

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0490.17
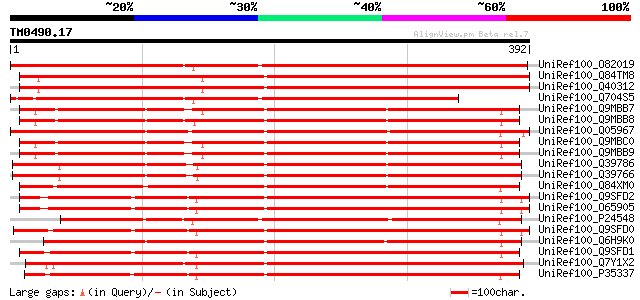
(392 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O82019 Polygalacturonase precursor [Medicago sativa] 547 e-154
UniRef100_Q84TM8 Polygalactorunase PG11 precursor [Medicago sativa] 512 e-144
UniRef100_Q40312 Polygalacturonase precursor [Medicago sativa] 494 e-138
UniRef100_Q704S5 Polygalacturonase precursor [Medicago truncatula] 452 e-126
UniRef100_Q9MBB7 Polygalacturonase [Salix gilgiana] 404 e-111
UniRef100_Q9MBB8 Polygalacturonase [Salix gilgiana] 402 e-111
UniRef100_Q05967 Polygalacturonase precursor [Nicotiana tabacum] 400 e-110
UniRef100_Q9MBC0 Polygalacturonase [Salix gilgiana] 399 e-110
UniRef100_Q9MBB9 Polygalacturonase [Salix gilgiana] 399 e-109
UniRef100_Q39786 Polygalacturonase precursor [Gossypium hirsutum] 378 e-103
UniRef100_Q39766 Polygalacturonase precursor [Gossypium barbadense] 377 e-103
UniRef100_Q84XM0 Putative pollen polygalacturonase [Turnera subu... 375 e-102
UniRef100_Q9SFD2 Polygalacturonase PGA3 [Arabidopsis thaliana] 373 e-102
UniRef100_O65905 Polygalacturonase [Arabidopsis thaliana] 373 e-102
UniRef100_P24548 Exopolygalacturonase [Oenothera organensis] 364 3e-99
UniRef100_Q9SFD0 Putative polygalacturonase [Arabidopsis thaliana] 363 4e-99
UniRef100_Q6H9K0 Exopolygalacturonase precursor [Platanus acerif... 362 1e-98
UniRef100_Q9SFD1 Putative polygalacturonase [Arabidopsis thaliana] 355 2e-96
UniRef100_Q7Y1X2 Polygalacturonase [Fragaria ananassa] 355 2e-96
UniRef100_P35337 Polygalacturonase precursor [Brassica napus] 355 2e-96
>UniRef100_O82019 Polygalacturonase precursor [Medicago sativa]
Length = 395
Score = 547 bits (1409), Expect = e-154
Identities = 259/393 (65%), Positives = 311/393 (78%), Gaps = 5/393 (1%)
Query: 1 MDMSLIRSTAIVLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKI 60
MD L +T ++ L G AQ +FDI++FGGA ++DIT AF NAWK AC ST+ SK+
Sbjct: 1 MDRKLSITTIVLSLLFATLGIAQPNIFDIKSFGGASHADITMAFTNAWKAACGSTTASKV 60
Query: 61 VISKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNG 120
+I + YKM+AV+VKGPCKAPIE+QVDGTIQAP +P+ LN A+QW+ GYVDYFTLSG G
Sbjct: 61 MIPRGVYKMHAVDVKGPCKAPIEVQVDGTIQAPQNPDELNDAYQWIKFGYVDYFTLSGKG 120
Query: 121 TFDGRGASAWKLQKACG--ATCSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNF 178
FDG G +AW+ Q CG +TC +RSMN GFNFLKHS VR +TSKDSK FHV +LGCTNF
Sbjct: 121 VFDGNGETAWR-QNDCGKNSTCKRRSMNFGFNFLKHSIVRDITSKDSKNFHVNVLGCTNF 179
Query: 179 TFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCG 238
TFD I AP SINTDGIHI +STDVK+LN+NIATGDDCVSLGDG++ IT+ NV CG
Sbjct: 180 TFDSVTITAP--VTSINTDGIHIGRSTDVKVLNTNIATGDDCVSLGDGSRQITVQNVNCG 237
Query: 239 PGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITM 298
PGHG S+GSLG P EE VE+++VKNCT+TNT NGVRIKTWP++PGTSP+T++ FEDI M
Sbjct: 238 PGHGISVGSLGKYPKEEAVEHVLVKNCTITNTDNGVRIKTWPSSPGTSPITDMHFEDIIM 297
Query: 299 VNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGV 358
VNV NPVIIDQ+YCP N CSK PSKIKIS V FKNI+GTS+TKDGV+L CS PC+ V
Sbjct: 298 VNVLNPVIIDQEYCPWNQCSKLVPSKIKISKVIFKNIRGTSKTKDGVVLICSKSFPCDAV 357
Query: 359 ELSDISLTFNGAPAMAKCANVKPIIIGKAPSCS 391
EL++++LTFNGAP AKC NVKPI+ G P C+
Sbjct: 358 ELNNVALTFNGAPVNAKCVNVKPILTGPTPKCT 390
>UniRef100_Q84TM8 Polygalactorunase PG11 precursor [Medicago sativa]
Length = 407
Score = 512 bits (1318), Expect = e-144
Identities = 246/392 (62%), Positives = 305/392 (77%), Gaps = 9/392 (2%)
Query: 8 STAIVLFLLLLSG--SAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVISKA 65
STAI++ L L+ + QS+V DI FGGAPNSDITQA NAWK+AC ST+ +KI+I
Sbjct: 4 STAIIVSFLFLANFCAVQSVVLDISKFGGAPNSDITQAIGNAWKEACLSTTAAKIMIPGG 63
Query: 66 TYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFDGR 125
TY+MN V +KGPCKAPIE+QVDGTIQAP DP+ + A QW+ Y+++ TLSG G FDG+
Sbjct: 64 TYRMNGVELKGPCKAPIEVQVDGTIQAPADPSQMKAADQWIKFLYMEHLTLSGKGVFDGQ 123
Query: 126 GASAWKLQKACGATCSKRS-----MNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTF 180
GA+ +K A K S MN+GF F+ +S V +TSKDSK FHV +LGC NFTF
Sbjct: 124 GATVYKKAAPAAAWSGKNSNVKIFMNLGFMFVNNSMVHGLTSKDSKNFHVMVLGCNNFTF 183
Query: 181 DGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPG 240
DGF + APGDS NTDGIH+ +STD+KILN+NIATGDDC+SLGDG++ IT+ NVKCGPG
