 131
Score = 159 bits (401), Expect = 1e-37
Identities = 76/118 (64%), Positives = 95/118 (80%), Gaps = 4/118 (3%)
Query: 26 HKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTS 85
++ +LPKRIIL+RHGES GN+D AY TTPDHKI LT +G QAR AG ++R +IS+ S
Sbjct: 3 NEKKMLPKRIILMRHGESAGNIDAGAYATTPDHKIPLTEEGRAQAREAGKKMRALISTQS 62
Query: 86 S----TDWRVYFYVSPYARTRSTLREIGRSFSKHRLLGVREECRIREQDFGNFQVQER 139
+WRVYFYVSPY RTR+TLRE+G+ FS+ R++GVREECRIREQDFGNFQV+E+
Sbjct: 63 GGACGENWRVYFYVSPYERTRTTLREVGKGFSRKRVIGVREECRIREQDFGNFQVEEK 120
>UniRef100_UPI000021962A UPI000021962A UniRef100 entry
Length = 633
Score = 158 bits (400), Expect = 2e-37
Identities = 93/224 (41%), Positives = 128/224 (56%), Gaps = 21/224 (9%)
Query: 32 PKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRV 91
P+ IIL+RH +S+GN + T PDH++ LT G QA AG R+R ++ + D +
Sbjct: 4 PRLIILIRHAQSEGNKKREIHQTIPDHRVKLTQDGWQQAYEAGRRLRSLLRA----DDTL 59
Query: 92 YFYVSPYARTRSTLREIGRSFS---------KHRLLGVREECRIREQDFGNFQ-VQERIN 141
F+ SPY RTR T I + + K + V EE R+REQDFGNFQ +
Sbjct: 60 QFFTSPYRRTRETTEGILATLTADEPDPSPFKREKIKVYEEPRLREQDFGNFQPCSAEME 119
Query: 142 ALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSH 201
+ + R +G FF+R P+GESAAD +DRVS F ESLWR D ++V+H
Sbjct: 120 RMWQERADYGHFFYRIPNGESAADAYDRVSGFNESLWRQFG------DEDFASVCVLVTH 173
Query: 202 GLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLGSG-GEYSL 244
GL SRVFLMKW+ ++VE F+ L N + EF +M+ G G+Y L
Sbjct: 174 GLMSRVFLMKWYHFSVEYFEDLRNVNHCEFLIMRQREGTGKYDL 217
>UniRef100_UPI0000235FC2 UPI0000235FC2 UniRef100 entry
Length = 651
Score = 156 bits (395), Expect = 6e-37
Identities = 94/222 (42%), Positives = 129/222 (57%), Gaps = 23/222 (10%)
Query: 35 IILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVISSTSSTDWRVYFY 94
IIL+RH +S+GN + + T PDH++ LTP+G QAR AG R+R ++ D ++F+
Sbjct: 82 IILIRHAQSEGNKNREIHQTIPDHRVKLTPEGHRQARDAGTRLRGLL----RPDDTIHFF 137
Query: 95 VSPYARTRSTLREIGRS----------FSKHRLLGVREECRIREQDFGNFQ-VQERINAL 143
SPY RTR T I S F +H + V EE R+REQDFGNFQ + +
Sbjct: 138 TSPYRRTRETTEGILESLTADTPSPSPFPRH-TIKVYEEPRLREQDFGNFQPCSAEMERM 196
Query: 144 KETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGL 203
R +G FF+R P+GESAAD +DRVS F ES+W RL + ++V+HGL
Sbjct: 197 WLERADYGHFFYRIPNGESAADAYDRVSGFNESMW------RLFGEKDFASVCVLVTHGL 250
Query: 204 ASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLG-SGGEYSL 244
+RVFLMKW+ VE F+ L N + EF +M+L G+Y L
Sbjct: 251 MTRVFLMKWYHSFVEYFEDLRNINHCEFVIMKLNEDSGKYVL 292
>UniRef100_Q6FRI4 Similar to tr|Q99288 Saccharomyces cerevisiae YDR051c [Candida
