

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.1
(309 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
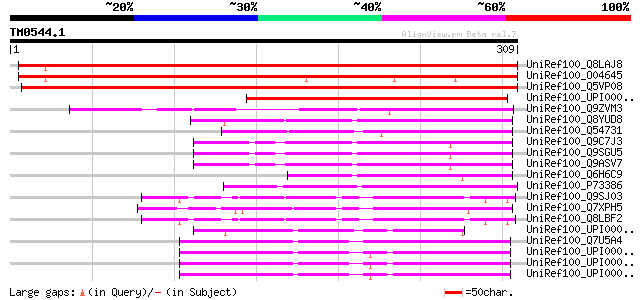
UniRef100_Q8LAJ8 Hypothetical protein [Arabidopsis thaliana] 446 e-124
UniRef100_O04645 A_TM021B04.11 protein [Arabidopsis thaliana] 405 e-112
UniRef100_Q5VP08 ATP-dependent Zn proteases-like protein [Oryza ... 382 e-105
UniRef100_UPI0000276473 UPI0000276473 UniRef100 entry 199 6e-50
UniRef100_Q9ZVM3 T22H22.11 protein [Arabidopsis thaliana] 148 2e-34
UniRef100_Q8YUD8 Alr2414 protein [Anabaena sp.] 112 1e-23
UniRef100_Q54731 Orf5; Method: conceptual translation supplied b... 91 5e-17
UniRef100_Q9C7J3 Hypothetical protein F14G9.20 [Arabidopsis thal... 88 3e-16
UniRef100_Q9SGU5 T6H22.2 protein [Arabidopsis thaliana] 88 3e-16
UniRef100_Q9ASV7 At1g56180/F14G9_20 [Arabidopsis thaliana] 88 3e-16
UniRef100_Q6H6C9 Hypothetical protein P0669G09.17 [Oryza sativa] 76 1e-12
UniRef100_P73386 Sll1738 protein [Synechocystis sp.] 70 5e-11
UniRef100_Q9SJ03 Expressed protein [Arabidopsis thaliana] 64 7e-09
UniRef100_Q7XPH5 OSJNBb0003B01.8 protein [Oryza sativa] 61 3e-08
UniRef100_Q8LBF2 Hypothetical protein [Arabidopsis thaliana] 61 3e-08
UniRef100_UPI0000254F1B UPI0000254F1B UniRef100 entry 54 5e-06
UniRef100_Q7U5A4 Hypothetical protein [Synechococcus sp.] 54 7e-06
UniRef100_UPI00002F471A UPI00002F471A UniRef100 entry 53 9e-06
UniRef100_UPI00002A5C78 UPI00002A5C78 UniRef100 entry 53 9e-06
UniRef100_UPI0000310B3E UPI0000310B3E UniRef100 entry 53 1e-05
>UniRef100_Q8LAJ8 Hypothetical protein [Arabidopsis thaliana]
Length = 341
Score = 446 bits (1147), Expect = e-124
Identities = 220/311 (70%), Positives = 263/311 (83%), Gaps = 7/311 (2%)
Query: 6 LMHCVGVTSCPGQLK------LVRVRIQCSTTNVGVS-RRQLLEKLDKELLKGDDRAALA 58
L+H VG+ C Q K R R ++ G+S RRQ LE++D +L GD+RAAL+
Sbjct: 4 LLHSVGLIPCSNQQKSFLFHSYYRYRCIVCSSETGLSIRRQALEQVDSKLSSGDERAALS 63
Query: 59 LVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAA 118
LVKDLQGKPDGLRCFGAARQVPQRLYTL+ELKLNGI A SLLSP D+TLGSIER LQIAA
Sbjct: 64 LVKDLQGKPDGLRCFGAARQVPQRLYTLEELKLNGINAASLLSPTDTTLGSIERNLQIAA 123
Query: 119 IAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRV 178
++GG+ AW AF +S QQLF+++LG +FLWTLD VS+ GG+G+LV+DT GH+FS++YHNRV
Sbjct: 124 VSGGIVAWKAFDLSSQQLFFLTLGFMFLWTLDLVSFNGGIGSLVLDTTGHTFSQRYHNRV 183
Query: 179 IQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAK 238
+QHEAGHFL+AYLV ILP+GYTLSSL+AL+K+GSLN+QAGSAFVD+EFLEEVN+GKVSA