Sbjct: 184 DGFTVTAPGDSP--NTDGIHMGRSTDIKILNTNIATGDDCISLGDGSRKITVQNVKCGPG 241
Query: 241 HGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVN 300
HG S+GSLG TE+ VE +IVKNCTLT TQNGVRIKTWP APGT V+++ FEDI M N
Sbjct: 242 HGISVGSLGKYTTEDNVEGVIVKNCTLTATQNGVRIKTWPDAPGTITVSDIHFEDIIMNN 301
Query: 301 VYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVEL 360
V NPVIIDQ+YCP N CSK++PSKIK+S ++FKNI+GTS T +GV+L CSSGVPC+GVEL
Sbjct: 302 VMNPVIIDQEYCPWNQCSKKNPSKIKLSKISFKNIQGTSGTPEGVVLICSSGVPCDGVEL 361
Query: 361 SDISLTFNGAPAMAKCANVKPIIIGKAPSCSA 392
+++ LTFNG PA+AKC+NVKP++ G AP+C A
Sbjct: 362 NNVGLTFNGKPAIAKCSNVKPLVTGTAPACQA 393
>UniRef100_Q40312 Polygalacturonase precursor [Medicago sativa]
Length = 421
Score = 494 bits (1271), Expect = e-138
Identities = 243/392 (61%), Positives = 291/392 (73%), Gaps = 9/392 (2%)
Query: 8 STAIVLFLLLLSG--SAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVISKA 65
STAI++ L ++ +AQS V DI FGG PNSDI QA +AW +ACAST+ +KIVI
Sbjct: 4 STAIIVSFLFIADFCAAQSGVLDISKFGGKPNSDIGQALTSAWNEACASTTAAKIVIPAG 63
Query: 66 TYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFDGR 125
TY++N + +KGPCKAPIE+QVDGTIQAP DP+ + G QW Y+D+ TLSG G FDG+
Sbjct: 64 TYQLNGIELKGPCKAPIELQVDGTIQAPADPSVIKGTEQWFKFLYMDHLTLSGKGVFDGQ 123
Query: 126 GASAWKLQKACGATCSKRS-----MNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTF 180
GA+ +K A K S MN GFNF+ +S VR VTSKDSK FHV + GC N TF
Sbjct: 124 GATVYKKAAPASAWSGKNSNSKVFMNFGFNFVNNSIVRGVTSKDSKNFHVMVFGCKNITF 183
Query: 181 DGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPG 240
DGF I APGDS NTDGIH+ KSTDVKILN+NI TGDDCVS+GDG+K IT+ V CGPG
Sbjct: 184 DGFTITAPGDSP--NTDGIHMGKSTDVKILNTNIGTGDDCVSIGDGSKQITVQGVNCGPG 241
Query: 241 HGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVN 300
HG S+GSLG TEE VE I VKNCTLT T NGVRIKTWP APGT V+++ FEDITM N
Sbjct: 242 HGLSVGSLGKFTTEENVEGITVKNCTLTATDNGVRIKTWPDAPGTITVSDIHFEDITMTN 301
Query: 301 VYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVEL 360
V NPVIIDQ+Y P N CSK++PSKIK+S ++FKN+KGTS T +GV+L CSS VPC+GVEL
Sbjct: 302 VKNPVIIDQEYYPWNQCSKKNPSKIKLSKISFKNVKGTSGTAEGVVLICSSAVPCDGVEL 361
Query: 361 SDISLTFNGAPAMAKCANVKPIIIGKAPSCSA 392
+++ L FNGAP AKC NVKP++ G AP C A
Sbjct: 362 NNVDLKFNGAPTTAKCTNVKPLVTGTAPVCQA 393
>UniRef100_Q704S5 Polygalacturonase precursor [Medicago truncatula]
Length = 343
Score = 452 bits (1163), Expect = e-126
Identities = 225/343 (65%), Positives = 269/343 (77%), Gaps = 9/343 (2%)
Query: 1 MDMSLIRSTAIVLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKI 60
MDM L TAIVL L G AQ +FDI++FGGA N+DIT AF NAWK AC ST+ SK+
Sbjct: 1 MDMKL-SITAIVLSFATL-GIAQPNIFDIKSFGGASNADITMAFTNAWKAACGSTTASKV 58
Query: 61 VISKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGA-FQWVTIGYVDYFTLSGN 119
+I + YK++AV+VKGPCKAPIE+QVDGTIQA +P+ LN A +QWV GYVDYFTLSG
Sbjct: 59 IIPRGIYKIHAVDVKGPCKAPIEVQVDGTIQASQNPDELNDASYQWVKFGYVDYFTLSGK 118
Query: 120 GTFDGRGASAWKLQKACG--ATCSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTN 177
G FDG G +AW Q CG +TC +RSMN GFNFLK S V+ + SKDSK FHV +LGCTN
Sbjct: 119 GVFDGNGETAWT-QNDCGKNSTCKRRSMNFGFNFLKLSIVQDIISKDSKNFHVNVLGCTN 177
Query: 178 FTFDGFKIIAPGDSISINTDGIHIAKSTD-VKILNSNIATGDDCVSLGDGNKHITIHNVK 236
FTFDG I AP + S NTDGIHI +STD VK+LN+NI+TGDDC+SLG G++ IT+ NV
Sbjct: 178 FTFDGLTITAP--ATSKNTDGIHIGRSTDDVKVLNTNISTGDDCISLGQGSRQITVQNVN 235
Query: 237 CGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDI 296
CGPGHG S+GSLG NP EE E+++VKNCT++NT NGVRIKTWP++PGTSP+T++ FED
Sbjct: 236 CGPGHGISVGSLGKNPKEEATEHVLVKNCTISNTDNGVRIKTWPSSPGTSPITDMHFEDT 295
Query: 297 TMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTS 339
MVNV NPVIIDQ+YCP N CSKQ PSKIKIS V FKNI+GT+
Sbjct: 296 IMVNVLNPVIIDQEYCPWNQCSKQVPSKIKISKVIFKNIRGTT 338
>UniRef100_Q9MBB7 Polygalacturonase [Salix gilgiana]
Length = 393
Score = 404 bits (1039), Expect = e-111
Identities = 210/391 (53%), Positives = 271/391 (68%), Gaps = 24/391 (6%)
Query: 8 STAIVLFLLLL----SGSAQSI-VFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVI 62
S+AI+ F LLL + AQS VFD+ +GG DIT+A NAWK ACAST+PSK+ I
Sbjct: 7 SSAIISFSLLLLLTSTAKAQSNGVFDVTKYGG--KEDITEALNNAWKDACASTNPSKVHI 64
Query: 63 SKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTF 122
TY + V + GPCKA IE+QVDG ++AP DPN +G WV +VD TLSG+GTF