glabrata]
Length = 319
Score = 150 bits (378), Expect = 5e-35
Identities = 96/288 (33%), Positives = 137/288 (47%), Gaps = 86/288 (29%)
Query: 32 PKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHVI---------- 81
P+ I+L+RHGES+ N D TP+H I LTP G QA AG ++ ++
Sbjct: 8 PRLIVLIRHGESESNKDKKINEVTPNHLIPLTPYGKKQAHNAGLKLLRLLNMADASVIEK 67
Query: 82 ---------SSTSSTDWRVY------------FYVSPYARTRSTLREIGRSFSKHRLL-- 118
+ST++ + R Y FY SPY RTR TL+ + ++ L
Sbjct: 68 LGEEYALPATSTNTLELRGYKPNGKHMDDNIIFYTSPYKRTRETLKGVLEVIDRYNELKS 127
Query: 119 ----------------------------GVRE----------------------ECRIRE 128
G+ E E R+RE
Sbjct: 128 GINMCAEQTYNPYGKQKHAIWPHDLENSGIYENDESTHCGPEREGTYIRYRIIDEPRLRE 187
Query: 129 QDFGNFQVQERINALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNH 188
QDFGN+Q + + E R+ +G FFFRFP+GESAADV+DRV+SF ++L+R + H
Sbjct: 188 QDFGNYQEVSSMQDVMEKRKTYGHFFFRFPEGESAADVYDRVASFQDTLYRHFQFRQ--H 245
Query: 189 DPSNDLNLIIVSHGLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQL 236
D+ +++V+HG+ SRVFLMKWF+WT E+F+ N NG VM+L
Sbjct: 246 TKGRDV-VVLVTHGIYSRVFLMKWFRWTFEEFESFTNVPNGSLMVMEL 292
>UniRef100_Q6FV76 Similar to tr|Q99288 Saccharomyces cerevisiae YDR051c [Candida
glabrata]
Length = 327
Score = 142 bits (358), Expect = 1e-32
Identities = 101/297 (34%), Positives = 139/297 (46%), Gaps = 91/297 (30%)
Query: 27 KPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARI--------- 77
KP P+ IILVRHGES+ N D P+H I LT +G ++AR AG +
Sbjct: 14 KPLKKPRLIILVRHGESESNRDKGVNEKIPNHLIPLTSRGWSEARNAGVELLKLLNMDGN 73
Query: 78 -RHVISSTS-----------------------STDWRVYFYVSPYARTRSTLR---EIGR 110
+ VI S S D + FY SPY RT+ TL+ EI
Sbjct: 74 CKSVIESLSQKYRVEDDRRNMLKTSECNNMKKKIDRSIVFYTSPYRRTKETLKGILEIID 133
Query: 111 SFSK---------------------------------------HRLLGVREEC------- 124
F++ H L ++ C
Sbjct: 134 DFNEISAEIKLTEDEKYEPCSKKKHAIWPNNMMIPVGEYENNTHTALKHKDTCCYLRYKV 193
Query: 125 ----RIREQDFGNFQVQERINALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRD 180
RIREQDFGN+Q +N + E R+ +G FF+RFP GESAADV+DRV+SF ESL+R
Sbjct: 194 KDDPRIREQDFGNYQNVSSMNDVMEERKNYGHFFYRFPQGESAADVYDRVASFQESLFRK 253
Query: 181 ID-MNRLNHDPSNDLNLIIVSHGLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQL 236
+ NH D+ +++V+HG+ +R+FLMKWF+WT E+++ N NG VM+L
Sbjct: 254 FERRTEANH---RDV-VVLVTHGIFARIFLMKWFRWTYEEYESFINVPNGCLIVMEL 306
>UniRef100_Q99288 S.cerevisiae chromosome IV reading frame ORF YDR051c [Saccharomyces
cerevisiae]
Length = 334
Score = 141 bits (355), Expect = 2e-32
Identities = 98/310 (31%), Positives = 137/310 (43%), Gaps = 87/310 (28%)