Sbjct: 184 VQHEAGHFLVAYLVEILPRGYTLSSLEALQKEGSLNIQAGSAFVDYEFLEEVNSGKVSAT 243
Query: 239 TLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVL 298
LN+FSCIALAGV TEYL+YG++EGGLDDI KLD L+ LGFTQKK DSQVRWSVLNT+L
Sbjct: 244 MLNRFSCIALAGVATEYLLYGYAEGGLDDISKLDGLVKSLGFTQKKADSQVRWSVLNTIL 303
Query: 299 LLRRHEAARAK 309
LLRRHE AR+K
Sbjct: 304 LLRRHEIARSK 314
>UniRef100_O04645 A_TM021B04.11 protein [Arabidopsis thaliana]
Length = 386
Score = 405 bits (1041), Expect = e-112
Identities = 215/356 (60%), Positives = 258/356 (72%), Gaps = 52/356 (14%)
Query: 6 LMHCVGVTSCPGQLK------LVRVRIQCSTTNVGVS-RRQLLEKLDKELLKGDDRAALA 58
L+H VG+ C Q K R R ++ G+S RRQ LE++D +L GD+RAAL+
Sbjct: 4 LLHSVGLIPCSNQQKSFLFHSYYRYRCIVCSSETGLSIRRQALEQVDSKLSSGDERAALS 63
Query: 59 LVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAA 118
LVKDLQGKPDGLRCFGAARQVPQRLYTL+ELKLNGI A SLLSP D+TLGSIER LQIAA
Sbjct: 64 LVKDLQGKPDGLRCFGAARQVPQRLYTLEELKLNGINAASLLSPTDTTLGSIERNLQIAA 123
Query: 119 IAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRV 178
++GG+ AW AF +S QQLF+++LG +FLWTLD VS+ GG+G+LV+DT GH+FS++YHNRV
Sbjct: 124 VSGGIVAWKAFDLSSQQLFFLTLGFMFLWTLDLVSFNGGIGSLVLDTTGHTFSQRYHNRV 183
Query: 179 I----------------QHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFV 222
+ QHEAGHFL+AYLVGILP+GYTLSSL+AL+K+GSLN+QAGSAFV
Sbjct: 184 VQKHYIIFHWTYCELRSQHEAGHFLVAYLVGILPRGYTLSSLEALQKEGSLNIQAGSAFV 243
Query: 223 DFEFLEEVNAG---KVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRK--------- 270
D+EFLEE N + LN+FSCIALAGV TEYL+YG++EGGLDDI K
Sbjct: 244 DYEFLEEPNKKLCLLFQNQMLNRFSCIALAGVATEYLLYGYAEGGLDDISKVSFLLPLKN 303
Query: 271 -----------------LDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAARAK 309
LD L+ LGFTQKK DSQVRWSVLNT+LLLRRHE AR+K
Sbjct: 304 SSDYVNMLYGFVVLMEQLDGLVKSLGFTQKKADSQVRWSVLNTILLLRRHEIARSK 359
>UniRef100_Q5VP08 ATP-dependent Zn proteases-like protein [Oryza sativa]
Length = 346
Score = 382 bits (980), Expect = e-105
Identities = 195/303 (64%), Positives = 242/303 (79%), Gaps = 1/303 (0%)
Query: 8 HCVGVTSCPGQLKLVRVRIQCSTTNVGVSR-RQLLEKLDKELLKGDDRAALALVKDLQGK 66
H G + +L+L+ +T++ +R R +LE++D+EL KG+D AAL+LV+ QG
Sbjct: 17 HPAGGVAAAVRLRLLPPARAANTSSEPAARLRAVLEQVDEELRKGNDEAALSLVRGSQGA 76
Query: 67 PDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAAIAGGLTAW 126
GLR FGAARQVPQRLYTLDELKLNGI+ + LSPVD TLGSIER LQIAA+ GGL+
Sbjct: 77 DGGLRFFGAARQVPQRLYTLDELKLNGIDTSAFLSPVDLTLGSIERNLQIAAVLGGLSVS 136
Query: 127 NAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHF 186
AF +S Q+ ++ LGLL LW++D V +GGG+ NL++DTIGH+ S+KY NRVIQHEAGHF