Sbjct: 65 PSGTYSLRQVTLAGPCKAAIELQVDGILKAPVDPNQFSGG-HWVDFSHVDQLTLSGSGTF 123
Query: 123 DGRGASAWKLQKACGATCSKRS------MNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCT 176
DG+G+ AW +TCSK MNI F+F+ + VR +T++DSK FHV +L C
Sbjct: 124 DGQGSVAWSK-----STCSKDKNCEGFPMNIRFDFITNGFVRDITTRDSKNFHVNVLECK 178
Query: 177 NFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVK 236
N TF F + AP +SI NTDGIHI +ST + I++S I TGDDC+S+GDG + + + V
Sbjct: 179 NLTFQHFTVTAPAESI--NTDGIHIGRSTGIYIIDSKIGTGDDCISVGDGTEDLHVTGVT 236
Query: 237 CGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDI 296
CGPGHG S+GSLG P E+PV I VKNCT++NT NGVRIK+WP G +N+ FEDI
Sbjct: 237 CGPGHGISVGSLGRYPNEKPVSGIFVKNCTISNTANGVRIKSWPDLYG-GDASNMHFEDI 295
Query: 297 TMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCE 356
M NV NPV +DQ+YCP N CS ++PSK+KIS+V+FKNI+GTS T V L CSSG+PCE
Sbjct: 296 VMNNVQNPVAVDQEYCPWNQCSLKAPSKVKISDVSFKNIRGTSATPVVVKLACSSGIPCE 355
Query: 357 GVELSDISLTFNGA--PAMAKCANVKPIIIG 385
VEL+DI+L ++G+ PA ++C+NVKPII G
Sbjct: 356 KVELADINLVYSGSEGPAKSQCSNVKPIISG 386
>UniRef100_Q9MBB8 Polygalacturonase [Salix gilgiana]
Length = 393
Score = 402 bits (1033), Expect = e-111
Identities = 206/387 (53%), Positives = 269/387 (69%), Gaps = 16/387 (4%)
Query: 8 STAIVLFLLLL----SGSAQSI-VFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVI 62
S+AI+ F LLL + AQS V D+ + G DIT+A NAWK ACAST+PSK++I
Sbjct: 7 SSAIISFSLLLLLASTAKAQSNGVLDVTKYDG--KEDITEALNNAWKDACASTNPSKVLI 64
Query: 63 SKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTF 122
TY + V + GPCKA IE+QVDG ++AP +P+ +G WV +VD TLSG+GTF
Sbjct: 65 PSGTYSLRQVTLAGPCKAAIELQVDGILKAPVNPDQFSGG-SWVDFRHVDQLTLSGSGTF 123
Query: 123 DGRGASAWKLQKACGA--TCSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTF 180
DG+G AW + C C MNI F+F+ + VR +T++DSK FHV +LGC N TF
Sbjct: 124 DGQGNVAWS-KSTCNKDKNCEGLPMNIRFDFITNGLVRDITTRDSKNFHVNVLGCKNLTF 182
Query: 181 DGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPG 240
F + APG+SI NTDGIHI +ST + I++SNI TGDDC+S+GDG + + + V CGPG
Sbjct: 183 QHFTMTAPGESI--NTDGIHIGQSTGIYIIDSNIGTGDDCISVGDGTEELHVTGVTCGPG 240
Query: 241 HGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVN 300
HG S+GSLG P E+PV I VKNCT++NT NGVRIK+WP G +N+ FEDI M N
Sbjct: 241 HGISVGSLGRYPNEKPVSGIFVKNCTISNTANGVRIKSWPDLYG-GAASNMHFEDIVMNN 299
Query: 301 VYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVEL 360
V NPV++DQ+YCP N CS +SPSK+KIS+V+FKNI+GTS T V L CS G+PCE VEL
Sbjct: 300 VQNPVVVDQEYCPWNQCSLKSPSKVKISDVSFKNIRGTSATPVVVKLACSGGIPCEKVEL 359
Query: 361 SDISLTFNGA--PAMAKCANVKPIIIG 385
+DI+L ++G+ PA ++C+NVKP I G
Sbjct: 360 ADINLVYSGSEGPAKSQCSNVKPTISG 386
>UniRef100_Q05967 Polygalacturonase precursor [Nicotiana tabacum]
Length = 396
Score = 400 bits (1028), Expect = e-110
Identities = 199/397 (50%), Positives = 275/397 (69%), Gaps = 12/397 (3%)
Query: 1 MDMSLIRSTAIVLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKI 60
MD+ A+VL L G +Q+ VFDI +G N+DI++A LNA+K+AC STSPS I
Sbjct: 1 MDLKFKVHFALVLLFLAHFGESQTGVFDITKYGANSNADISEALLNAFKEACQSTSPSTI 60
Query: 61 VISKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNG 120
VI K T+ MN V ++GPCK+P+E+Q+ T++AP+DP+ L +W+T+ +D FT+SG G
Sbjct: 61 VIPKGTFTMNQVKLEGPCKSPLELQIQATLKAPSDPSQLKVG-EWLTVNKLDQFTMSGGG 119
Query: 121 TFDGRGASAWKLQKACGATCSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTF 180
DG+ A+AW+ +++ C+K N+ FN L +ST++ +T+ DSK FHV + C N TF
Sbjct: 120 ILDGQAAAAWECKQS--KKCNKLPNNLSFNSLTNSTIKDITTLDSKSFHVNVNQCKNLTF 177
Query: 181 DGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPG 240
F + AP +S NTDGIH+++S+ V I +SN +TGDDC+S+GD + + I V CGPG
Sbjct: 178 IRFNVSAPANSP--NTDGIHVSRSSSVNITDSNFSTGDDCISVGDETEQLYITRVTCGPG 235
Query: 241 HGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTA-PGTSPVTNLTFEDITMV 299
HG S+GSLG NP E+PV + V+NCT TNT NGVRIKTWP + PG V ++ FEDI +
Sbjct: 236 HGISVGSLGGNPDEKPVVGVFVRNCTFTNTDNGVRIKTWPASHPGV--VNDVHFEDIIVQ 293
Query: 300 NVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVE 359
NV NPV+IDQ YCP N C+K PS++KIS V+F+NIKGTS T+D V L S GVPCEG+E
Sbjct: 294 NVSNPVVIDQVYCPFNKCNKDLPSQVKISKVSFQNIKGTSRTQDAVSLLRSKGVPCEGIE 353
Query: 360 LSDISLTFNG--APAMAKCANVKPIIIGK--APSCSA 392
+ DI +T++G PA + C N+KP + GK P C+A
Sbjct: 354 VGDIDITYSGKEGPAKSSCENIKPSLKGKQNPPVCTA 390