Query: 11 DNNNHHQQQIPNQNNHKPSVLPKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQA 70
+ N H + + + + P+ I+L+RHGES+ N + P+H ISLT G QA
Sbjct: 3 EENVHVSEDVAGSHGSFTNARPRLIVLIRHGESESNKNKEVNGYIPNHLISLTKTGQIQA 62
Query: 71 RIAGARIRHVIS-------------------------------STSSTDWRVYFYVSPYA 99
R AG + V++ + D + FY SPY
Sbjct: 63 RQAGIDLLRVLNVDDHNLVEDLAKKYIKDESSRRTLPLKDYTRLSREKDTNIVFYTSPYR 122
Query: 100 RTRSTLREIGRSFSKHRLL--GVR------------------------------------ 121
R R TL+ I ++ L GVR
Sbjct: 123 RARETLKGILDVIDEYNELNSGVRICEDMRYDPHGKQKHAFWPRGLNNTGGVYENNEDNI 182
Query: 122 ---------------EECRIREQDFGNFQVQERINALKETRQRFGRFFFRFPDGESAADV 166
+E RIREQDFGNFQ + + + R +G FFFRFP GESAADV
Sbjct: 183 CEGKPGKCYLQYRVKDEPRIREQDFGNFQKINSMQDVMKKRSTYGHFFFRFPHGESAADV 242
Query: 167 FDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFLMKWFKWTVEQFDLLNNF 226
+DRV+SF E+L+R +R P + +++V+HG+ SRVFLMKWF+WT E+F+ N
Sbjct: 243 YDRVASFQETLFRHFH-DRQERRPRD--VVVLVTHGIYSRVFLMKWFRWTYEEFESFTNV 299
Query: 227 GNGEFRVMQL 236
NG VM+L
Sbjct: 300 PNGSVMVMEL 309
>UniRef100_Q6BMB7 Similar to CA5949|IPF3498 Candida albicans IPF3498 [Debaryomyces
hansenii]
Length = 271
Score = 126 bits (317), Expect = 6e-28
Identities = 71/181 (39%), Positives = 106/181 (58%), Gaps = 15/181 (8%)
Query: 91 VYFYVSPYARTRSTLREI--GRSFSKHRLLGVREECRIREQDFGNFQ-VQERINALKETR 147
+ FY SPY R R T I G + + V EE R+REQDFGNFQ E++ + + R
Sbjct: 1 ILFYTSPYLRARQTCNNIIDGIKYVPGVMYKVHEEPRMREQDFGNFQSTSEQMEKIWQER 60
Query: 148 QRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRV 207
+G FF+R P GESAADV+DR++SF E+L+R + + LI+V+HG+ +RV
Sbjct: 61 AHYGHFFYRIPHGESAADVYDRIASFNETLFRQFQQDNFPNI------LILVTHGIWARV 114
Query: 208 FLMKWFKWTVEQFDLLNNFGNGEFRVMQLGS-GGEYSLAVHHTDREMHEWGLSPDMIADQ 266
FLMKWF+W+ E+F+ L N + ++ +M+ S GG++ L + W PD D+
Sbjct: 115 FLMKWFRWSYEEFESLRNIPHCQYLIMKKQSEGGKFRLKT-----PLKTWDDLPDSDIDE 169
Query: 267 K 267
+
Sbjct: 170 E 170
>UniRef100_Q6CJG6 Similar to sgd|S0002458 Saccharomyces cerevisiae YDR051c
[Kluyveromyces lactis]
Length = 313
Score = 122 bits (307), Expect = 9e-27
Identities = 94/300 (31%), Positives = 129/300 (42%), Gaps = 101/300 (33%)
Query: 32 PKRIILVRHGESQGNLDPSAYTTTPDHKISLTPQGITQARIAGARIRHV----------- 80
P+ I+L+RHGES+ N D TP+H I LT G QAR+AG I +
Sbjct: 3 PRLILLIRHGESESNKDKKVNEHTPNHLIPLTKNGRYQARMAGIEILKLLNVDDDSITKD 62
Query: 81 --------------------ISSTSSTDWRVYFYVSPYARTRSTLREIGRSFSKHRLL-- 118
+ ST TD + FY SPY RTR TL+ I ++ L
Sbjct: 63 IEAEEDYVEDRPQLEQNYQRVKSTKDTD--LVFYTSPYRRTRETLKGIIEMIDRYNELNC 120