Sbjct: 137 AAFELSKLQVLFLFLGLLSLWSVDLVYFGGGVRNLILDTIGHNLSQKYRNRVIQHEAGHF 196
Query: 187 LIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCI 246
LIAYL+G+LPKGYT++SLD K GSLNVQAG+AFVDFEFL+EVN+GK+SA LNKFSCI
Sbjct: 197 LIAYLLGVLPKGYTITSLDTFIKKGSLNVQAGTAFVDFEFLQEVNSGKLSATMLNKFSCI 256
Query: 247 ALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAA 306
ALAGV TEYL+YG++EGGL DI +LD LL GLGFTQKK DSQVRW+VLNTV LRRH+ A
Sbjct: 257 ALAGVATEYLLYGYAEGGLADIGQLDGLLKGLGFTQKKADSQVRWAVLNTVPALRRHKKA 316
Query: 307 RAK 309
R++
Sbjct: 317 RSQ 319
>UniRef100_UPI0000276473 UPI0000276473 UniRef100 entry
Length = 213
Score = 199 bits (507), Expect = 6e-50
Identities = 92/159 (57%), Positives = 121/159 (75%)
Query: 145 FLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSL 204
F D +++GGG+ L +DT+ + S +Y R+ +HEA HFL AYLVGILPKGYTLSSL
Sbjct: 17 FCSAYDQIAFGGGVSALALDTVAQATSNEYVTRLRRHEAAHFLTAYLVGILPKGYTLSSL 76
Query: 205 DALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGG 264
DA K G+ N+QAG AF D EF EV GK+++ +L++F+C+A+AG+C EY+++GF+EGG
Sbjct: 77 DAFKTYGAFNIQAGCAFCDGEFQREVQQGKITSTSLDRFACVAMAGICMEYILFGFAEGG 136
Query: 265 LDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRH 303
L D+R+LD LL L FTQKK DSQVRW+VLNT LLRRH
Sbjct: 137 LSDVRQLDGLLQALAFTQKKSDSQVRWAVLNTTTLLRRH 175
>UniRef100_Q9ZVM3 T22H22.11 protein [Arabidopsis thaliana]
Length = 289
Score = 148 bits (373), Expect = 2e-34
Identities = 96/286 (33%), Positives = 152/286 (52%), Gaps = 63/286 (22%)
Query: 37 RRQLLEKLDKELLKGDDRAALALVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEA 96
RR+ LE++DKELL+G+ AL+LVK L+ K L FG+A+ +P+ KL+
Sbjct: 23 RRKSLERVDKELLRGNYETALSLVKQLKSKHGCLSAFGSAKLLPK--------KLDMSSK 74
Query: 97 ESLLSPVDSTLGSIERTLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGG 156
L S +DS SIE ++ + + + SP++ ++
Sbjct: 75 TDLRSLIDSVSRSIE-SVYVQEDSVRTSKEMEIKTSPEEDWF------------------ 115
Query: 157 GLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQ 216
V+QHE+GHFL+ YL+G+LP+ Y + +L+A++++ S NV
Sbjct: 116 --------------------SVVQHESGHFLVGYLLGVLPRHYEIPTLEAVRQNVS-NVT 154
Query: 217 AGSAFVDFEFLEEV---------------NAGKVSAKTLNKFSCIALAGVCTEYLIYGFS 261
FV FEFL++V N G +S+KTLN FSC+ L G+ TE++++G+S
Sbjct: 155 GRVEFVGFEFLKQVGAANQLMKDDVDGQMNQGNISSKTLNNFSCVILGGMVTEHILFGYS 214
Query: 262 EGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAAR 307
EG DI KL+ +L LGFT+ + ++ ++W+V NTV LL H+ AR
Sbjct: 215 EGLYSDIVKLNDVLRWLGFTESEKEAHIKWAVSNTVSLLHSHKEAR 260
>UniRef100_Q8YUD8 Alr2414 protein [Anabaena sp.]