>UniRef100_Q9MBC0 Polygalacturonase [Salix gilgiana]
Length = 393
Score = 399 bits (1025), Expect = e-110
Identities = 206/391 (52%), Positives = 270/391 (68%), Gaps = 24/391 (6%)
Query: 8 STAIVLFLLLL----SGSAQSI-VFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVI 62
S+AIVLF LLL + AQS VFD+ +GG DIT A NAWK ACAST PSK+ I
Sbjct: 7 SSAIVLFSLLLLLASTAKAQSNGVFDVTKYGG--KQDITAALTNAWKDACASTKPSKVRI 64
Query: 63 SKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTF 122
TY + V + GPCK+ IE+QV+G ++AP +P+ +G WV Y+D+ TLSG+GTF
Sbjct: 65 PSGTYSLRQVTLAGPCKSAIELQVNGILKAPVNPDQFSGG-HWVNFRYIDHLTLSGSGTF 123
Query: 123 DGRGASAWKLQKACGATCSKRS------MNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCT 176
DG+G AW +TCSK MNI F+F+ + VR +T++DSK FHV +LGC
Sbjct: 124 DGQGNVAWSK-----STCSKNKNCEGLPMNIRFDFITNGLVRDITTRDSKNFHVNVLGCK 178
Query: 177 NFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVK 236
N TF F + AP +SI NTDGIHI +ST + I++S I TGDDC+S+GDG + + + V
Sbjct: 179 NLTFQHFTVTAPAESI--NTDGIHIGRSTGIYIIDSKIGTGDDCISVGDGTEELHVTRVT 236
Query: 237 CGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDI 296
CGPGHG S+GSLG P E+PV I VKNCT++NT NGVRIK+WP G +N+ FEDI
Sbjct: 237 CGPGHGISVGSLGRYPNEKPVSGIFVKNCTISNTANGVRIKSWPDLYG-GVASNMHFEDI 295
Query: 297 TMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCE 356
M NV NP+++DQ YCP N CS ++PSK+KIS+V+FKNI+GTS T V L CSSG+PCE
Sbjct: 296 VMNNVQNPILLDQVYCPWNQCSLKAPSKVKISDVSFKNIRGTSATPVVVKLACSSGIPCE 355
Query: 357 GVELSDISLTFNGA--PAMAKCANVKPIIIG 385
VEL++I+L ++G+ PA ++C+NVKP I G
Sbjct: 356 KVELANINLVYSGSEGPAKSQCSNVKPKISG 386
>UniRef100_Q9MBB9 Polygalacturonase [Salix gilgiana]
Length = 393
Score = 399 bits (1024), Expect = e-109
Identities = 205/391 (52%), Positives = 272/391 (69%), Gaps = 24/391 (6%)
Query: 8 STAIVLFLLLL----SGSAQSI-VFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVI 62
S+AI+ F LLL + AQS VFD+ +GG DIT+A NAWK ACAST+PSK++I
Sbjct: 7 SSAIISFSLLLLLASTAKAQSNGVFDVTKYGG--KEDITEALNNAWKDACASTNPSKVLI 64
Query: 63 SKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTF 122
TY + V + GPCKA IE+QV+G ++AP +P+ +G+ WV Y+D TLSG+GTF
Sbjct: 65 PSGTYSLRQVTLAGPCKAAIELQVNGILKAPVNPDQFSGS-HWVNFRYIDQLTLSGSGTF 123
Query: 123 DGRGASAWKLQKACGATCSKRS------MNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCT 176
DG+G AW +TCSK MNI F+F+ + VR +T++DSK FHV +LGC
Sbjct: 124 DGQGNVAWSK-----STCSKNKNCEGLPMNIRFDFITNGLVRDITTRDSKNFHVNVLGCK 178
Query: 177 NFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVK 236
N TF F + AP +SI NTDGIHI +ST + I++S I TGDDC+S+GDG + + + V
Sbjct: 179 NLTFQHFTVTAPAESI--NTDGIHIGRSTGIYIIDSKIGTGDDCISVGDGTEELHVTGVT 236
Query: 237 CGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDI 296
CGPGHG S+GSLG P E+PV I VKNCT++NT NGVRIK+WP G +N+ FEDI
Sbjct: 237 CGPGHGISVGSLGRYPNEKPVSGIFVKNCTISNTANGVRIKSWPDLYG-GVASNMHFEDI 295
Query: 297 TMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCE 356
M NV NP+++DQ YCP N CS ++PSK+KIS+V+FKNI+GTS T V L CSSG+PCE
Sbjct: 296 VMNNVQNPILLDQVYCPWNQCSLKAPSKVKISDVSFKNIRGTSATPVVVKLACSSGIPCE 355
Query: 357 GVELSDISLTFNGA--PAMAKCANVKPIIIG 385
VEL++I+L ++G+ PA ++C+NVKP I G
Sbjct: 356 KVELANINLLYSGSEGPAKSQCSNVKPKISG 386
>UniRef100_Q39786 Polygalacturonase precursor [Gossypium hirsutum]
Length = 407
Score = 378 bits (970), Expect = e-103
Identities = 193/394 (48%), Positives = 259/394 (64%), Gaps = 19/394 (4%)
Query: 3 MSLIRSTAIVLFLLLLSGSAQSIVFDIRTFGGAP---NSDITQAFLNAWKQACASTSPSK 59
++++ S ++L L + + QS FD+ GA +D+++ FL+AWK+ACAS +PS
Sbjct: 5 LNIVPSMFVLLLLFISASKVQSDAFDVVAKFGAKADGKTDLSKPFLDAWKEACASVTPST 64
Query: 60 IVISKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGN 119
+VI K TY ++ VN++GPCKAPIEI V GTIQAP DP+A WV V+ F + G
Sbjct: 65 VVIPKGTYLLSKVNLEGPCKAPIEINVQGTIQAPADPSAFKDP-NWVRFYSVENFKMFGG 123
Query: 120 GTFDGRGASAWKLQKACGATC------SKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLL 173
G FDG+G+ A++ TC SK +NI F+FL ++ ++ +TSKDSK FH+ +
Sbjct: 124 GIFDGQGSIAYEKN-----TCENREFRSKLPVNIRFDFLTNALIQDITSKDSKLFHINVF 178
Query: 174 GCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIH 233
C N T + KI AP +S NTDGIH+ KS V I+ S+I TGDDC+S+GDG K++ I
Sbjct: 179 ACKNITLERLKIEAPDESP--NTDGIHMGKSEGVNIIASDIKTGDDCISIGDGTKNMVIK 236