Query: 119 GVRE-----------------ECRI-----------------------------REQDFG 132
G+R +C + E+ F
Sbjct: 121 GIRVCEGETYQPLGTRRLAHWQCPVFPSAVFENNESVTNFKLINNTETNPSLIHEEKHFL 180
Query: 133 NFQVQER----------------INALKETRQRFGRFFFRFPDGESAADVFDRVSSFLES 176
+++V+E + + TR +G FFFRFP GESAADV+DR S F ES
Sbjct: 181 HYRVKEEPRIREQDFGNFQKTASMTEVMNTRANYGHFFFRFPQGESAADVYDRCSGFQES 240
Query: 177 LWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQL 236
L+R + + D+ + IV+HG+ RVFLMKWF+WT E+F+ L N NG VM+L
Sbjct: 241 LFRHFEK---SGGKKRDV-VAIVTHGIFLRVFLMKWFRWTYEEFESLTNVPNGSVIVMEL 296
>UniRef100_UPI000042C7BD UPI000042C7BD UniRef100 entry
Length = 240
Score = 120 bits (301), Expect = 5e-26
Identities = 68/146 (46%), Positives = 94/146 (63%), Gaps = 9/146 (6%)
Query: 93 FYVSPYARTRSTLREIGRSFSKHRLLG--VREECRIREQDFGNFQVQ-ERINALKETRQR 149
F SPY RTR T I + VREE R+REQDFGNFQ + + + + R +
Sbjct: 3 FITSPYLRTRQTCNNIIEGIKNVPGVEYKVREEPRMREQDFGNFQPSSDEMELIWKERAQ 62
Query: 150 FGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFL 209
+G FF+R P GESAADV+DRV+SF+ESL+R R + P+ L++VSHG+ SRVF+
Sbjct: 63 YGHFFYRIPHGESAADVYDRVASFMESLFRQF---RSDDFPNI---LVLVSHGIWSRVFI 116
Query: 210 MKWFKWTVEQFDLLNNFGNGEFRVMQ 235
MKWFKW+ E+F+ L N + ++ VM+
Sbjct: 117 MKWFKWSYEEFESLKNVPHCKYLVMK 142
>UniRef100_Q8L667 Hypothetical protein OSJNBb0091B12.12 [Oryza sativa]
Length = 188
Score = 68.2 bits (165), Expect = 3e-10
Identities = 35/86 (40%), Positives = 50/86 (57%), Gaps = 7/86 (8%)
Query: 183 MNRLNHDPSNDLNLIIVSHGLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEY 242
M R++ ND+ + V + M+W+KW V QF+ L N N VMQ G+GG Y
Sbjct: 96 MMRVDRLRQNDVTFLTV-------LSAMRWYKWIVTQFEGLANLSNDGALVMQTGAGGRY 148
Query: 243 SLAVHHTDREMHEWGLSPDMIADQKW 268
SL VHH+ E+ +GL+ DMI DQ++
Sbjct: 149 SLLVHHSVEELRVFGLTDDMIEDQRF 174
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.135 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 562,954,148
Number of Sequences: 2790947
Number of extensions: 23930143
Number of successful extensions: 198347
Number of sequences better than 10.0: 937
Number of HSP's better than 10.0 without gapping: 568
Number of HSP's successfully gapped in prelim test: 380
Number of HSP's that attempted gapping in prelim test: 153294
Number of HSP's gapped (non-prelim): 22738
length of query: 306
length of database: 848,049,833
effective HSP length: 127
effective length of query: 179
effective length of database: 493,599,564
effective search space: 88354321956
effective search space used: 88354321956
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0544.10