Length = 228
Score = 112 bits (280), Expect = 1e-23
Identities = 68/201 (33%), Positives = 112/201 (54%), Gaps = 8/201 (3%)
Query: 111 ERTLQIAAIAGGLTAWNAF-----GISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDT 165
+ L + AI+ L +A +SP + +L + T D+ S G G +++D
Sbjct: 3 QTALNLVAISVFLITMSALLGPLINLSPAIPAIATFTILGIATFDSFSLQGKGGTILLDW 62
Query: 166 IGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFE 225
I FS ++ +R+I HEAGHFL+AYL+G+ GYTLS+ +A ++ L Q G F D E
Sbjct: 63 IA-GFSPQHRDRIIHHEAGHFLVAYLLGVPVTGYTLSAWEAWRQ--GLPGQGGVTFDDVE 119
Query: 226 FLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKV 285
+ +V GK+S + L ++ I +AG+ E L++ +EGG+DD KL ++ LGF++
Sbjct: 120 LMSQVQQGKISNQVLERYCTICMAGIAAETLVFERAEGGIDDKSKLATIFKVLGFSESVC 179
Query: 286 DSQVRWSVLNTVLLLRRHEAA 306
+ R+ VL LL+ + A+
Sbjct: 180 QQKQRFHVLQAKTLLQNNWAS 200
>UniRef100_Q54731 Orf5; Method: conceptual translation supplied by author
[Synechococcus sp.]
Length = 233
Score = 90.5 bits (223), Expect = 5e-17
Identities = 59/180 (32%), Positives = 92/180 (50%), Gaps = 9/180 (5%)
Query: 130 GISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIA 189
G SP + LL L++LD V++ G L++D I S +Y R++ HEAGH+L+A
Sbjct: 27 GSSPLLPAGLGFSLLVLFSLDAVTWQGRGATLLLDGIQQR-SPEYRQRILHHEAGHYLVA 85
Query: 190 YLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFE---FLEEVNAGKVSAKTLNKFSCI 246
+G+ GYTLS+ +AL++ Q G V F+ E G++S ++L ++ +
Sbjct: 86 TALGLPVTGYTLSAWEALRQG-----QPGRGGVQFQAAALEAEAAQGQLSQRSLEQWCQV 140
Query: 247 ALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAA 306
+AG E L+YG EGG DD + L L + D + RW +L LL + A
Sbjct: 141 LMAGAAAEQLVYGNVEGGADDRAQWKQLWRQLDRNPAEADLRSRWGLLRAKTLLEQQRPA 200
>UniRef100_Q9C7J3 Hypothetical protein F14G9.20 [Arabidopsis thaliana]
Length = 389
Score = 87.8 bits (216), Expect = 3e-16
Identities = 60/196 (30%), Positives = 106/196 (53%), Gaps = 12/196 (6%)
Query: 113 TLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSK 172
++ +AA+ GG++ + I + + LGL +L D+V GG V +
Sbjct: 175 SIALAALLGGVSYLLSQEIDVRPNLAVILGLAYL---DSVFLGGTCLAQV-----SCYWP 226
Query: 173 KYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNA 232
+ R++ HEAGH L+AYL+G +G L + A++ + QAG+ F D + E+
Sbjct: 227 PHKRRIVVHEAGHLLVAYLMGCPIRGVILDPVVAMQM--GVQGQAGTQFWDQKMESEIAE 284
Query: 233 GKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDD--IRKLDSLLSGLGFTQKKVDSQVR 290
G++S + +++S + AG+ E L+YG +EGG +D + + S+L + ++ +Q R
Sbjct: 285 GRLSGSSFDRYSMVLFAGIAAEALVYGEAEGGENDENLFRSISVLLEPPLSVAQMSNQAR 344
Query: 291 WSVLNTVLLLRRHEAA 306
WSVL + LL+ H+AA