Query: 234 NVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTF 293
+ CGPGHG SIGSLG EEPVE I + NCT+TNT NG RIKTWP G V+ + F
Sbjct: 237 EITCGPGHGISIGSLGKFQNEEPVEGIKISNCTITNTSNGARIKTWPGEHG-GAVSEIHF 295
Query: 294 EDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGV 353
EDITM NV +P++IDQQYCP N C K SK+K+SN++FKNI+GTS + + CS
Sbjct: 296 EDITMNNVSSPILIDQQYCPWNKCKKNEESKVKLSNISFKNIRGTSALPEAIKFICSGSS 355
Query: 354 PCEGVELSDISLTFNGA-PAMAKCANVKPIIIGK 386
PC+ VEL+DI + NGA PA ++C NVKPI GK
Sbjct: 356 PCQNVELADIDIKHNGAEPATSQCLNVKPITSGK 389
>UniRef100_Q39766 Polygalacturonase precursor [Gossypium barbadense]
Length = 407
Score = 377 bits (968), Expect = e-103
Identities = 192/394 (48%), Positives = 259/394 (65%), Gaps = 19/394 (4%)
Query: 3 MSLIRSTAIVLFLLLLSGSAQSIVFDIRTFGGAP---NSDITQAFLNAWKQACASTSPSK 59
++++ S ++L L + + Q FD+ GA +D+++ FL+AWK+ACAS +PS
Sbjct: 5 LNIVPSMFVLLLLFISASKVQPDAFDVVAKFGAKADGKTDLSKPFLDAWKEACASVTPST 64
Query: 60 IVISKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGN 119
+VI K TY ++ VN++GPCKAPIEI V GTIQAP DP+A WV V+ F + G
Sbjct: 65 VVIPKGTYLLSKVNLEGPCKAPIEINVQGTIQAPADPSAFKDP-NWVRFYSVENFKMFGG 123
Query: 120 GTFDGRGASAWKLQKACGATC------SKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLL 173
G FDG+G+ A++ TC SK +NI F+F+ ++ ++ +TSKDSK FH+ +
Sbjct: 124 GIFDGQGSIAYEKN-----TCENREFRSKLPVNIRFDFVTNALIQDITSKDSKLFHINVF 178
Query: 174 GCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIH 233
C N T + KI AP +S NTDGIH+ KS V I+ S+I TGDDC+S+GDG K++ I
Sbjct: 179 ACKNITLERLKIEAPDESP--NTDGIHMGKSEGVNIIASDIKTGDDCISIGDGTKNMVIK 236
Query: 234 NVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTF 293
+ CGPGHG SIGSLG EEPVE I + NCT+TNT NG RIKTWP G V+ + F
Sbjct: 237 EITCGPGHGISIGSLGKFQNEEPVEGIKISNCTITNTSNGARIKTWPGEHG-GAVSEIHF 295
Query: 294 EDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGV 353
EDITM NV +P++IDQQYCP N C K SK+K+SN++FKNI+GTS + + CS
Sbjct: 296 EDITMNNVSSPILIDQQYCPWNKCKKNEESKVKLSNISFKNIRGTSALPEAIKFICSGSS 355
Query: 354 PCEGVELSDISLTFNGA-PAMAKCANVKPIIIGK 386
PC+ VEL+DI + NGA PA ++C NVKPI IGK
Sbjct: 356 PCQNVELADIDIQHNGAEPATSQCLNVKPITIGK 389
>UniRef100_Q84XM0 Putative pollen polygalacturonase [Turnera subulata]
Length = 382
Score = 375 bits (962), Expect = e-102
Identities = 189/382 (49%), Positives = 250/382 (64%), Gaps = 12/382 (3%)
Query: 8 STAIVLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAW-KQACASTSPSKIVISKAT 66
S +L LL S SAQ VFDI+ + P +DI +A A+ K ACAS +PSK+V+
Sbjct: 8 SAISLLLLLASSASAQDGVFDIKKYN--PKADIAEALSAAFQKDACASATPSKVVVPAGE 65
Query: 67 YKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFDGRG 126
Y M V++KGPCK+ +E+QVDG ++AP +P G WV+ Y+D T SG G FDG+G
Sbjct: 66 YTMGPVDMKGPCKSTVEVQVDGNLKAPAEPKGEPG---WVSFEYIDGLTFSGKGVFDGQG 122
Query: 127 ASAWKLQKAC-GATCSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFDGFKI 185
AW C +N F LKH+ ++ +TSKDSK FHV ++ C N TF F I
Sbjct: 123 HIAWANNNCNKNPNCKSFPLNFRFTSLKHALIKDLTSKDSKNFHVNVINCENVTFQNFVI 182
Query: 186 IAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGHGFSI 245
AP +S+ NTDGIH+ +S +KI++S I TGDDC+S+GDG + +T+ V CGPGHG SI
Sbjct: 183 DAPAESL--NTDGIHMGRSKGIKIIDSKIGTGDDCISIGDGTEQVTVTGVTCGPGHGISI 240
Query: 246 GSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNVYNPV 305
GSLG E+PV ++VKNC L NT NGVRIK+WP G +++ FEDITM NV NPV
Sbjct: 241 GSLGRYDNEQPVRGVLVKNCILKNTDNGVRIKSWPAMKG-GEASDIHFEDITMENVTNPV 299
Query: 306 IIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISL 365
IIDQ+YCP N C+K +PSK+KISNV+FKNIKGT+ T + V + CSS +PCE V+L+ I L
Sbjct: 300 IIDQEYCPWNQCTKDAPSKVKISNVSFKNIKGTTNTPEAVKIICSSALPCEQVQLNGIDL 359
Query: 366 TFNG--APAMAKCANVKPIIIG 385
+ G PA ++C N K + G
Sbjct: 360 KYTGTQGPAKSECKNAKLTVTG 381
>UniRef100_Q9SFD2 Polygalacturonase PGA3 [Arabidopsis thaliana]
Length = 391
Score = 373 bits (958), Expect = e-102
Identities = 189/391 (48%), Positives = 262/391 (66%), Gaps = 16/391 (4%)
Query: 8 STAIVLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVISKATY 67
ST ++ LL +S +A+ + T G + SDITQA L A+ AC S+SPSK+VI K +
Sbjct: 8 STIFIICLLGISANAE-----VFTIGSSSGSDITQALLKAFTSACQSSSPSKVVIPKGEF 62
Query: 68 KMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFDGRGA 127
K+ + ++GPCKAPIE+ + GT++A D NA+ G +WV G +D F L+G G FDG G