Sbjct: 345 WSVLQSYNLLKWHKAA 360
>UniRef100_Q9SGU5 T6H22.2 protein [Arabidopsis thaliana]
Length = 368
Score = 87.8 bits (216), Expect = 3e-16
Identities = 60/196 (30%), Positives = 106/196 (53%), Gaps = 12/196 (6%)
Query: 113 TLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSK 172
++ +AA+ GG++ + I + + LGL +L D+V GG V +
Sbjct: 154 SIALAALLGGVSYLLSQEIDVRPNLAVILGLAYL---DSVFLGGTCLAQV-----SCYWP 205
Query: 173 KYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNA 232
+ R++ HEAGH L+AYL+G +G L + A++ + QAG+ F D + E+
Sbjct: 206 PHKRRIVVHEAGHLLVAYLMGCPIRGVILDPVVAMQM--GVQGQAGTQFWDQKMESEIAE 263
Query: 233 GKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDD--IRKLDSLLSGLGFTQKKVDSQVR 290
G++S + +++S + AG+ E L+YG +EGG +D + + S+L + ++ +Q R
Sbjct: 264 GRLSGSSFDRYSMVLFAGIAAEALVYGEAEGGENDENLFRSISVLLEPPLSVAQMSNQAR 323
Query: 291 WSVLNTVLLLRRHEAA 306
WSVL + LL+ H+AA
Sbjct: 324 WSVLQSYNLLKWHKAA 339
>UniRef100_Q9ASV7 At1g56180/F14G9_20 [Arabidopsis thaliana]
Length = 389
Score = 87.8 bits (216), Expect = 3e-16
Identities = 60/196 (30%), Positives = 106/196 (53%), Gaps = 12/196 (6%)
Query: 113 TLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSK 172
++ +AA+ GG++ + I + + LGL +L D+V GG V +
Sbjct: 175 SIALAALLGGVSYLLSQEIDVRPNLAVILGLAYL---DSVFLGGPCLAQV-----SCYWP 226
Query: 173 KYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNA 232
+ R++ HEAGH L+AYL+G +G L + A++ + QAG+ F D + E+
Sbjct: 227 PHKRRIVVHEAGHLLVAYLMGCPIRGVILDPVVAMQM--GVQGQAGTQFWDQKMESEIAE 284
Query: 233 GKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDD--IRKLDSLLSGLGFTQKKVDSQVR 290
G++S + +++S + AG+ E L+YG +EGG +D + + S+L + ++ +Q R
Sbjct: 285 GRLSGSSFDRYSMVLFAGIAAEALVYGEAEGGENDENLFRSISVLLEPPLSVAQMSNQAR 344
Query: 291 WSVLNTVLLLRRHEAA 306
WSVL + LL+ H+AA
Sbjct: 345 WSVLQSYNLLKWHKAA 360
>UniRef100_Q6H6C9 Hypothetical protein P0669G09.17 [Oryza sativa]
Length = 374
Score = 76.3 bits (186), Expect = 1e-12
Identities = 47/139 (33%), Positives = 75/139 (53%), Gaps = 4/139 (2%)
Query: 170 FSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEE 229
F Y R++ HEAGH L AYL+G +G L AL+ + QAG+ F D + +E
Sbjct: 209 FWPPYKRRILVHEAGHLLTAYLMGCPIRGVILDPFVALRM--GIQGQAGTQFWDEKMEKE 266
Query: 230 VNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSL--LSGLGFTQKKVDS 287
+ G +S+ +++ I AG+ E L+YG +EGG +D SL L + ++ +
Sbjct: 267 LAEGHLSSTAFDRYCMILFAGIAAEALVYGEAEGGENDENLFRSLCILLDPPLSVAQMAN 326
Query: 288 QVRWSVLNTVLLLRRHEAA 306
+ RWSV+ + LL+ H+ A
Sbjct: 327 RARWSVMQSYNLLKWHKKA 345
>UniRef100_P73386 Sll1738 protein [Synechocystis sp.]