Sbjct: 63 KLGEIEMRGPCKAPIEVTLQGTVKA--DGNAIQGKEKWVVFGNIDGFKLNGGGAFDGEGN 120
Query: 128 SAWKLQKACGAT--CSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFDGFKI 185
+AW++ C T C K ++I F+F+ +S +R ++S D+K FH+ +LG N T + KI
Sbjct: 121 AAWRVNN-CHKTFECKKLPISIRFDFILNSEIRDISSIDAKNFHINVLGAKNMTMNNIKI 179
Query: 186 IAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGHGFSI 245
+AP DS NTDGIH+ +S VKILNS I+TGDDC+S+GDG K++ + V CGPGHG S+
Sbjct: 180 VAPEDSP--NTDGIHLGRSDGVKILNSFISTGDDCISVGDGMKNLHVEKVTCGPGHGISV 237
Query: 246 GSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNVYNPV 305
GSLG E+ V I V NCTL T NG+RIKTWP+A ++ +++ FEDI + +V NP+
Sbjct: 238 GSLGRYGHEQDVSGIKVINCTLQETDNGLRIKTWPSAACSTTASDIHFEDIILKDVSNPI 297
Query: 306 IIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISL 365
+IDQ+YCP N C+KQ S IK+ N++FKNI+GTS KD V L CS G PC+ VE+ DI +
Sbjct: 298 LIDQEYCPWNQCNKQKASTIKLVNISFKNIRGTSGNKDAVKLLCSKGYPCQNVEIGDIDI 357
Query: 366 TFNGA--PAMAKCANVKPIIIG--KAPSCSA 392
+NGA PA C+NV P I+G +CSA
Sbjct: 358 KYNGADGPATFHCSNVSPKILGSQSPKACSA 388
>UniRef100_O65905 Polygalacturonase [Arabidopsis thaliana]
Length = 391
Score = 373 bits (957), Expect = e-102
Identities = 188/391 (48%), Positives = 262/391 (66%), Gaps = 16/391 (4%)
Query: 8 STAIVLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVISKATY 67
ST ++ LL +S +A+ + T G + SDITQA L A+ AC S+SPSK+VI K +
Sbjct: 8 STIFIICLLGISANAE-----VFTIGSSSGSDITQALLKAFTSACQSSSPSKVVIPKGEF 62
Query: 68 KMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFDGRGA 127
K+ + ++GPCKAPIE+ + GT++A D NA+ G +WV G +D F L+G G FDG G
Sbjct: 63 KLGEIEMRGPCKAPIEVTLQGTVKA--DGNAIQGKEKWVVFGNIDGFKLNGGGAFDGEGN 120
Query: 128 SAWKLQKACGAT--CSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFDGFKI 185
+AW++ C T C K ++I F+F+ +S +R ++S D+K FH+ +LG N T + KI
Sbjct: 121 AAWRVNN-CHKTFECKKLPISIRFDFILNSEIRDISSIDAKNFHINVLGAKNMTMNNIKI 179
Query: 186 IAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGHGFSI 245
+AP DS NTDGIH+ +S +KILNS I+TGDDC+S+GDG K++ + V CGPGHG S+
Sbjct: 180 VAPEDSP--NTDGIHLGRSDGIKILNSFISTGDDCISVGDGMKNLHVEKVTCGPGHGISV 237
Query: 246 GSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNVYNPV 305
GSLG E+ V I V NCTL T NG+RIKTWP+A ++ +++ FEDI + +V NP+
Sbjct: 238 GSLGRYGHEQDVSGIKVINCTLQETDNGLRIKTWPSAACSTTASDIHFEDIILKDVSNPI 297
Query: 306 IIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISL 365
+IDQ+YCP N C+KQ S IK+ N++FKNI+GTS KD V L CS G PC+ VE+ DI +
Sbjct: 298 LIDQEYCPWNQCNKQKASTIKLVNISFKNIRGTSGNKDAVKLLCSKGYPCQNVEIGDIDI 357
Query: 366 TFNGA--PAMAKCANVKPIIIG--KAPSCSA 392
+NGA PA C+NV P I+G +CSA
Sbjct: 358 KYNGADGPATFHCSNVSPKILGSQSPKACSA 388
>UniRef100_P24548 Exopolygalacturonase [Oenothera organensis]
Length = 362
Score = 364 bits (934), Expect = 3e-99
Identities = 181/354 (51%), Positives = 240/354 (67%), Gaps = 12/354 (3%)
Query: 39 DITQAFLNAWKQACASTSPSKIVISKATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNA 98
D TQA AWK+ACAS SPS I++ K + + + ++GPCK+ I +Q+ GT++AP DP+
Sbjct: 1 DSTQALTTAWKEACASASPSTILVPKGNFAVGLITLEGPCKSSIGLQLQGTLKAPADPSK 60
Query: 99 LNGAFQWVTIGYVDYFTLSGNGTFDGRGASAWKLQKAC---GATCSKRSMNIGFNFLKHS 155
+ G W+ + +D T+ G G FDG+G SAW +Q C G C SMN+ + +S
Sbjct: 61 IKG-LGWINLNKIDLLTIFGGGVFDGQGKSAW-VQNDCHKNGPICKTLSMNLRLYAVTNS 118
Query: 156 TVRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAKSTDVKILNSNIA 215
+R VT+ DSK FHV ++GC N TF+ FKI A SINTDGIHI +S V I+N+ I
Sbjct: 119 ILRDVTTLDSKNFHVNVIGCKNLTFERFKISAA--ETSINTDGIHIGRSDGVNIINTEIK 176
Query: 216 TGDDCVSLGDGNKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVR 275
TGDDC+SLGDG+K+I I N+ CGPGHG S+GSLG EE V I VKNCT+T +QNGVR
Sbjct: 177 TGDDCISLGDGSKNINITNITCGPGHGISVGSLGRYKNEESVVGIYVKNCTITGSQNGVR 236
Query: 276 IKTWP-TAPGTSPVTNLTFEDITMVNVYNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKN 334
IKTWP + PG + + + F+DITM +V P++IDQ YCP N C+ + PS +K+S ++FKN
Sbjct: 237 IKTWPKSEPGEA--SEMHFQDITMNSVGTPILIDQGYCPYNQCTAEVPSSVKLSKISFKN 294
Query: 335 IKGTSETKDGVILNCSSGVPCEGVELSDISLTFN--GAPAMAKCANVKPIIIGK 386
IKGTS TK+ V L CS PC GVEL+DI LT++ G PA + C N+KP I GK
Sbjct: 295 IKGTSTTKEAVKLVCSKSFPCNGVELADIDLTYSGKGGPATSVCENIKPTIKGK 348
>UniRef100_Q9SFD0 Putative polygalacturonase [Arabidopsis thaliana]
Length = 401