Length = 231
Score = 70.5 bits (171), Expect = 5e-11
Identities = 49/179 (27%), Positives = 85/179 (47%), Gaps = 5/179 (2%)
Query: 131 ISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAY 190
+SP +LG+L + T D +S+ G + V G +K R++ HEAGHFL+A+
Sbjct: 28 LSPAIPAAATLGILGIITADQISWQGKGTDFFV---GLFQTKAEKERILCHEAGHFLVAH 84
Query: 191 LVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAG 250
+ I Y+LS + L++ AG F + + + L +++ + +AG
Sbjct: 85 CLQIPITNYSLSPWEVLRQGAG--GMAGIQFDTTNLENQCRDWHLRPQALERWATVWMAG 142
Query: 251 VCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAARAK 309
+ E +IYG S+GG D ++L G + K+ + W+ L LL +H A +
Sbjct: 143 IAAEKIIYGESQGGNGDRQQLRQAFRRAGLPEIKLQQKESWAFLQAKNLLEQHRQAHGQ 201
>UniRef100_Q9SJ03 Expressed protein [Arabidopsis thaliana]
Length = 332
Score = 63.5 bits (153), Expect = 7e-09
Identities = 73/247 (29%), Positives = 115/247 (46%), Gaps = 44/247 (17%)
Query: 81 QRLYTLDELKLNGIE-AESLLSP--------VDSTLGSIERTLQIAAIAGGLTA-WNAFG 130
+R +L EL GI+ AE+L P + + +GS T IA +AG L W F
Sbjct: 92 RRTTSLRELTTLGIKNAETLAIPSVRNDAAFLFTVVGS---TGFIAVLAGQLPGDWGFF- 147
Query: 131 ISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAY 190
+ Y+ +G + L L S GL + +F Y R+ HEA HFL+AY
Sbjct: 148 -----VPYL-VGSISLVVLAVGSVSPGLLQAAISGFS-TFFPDYQERIAAHEAAHFLVAY 200
Query: 191 LVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAG 250
L+G+ GY SLD K+ +L +D + + +GK+ +K L++ + +A+AG
Sbjct: 201 LIGLPILGY---SLDIGKEHVNL--------IDERLAKLIYSGKLDSKELDRLAAVAMAG 249
Query: 251 VCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQ-----VRWSVLNTVLLLRR--- 302
+ E L Y G D+ L ++ +Q K+ ++ RW+VL + LL+
Sbjct: 250 LAAEGLKYDKVIGQSADLFSLQRFINR---SQPKISNEQQQNLTRWAVLYSASLLKNNKT 306
Query: 303 -HEAARA 308
HEA A
Sbjct: 307 IHEALMA 313
>UniRef100_Q7XPH5 OSJNBb0003B01.8 protein [Oryza sativa]
Length = 325
Score = 61.2 bits (147), Expect = 3e-08
Identities = 64/238 (26%), Positives = 109/238 (44%), Gaps = 30/238 (12%)
Query: 79 VPQRLYTLDELKLNGIE-AESLLSPVDSTLGSIERTLQIAAIAGGLTAWNAFGISPQQL- 136
V +R +L EL GI+ AE+L P S+ G T + G+ QL
Sbjct: 83 VSRRTTSLRELTTLGIKNAENLAIP------SVRNDAAFLFTVVGSTGF--LGVLAGQLP 134
Query: 137 ----FYIS--LGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAY 190
F++ +G + L L S GL + F Y R+ +HEA HFL+AY
Sbjct: 135 GDWGFFVPYLIGSISLIVLAIGSISPGLLQAAIGAFSTVFPD-YQERIARHEAAHFLVAY 193
Query: 191 LVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAG 250
L+G+ GY SLD K+ +L +D + + + +G++ K +++ + +++AG
Sbjct: 194 LIGLPILGY---SLDIGKEHVNL--------IDDQLQKLIYSGQLDQKEIDRLAVVSMAG 242
Query: 251 VCTEYLIYGFSEGGLDDIRKLDSLLSGL--GFTQKKVDSQVRWSVLNTVLLLRRHEAA 306
+ E L Y G D+ L ++ T+ + + RW+VL + LL+ ++AA
Sbjct: 243 LAAEGLEYDKVVGQSADLFTLQRFINRTKPPLTKDQQQNLTRWAVLFSASLLKNNKAA 300
>UniRef100_Q8LBF2 Hypothetical protein [Arabidopsis thaliana]
Length = 332
Score = 61.2 bits (147), Expect = 3e-08
Identities = 72/247 (29%), Positives = 114/247 (46%), Gaps = 44/247 (17%)