Score = 363 bits (932), Expect = 4e-99
Identities = 184/395 (46%), Positives = 262/395 (65%), Gaps = 14/395 (3%)
Query: 4 SLIRSTAIVLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVIS 63
S + +A++ + LL SA + VF++ GG+P SDITQA L A+ AC + + SK+VI+
Sbjct: 3 SYLGISAVLFVISLLGFSANAEVFNV---GGSPGSDITQALLKAFTSACQAPTASKVVIT 59
Query: 64 KATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFD 123
K +K+ + + GPCKAP+EI + GT++A D A+ G +WV ++ F L+G G FD
Sbjct: 60 KGEFKLGEIEMTGPCKAPVEINLQGTLKA--DGKAIQGKERWVVFLRINGFKLNGGGIFD 117
Query: 124 GRGASAWKLQKACGAT--CSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFD 181
G G +AW++ C T C K ++I F+F++++ +R ++S D+K FH+ +LG N TFD
Sbjct: 118 GEGNAAWRVNN-CHKTFECKKLPISIRFDFVENAEIRDISSIDAKNFHINVLGAKNMTFD 176
Query: 182 GFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGH 241
K+IAP +S NTDGIH+ +S VKILNS IATGDDC+S+GDG K++ + NV CGPGH
Sbjct: 177 NVKVIAPAESP--NTDGIHLGRSEGVKILNSKIATGDDCISVGDGMKNLHVENVMCGPGH 234
Query: 242 GFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNV 301
G S+GSLG E+ V I V NCTL T NG+RIKTWP+A + + + FE+I + NV
Sbjct: 235 GISVGSLGRYVHEQDVTGITVVNCTLQGTDNGLRIKTWPSAACATTASGIHFENIILNNV 294
Query: 302 YNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELS 361
NP++IDQ+YCP N C+KQ PS IK+ +++FKNI+GTS KD V L CS PC VE+
Sbjct: 295 SNPILIDQEYCPWNQCNKQKPSTIKLVDISFKNIRGTSGNKDAVKLLCSKAHPCANVEIG 354
Query: 362 DISLTFNGA--PAMAKCANVKPIIIG--KAPSCSA 392
+I+L + GA P C+NV P ++G +CSA
Sbjct: 355 NINLEYKGADGPPTFMCSNVSPKLVGTQNPKACSA 389
>UniRef100_Q6H9K0 Exopolygalacturonase precursor [Platanus acerifolia]
Length = 377
Score = 362 bits (928), Expect = 1e-98
Identities = 176/365 (48%), Positives = 240/365 (65%), Gaps = 8/365 (2%)
Query: 26 VFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVISKATYKMNAVNVKGPCK-APIEI 84
VF++ +G DI+QA + AWK ACAS PS ++I K Y M V ++GPCK + I
Sbjct: 9 VFNVNDYGAKGAGDISQAVMKAWKAACASQGPSTVLIPKGNYNMGEVAMQGPCKGSKIGF 68
Query: 85 QVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFDGRGASAWKLQKAC-GATCSKR 143
Q+DG ++AP DP+ WV+ +D T+SG GT DG+G +AW C
Sbjct: 69 QIDGVVKAPADPSKFKSD-GWVSFYRIDGLTVSGTGTLDGQGQTAWAKNNCDKNPNCKHA 127
Query: 144 SMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPGDSISINTDGIHIAK 203
+MN+ F+FLKH+ VR +TS +SK FH+ +L C + TF + APG SI NTDGIH+
Sbjct: 128 AMNLRFDFLKHAMVRDITSLNSKMFHINVLECEDITFQHVTVTAPGTSI--NTDGIHVGI 185
Query: 204 STDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGHGFSIGSLGTNPTEEPVENIIVK 263
S V I N+ IATGDDC+S+G G++++TI V CGPGHG SIGSLG E+ V I VK
Sbjct: 186 SKGVTITNTKIATGDDCISIGPGSQNVTITQVNCGPGHGISIGSLGRYNNEKEVRGITVK 245
Query: 264 NCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNVYNPVIIDQQYCPSNLCSKQSPS 323
CT + T NGVR+KTWP +P T+LTF+D+TM NV NPVI+DQ+YCP CS+Q+PS
Sbjct: 246 GCTFSGTMNGVRVKTWPNSP-PGAATDLTFQDLTMNNVQNPVILDQEYCPYGQCSRQAPS 304
Query: 324 KIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISLTFNGA--PAMAKCANVKP 381
+IK+SN+ F NI+GTS K V++ CS G+PC +++ +I+L++ GA PA + C+NVKP
Sbjct: 305 RIKLSNINFNNIRGTSTGKVAVVIACSHGMPCSNMKIGEINLSYRGAGGPATSTCSNVKP 364
Query: 382 IIIGK 386
GK
Sbjct: 365 TFSGK 369
>UniRef100_Q9SFD1 Putative polygalacturonase [Arabidopsis thaliana]
Length = 397
Score = 355 bits (910), Expect = 2e-96
Identities = 177/382 (46%), Positives = 254/382 (66%), Gaps = 13/382 (3%)
Query: 8 STAIVLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVISKATY 67
ST V+ LL S +A ++ +P SDITQA L A+ AC S +P K+VI K +
Sbjct: 8 STIFVICLLGFSANAHEVL----RLTSSPGSDITQALLRAFTTACQSPTPRKVVIPKGQF 63
Query: 68 KMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFDGRGA 127
K+ + + GPCK+P+EI + GT+ A D N+++G +WV +D F L+G GTFDG G
Sbjct: 64 KLGEIMMSGPCKSPVEITLLGTVLA--DGNSIHGKEKWVVFQRMDGFRLNGGGTFDGEGN 121
Query: 128 SAWKLQKACGAT--CSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFDGFKI 185
+AW++ C T C K ++I F+F+ ++ +R ++S D+K FH+ ++G N TFD KI
Sbjct: 122 AAWRVNN-CHKTFECKKLPISIRFDFVTNAEIRDISSIDAKNFHINVIGAKNMTFDNVKI 180
Query: 186 IAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGHGFSI 245
+AP +S NTDGIH+ +S V I+NS I+TGDDCVS+GDG ++ + NV CGPGHG S+
Sbjct: 181 MAPAESP--NTDGIHLGRSVGVSIINSRISTGDDCVSVGDGMVNLLVKNVVCGPGHGISV 238
Query: 246 GSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNVYNPV 305