Query: 81 QRLYTLDELKLNGIE-AESLLSP--------VDSTLGSIERTLQIAAIAGGLTA-WNAFG 130
+R +L EL GI+ AE+L P + + +GS T IA +AG L W F
Sbjct: 92 RRTTSLRELTTLGIKNAETLAIPSVRNDAAFLFTVVGS---TGFIAVLAGQLPGDWGFF- 147
Query: 131 ISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAY 190
+ Y+ +G + L L S GL + +F Y R+ HEA HFL+AY
Sbjct: 148 -----VPYL-VGSISLVVLAMGSVSPGLLQAAISGFS-TFFPDYQERIAAHEAAHFLVAY 200
Query: 191 LVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAG 250
L+G+ GY SLD K+ +L +D + + +GK+ +K L++ + +A+AG
Sbjct: 201 LIGLPILGY---SLDIGKEHVNL--------IDERLAKLIYSGKLDSKELDRLAAVAMAG 249
Query: 251 VCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQ-----VRWSVLNTVLLLRR--- 302
+ E L Y G D+ L ++ +Q K+ ++ RW+ L + LL+
Sbjct: 250 LAAEGLKYDKVIGQSADLFSLQRFINR---SQPKISNEQQQNLTRWAXLYSASLLKNNKT 306
Query: 303 -HEAARA 308
HEA A
Sbjct: 307 IHEALMA 313
>UniRef100_UPI0000254F1B UPI0000254F1B UniRef100 entry
Length = 214
Score = 53.9 bits (128), Expect = 5e-06
Identities = 49/171 (28%), Positives = 78/171 (44%), Gaps = 16/171 (9%)
Query: 113 TLQIAAIAGGLTAWNAFG----ISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGH 168
TL+ A + +T AFG ISP + + G L ++D S+ G G+L+ + +
Sbjct: 10 TLRAAIVVAAITGLGAFGPVMGISPAWIVVVVGGGLVALSVDAASWQGMGGHLLAEAMPG 69
Query: 169 SFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLE 228
+ R+ HEAGH LIA + + + +L L+ S +F E
Sbjct: 70 GEGRL--RRIAVHEAGHLLIAEAEELPVQRVMVGTLACLRAG-----LRSSGATEFTVPE 122
Query: 229 EVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSL--LSG 277
V ++ + L ++S + AG+ E +IYG + GG DD L L LSG
Sbjct: 123 SV---RMPLEDLRRWSRVLQAGIAAETVIYGSARGGADDRALLGRLWGLSG 170
>UniRef100_Q7U5A4 Hypothetical protein [Synechococcus sp.]
Length = 215
Score = 53.5 bits (127), Expect = 7e-06
Identities = 50/202 (24%), Positives = 86/202 (41%), Gaps = 10/202 (4%)
Query: 104 DSTLGSIERTLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVV 163
+ + G++ L +AAI G G+SP + G L + ++D S+ G G+++
Sbjct: 6 EGSTGTLRAGLVVAAITGLGAFGPVLGLSPAWIVVAVGGGLVMLSVDAASWQGMGGHVLA 65
Query: 164 DTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVD 223
+ + + R+ HEAGH LIA + + + +L L QAG
Sbjct: 66 EALPGGEGRL--RRIAVHEAGHLLIAEQEQMPVQRVLVGTLACL--------QAGLRSRG 115
Query: 224 FEFLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQK 283
N+ ++ + L ++S + AG+ E ++YG + GG DD L L G
Sbjct: 116 ATEFPVPNSVRMPLEDLRRWSRVLQAGIAAETVVYGKARGGADDRALLGQLWGLSGHDVA 175
Query: 284 KVDSQVRWSVLNTVLLLRRHEA 305
+ R + LLRR +A
Sbjct: 176 TAQREQRRARREIEQLLRRDQA 197
>UniRef100_UPI00002F471A UPI00002F471A UniRef100 entry
Length = 215
Score = 53.1 bits (126), Expect = 9e-06
Identities = 52/205 (25%), Positives = 89/205 (43%), Gaps = 16/205 (7%)
Query: 104 DSTLGSIERTLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVV 163
+ + G++ L +AAI G G+SP + G L + ++D S+ G G+++
Sbjct: 6 EGSTGTLRAGLVVAAITGLGAFGPVLGLSPAWIVVAVGGGLVMLSVDAASWQGMGGHVLA 65
Query: 164 DTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAG---SA 220
+ + + R+ HEAGH LIA + + + +L L QAG S
Sbjct: 66 EALPGGEGRL--RRIAVHEAGHLLIAEQEQMPVQRVLVGTLACL--------QAGLRSSG 115
Query: 221 FVDFEFLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGF 280
+F + V ++ + L ++S + AG+ E ++YG + GG DD L L G
Sbjct: 116 ATEFPVPDSV---RMPLEDLRRWSRVLQAGIAAETVVYGKARGGADDRALLGQLWGLSGH 172
Query: 281 TQKKVDSQVRWSVLNTVLLLRRHEA 305
+ R + LLRR +A
Sbjct: 173 DVATAQREQRRARREIEQLLRRDQA 197
>UniRef100_UPI00002A5C78 UPI00002A5C78 UniRef100 entry
Length = 215
Score = 53.1 bits (126), Expect = 9e-06
Identities = 52/205 (25%), Positives = 89/205 (43%), Gaps = 16/205 (7%)
Query: 104 DSTLGSIERTLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVV 163
+ + G++ L +AAI G G+SP + G L + ++D S+ G G+++
Sbjct: 6 EGSTGTLRAGLVVAAITGLGAFGPVLGLSPAWIVVAVGGGLVMLSVDAASWQGMGGHVLA 65
Query: 164 DTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAG---SA 220
+ + + R+ HEAGH LIA + + + +L L QAG S
Sbjct: 66 EALPGGEGRL--RRIAVHEAGHLLIAEQEQMPVQRVLVGTLACL--------QAGLRSSG 115
Query: 221 FVDFEFLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGF 280
+F + V ++ + L ++S + AG+ E ++YG + GG DD L L G
Sbjct: 116 ATEFPVPDSV---RMPLEDLRRWSRVLQAGIAAETVVYGKARGGADDRALLGQLWGLSGH 172
Query: 281 TQKKVDSQVRWSVLNTVLLLRRHEA 305
+ R + LLRR +A
Sbjct: 173 DVATAQREQRRARREIEQLLRRDQA 197
>UniRef100_UPI0000310B3E UPI0000310B3E UniRef100 entry
Length = 215
Score = 52.8 bits (125), Expect = 1e-05
Identities = 52/205 (25%), Positives = 89/205 (43%), Gaps = 16/205 (7%)
Query: 104 DSTLGSIERTLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVV 163
+ + G++ L +AAI G G+SP + G L + ++D S+ G G+++
Sbjct: 6 EGSTGTLRAGLVVAAITGLGAFGPVLGLSPAWIVVAVGGGLVMLSVDAASWQGMGGHVLA 65
Query: 164 DTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAG---SA 220
+ + + R+ HEAGH LIA + + + +L L QAG S
Sbjct: 66 EALPGGEGRL--RRIAVHEAGHLLIAEQEQMPVQRVLVGTLACL--------QAGLRSSG 115
Query: 221 FVDFEFLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGF 280
+F + V ++ + L ++S + AG+ E ++YG + GG DD L L G
Sbjct: 116 ATEFPVPDSV---RMPLEDLRRWSRLLQAGIAAETVVYGKARGGADDRALLGQLWGLSGH 172
Query: 281 TQKKVDSQVRWSVLNTVLLLRRHEA 305
+ R + LLRR +A
Sbjct: 173 DVATAQREQRRARREIEQLLRRDQA 197
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.139 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 495,388,444
Number of Sequences: 2790947
Number of extensions: 20465240
Number of successful extensions: 49978
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 49923
Number of HSP's gapped (non-prelim): 46
length of query: 309
length of database: 848,049,833
effective HSP length: 127
effective length of query: 182
effective length of database: 493,599,564
effective search space: 89835120648
effective search space used: 89835120648
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0544.1