GSLG E+ V I V NCTL T NG+RIKTWP+A ++ +N+ FE+I + NV NP+
Sbjct: 239 GSLGRYGHEQDVSGIRVINCTLQETDNGLRIKTWPSAACSTTASNIHFENIILRNVSNPI 298
Query: 306 IIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISL 365
+IDQ+YCP N C+KQ S IK++N++F+ I+GTS KD V L CS G PCE V++ DI++
Sbjct: 299 LIDQEYCPWNQCNKQKSSSIKLANISFRRIRGTSGNKDAVKLLCSKGYPCENVQVGDINI 358
Query: 366 TFNGA--PAMAKCANVKPIIIG 385
+ GA PA C+NV+P ++G
Sbjct: 359 QYTGADGPATFMCSNVRPKLVG 380
>UniRef100_Q7Y1X2 Polygalacturonase [Fragaria ananassa]
Length = 405
Score = 355 bits (910), Expect = 2e-96
Identities = 181/388 (46%), Positives = 252/388 (64%), Gaps = 15/388 (3%)
Query: 13 LFLLLLSGSAQSIV-------FDIRT--FGGAPNSDITQAFLNAWKQACASTSPSKIVIS 63
+FLL S A + FD+ + +GG PN+DI+Q NAWK ACA+T+PS++++
Sbjct: 13 VFLLSSSAYASRVAQFLAPGDFDVTSPKYGGKPNTDISQPLANAWKDACAATTPSRVIVP 72
Query: 64 KATYKMNAVNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFD 123
K T+++ KGPCKAPI +QVDG +QAP L WV V+ T+SG GTFD
Sbjct: 73 KGTFQLKGAVFKGPCKAPITVQVDGILQAPPIEAQLANKEFWVQFLEVERLTVSGTGTFD 132
Query: 124 GRGASAWKLQKACGAT--CSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFD 181
G+G ++WK C C ++N+ F+ +K+S VR VTS +SK FH +LGC + TF
Sbjct: 133 GQGQNSWK-DNDCNKNPNCGGLAINVRFDRVKNSLVRDVTSLNSKNFHFNILGCEHLTFQ 191
Query: 182 GFKIIAPGDSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGH 241
+ APGDS NTDGIH+ +ST + I ++NI TGDDC+S+GDG + +T+ V CGPGH
Sbjct: 192 HVIVKAPGDSP--NTDGIHMGRSTRINITDTNIGTGDDCISVGDGTRQLTVSKVSCGPGH 249
Query: 242 GFSIGSLGTNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNV 301
G SIGSLG E+ V + +++CTL+NT NGVRIKT+P +P + +++ FE ITM V
Sbjct: 250 GISIGSLGRYDNEDDVSGLNIRDCTLSNTLNGVRIKTFPASPKATTASDIHFEKITMNYV 309
Query: 302 YNPVIIDQQYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELS 361
NPV+IDQ+YCP C+KQ PSK+KISNV+FKNI GT+ T + + + C+ G+ C+ V LS
Sbjct: 310 ANPVLIDQEYCPWGQCNKQIPSKVKISNVSFKNIIGTTSTAEALKIVCAKGLHCDQVVLS 369
Query: 362 DISLTFNGAPAM-AKCANVKPIIIGKAP 388
DI L +G + + CANV+P I P
Sbjct: 370 DIDLKLSGKGTLTSHCANVQPTITRVPP 397
>UniRef100_P35337 Polygalacturonase precursor [Brassica napus]
Length = 397
Score = 355 bits (910), Expect = 2e-96
Identities = 180/378 (47%), Positives = 245/378 (64%), Gaps = 12/378 (3%)
Query: 12 VLFLLLLSGSAQSIVFDIRTFGGAPNSDITQAFLNAWKQACASTSPSKIVISKATYKMNA 71
+L L LL SA + VF T GG PNSDIT A L A+ AC + +PS+++I K +K+
Sbjct: 10 ILVLCLLGYSANAEVF---TAGGPPNSDITAAVLKAFTSACQAPAPSQVLIPKGDFKLGE 66
Query: 72 VNVKGPCKAPIEIQVDGTIQAPTDPNALNGAFQWVTIGYVDYFTLSGNGTFDGRGASAWK 131
+ GPCK+PIE + G ++ TD + G +WV ++ F L+G GTFDG G +AWK
Sbjct: 67 TVMTGPCKSPIEFTLQGNVK--TDGGSTQGKDRWVVFEKINGFKLNGGGTFDGEGNAAWK 124
Query: 132 LQKACGAT--CSKRSMNIGFNFLKHSTVRSVTSKDSKFFHVTLLGCTNFTFDGFKIIAPG 189
C T C K +++ F+F+ ++ ++ VTS D+K FH ++ N TFD KIIAP
Sbjct: 125 ANN-CHKTFECKKLPISVRFDFVDNAEIKDVTSLDAKNFHFNVISGKNMTFDNIKIIAPA 183
Query: 190 DSISINTDGIHIAKSTDVKILNSNIATGDDCVSLGDGNKHITIHNVKCGPGHGFSIGSLG 249
+S NTDGIH+ + VKILN+ IATGDDC+S+GDG K++ I V CGPGHG S+GSLG
Sbjct: 184 ESP--NTDGIHLGRCEGVKILNTKIATGDDCISVGDGMKNLLIEKVVCGPGHGISVGSLG 241
Query: 250 TNPTEEPVENIIVKNCTLTNTQNGVRIKTWPTAPGTSPVTNLTFEDITMVNVYNPVIIDQ 309
E+ V +I VKNCTL T NG+RIKTWP+A T+ + FEDI + V NP++IDQ
Sbjct: 242 RYGWEQDVTDITVKNCTLEGTSNGLRIKTWPSAACTTTAAGIHFEDIILNKVSNPILIDQ 301
Query: 310 QYCPSNLCSKQSPSKIKISNVTFKNIKGTSETKDGVILNCSSGVPCEGVELSDISLTFNG 369
+YCP N C+K PS IK+ ++TF+NI+GTSE KD V L CS G PCE VE+ DI++ + G
Sbjct: 302 EYCPWNQCNKNKPSTIKLVDITFRNIRGTSENKDAVKLLCSKGHPCENVEIGDINIEYTG 361
Query: 370 --APAMAKCANVKPIIIG 385
P +C NV P ++G
Sbjct: 362 PDGPPTFECTNVTPKLVG 379
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 692,034,415
Number of Sequences: 2790947
Number of extensions: 30872700
Number of successful extensions: 66690
Number of sequences better than 10.0: 447
Number of HSP's better than 10.0 without gapping: 257
Number of HSP's successfully gapped in prelim test: 190
Number of HSP's that attempted gapping in prelim test: 65055
Number of HSP's gapped (non-prelim): 523
length of query: 392
length of database: 848,049,833
effective HSP length: 129
effective length of query: 263
effective length of database: 488,017,670
effective search space: 128348647210
effective search space used: 128348647210
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0490.17


