

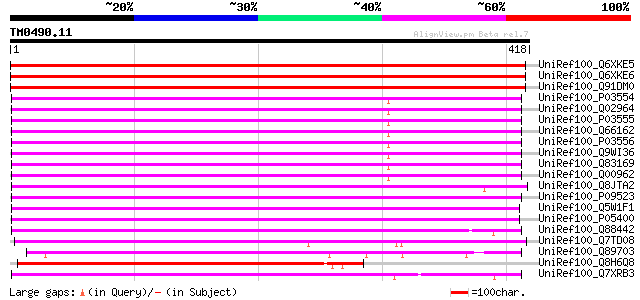
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0490.11
(418 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6XKE5 Polyprotein 1 [Petunia vein clearing virus] 504 e-141
UniRef100_Q6XKE6 Polyprotein 1 [Petunia vein clearing virus] 504 e-141
UniRef100_Q91DM0 ORF I polyprotein [Petunia vein clearing virus] 504 e-141
UniRef100_P03554 Enzymatic polyprotein [Contains: Aspartic prote... 318 2e-85
UniRef100_Q02964 Enzymatic polyprotein [Contains: Aspartic prote... 318 2e-85
UniRef100_P03555 Enzymatic polyprotein [Contains: Aspartic prote... 318 2e-85
UniRef100_Q66162 ORF V [Cauliflower mosaic virus] 317 3e-85
UniRef100_P03556 Enzymatic polyprotein [Contains: Aspartic prote... 315 1e-84
UniRef100_Q9WI36 Reverse transcriptase [Cauliflower mosaic virus] 315 2e-84
UniRef100_Q83169 Reverse transcriptase [Cauliflower mosaic virus] 315 2e-84
UniRef100_Q00962 Enzymatic polyprotein [Contains: Aspartic prote... 313 4e-84
UniRef100_Q8JTA2 , complete genome [Mirabilis mosaic virus] 294 4e-78
UniRef100_P09523 Enzymatic polyprotein [Contains: Aspartic prote... 291 2e-77
UniRef100_Q5W1F1 Polyprotein [Carnation etched ring virus] 276 8e-73
UniRef100_P05400 Enzymatic polyprotein [Contains: Aspartic prote... 267 5e-70
UniRef100_Q88442 ORF V protein [Strawberry vein banding virus] 253 7e-66
UniRef100_Q7TD08 Putative multifunctional pol protein [Cestrum y... 238 2e-61
UniRef100_Q89703 ORF 3 [Cassava vein mosaic virus] 235 2e-60
UniRef100_Q8H6Q8 CTV.20 [Poncirus trifoliata] 232 2e-59
UniRef100_Q7XRB3 OSJNBa0006B20.7 protein [Oryza sativa] 228 2e-58
>UniRef100_Q6XKE5 Polyprotein 1 [Petunia vein clearing virus]
Length = 1886
Score = 504 bits (1299), Expect = e-141
Identities = 241/415 (58%), Positives = 309/415 (74%)
Query: 1 MTPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFI 60
M P L+ A +EC +L + LIEP+ S+WAC AFYVNKRSEQVR K RLV++Y+PLN F+
Sbjct: 1394 MNPEHLQLALKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFL 1453
Query: 61 KDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLP 120
+DDKFP+P +L+S++ A +FSKFD+KSGFWQLG+ P +RPKT FCIP+ H+QW V+P
Sbjct: 1454 QDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWKVMP 1513
Query: 121 FGLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVML 180
FGLK APSLFQKAM KIF+PIL ++LVYIDD+LLFS E H K L QF + K GVML
Sbjct: 1514 FGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFGVML 1573
Query: 181 SSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDF 240
S++K +A ++ FLGM F +G F P HI EL KFPD L++KQIQQFLGI+NYIRDF
Sbjct: 1574 SAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYIRDF 1633
Query: 241 IPHVSKYTSKLSKMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIPGDGKRILQTDASDH 300
IP V+++ S LS ML+K P WGK Q A+K LK+ AQ L IP +GK+ILQTDASD
Sbjct: 1634 IPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQVKSLHIPSEGKKILQTDASDQ 1693
Query: 301 YWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQ 360
YW+AVL+EE +G++ CG ASG+FK +E HYH+TFKE LAVK GIK+F+F L +F V+
Sbjct: 1694 YWSAVLLEEHNGKRKICGFASGKFKVSEQHYHSTFKEILAVKNGIKKFNFFLIHTNFLVE 1753
Query: 361 IDNSSFPKMLEFKNKTPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSRPKQ 415
+D +FPKM+ K PN Q+LR +WFS Y F VKH++G N++ D+LSRP +
Sbjct: 1754 MDMRAFPKMIRLNPKIVPNSQLLRWAQWFSPYQFEVKHLKGKDNILADFLSRPHE 1808
>UniRef100_Q6XKE6 Polyprotein 1 [Petunia vein clearing virus]
Length = 2180
Score = 504 bits (1299), Expect = e-141
Identities = 241/415 (58%), Positives = 309/415 (74%)
Query: 1 MTPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFI 60
M P L+ A +EC +L + LIEP+ S+WAC AFYVNKRSEQVR K RLV++Y+PLN F+
Sbjct: 1394 MNPEHLQLALKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFL 1453
Query: 61 KDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLP 120
+DDKFP+P +L+S++ A +FSKFD+KSGFWQLG+ P +RPKT FCIP+ H+QW V+P
Sbjct: 1454 QDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWKVMP 1513
Query: 121 FGLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVML 180
FGLK APSLFQKAM KIF+PIL ++LVYIDD+LLFS E H K L QF + K GVML
Sbjct: 1514 FGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFGVML 1573
Query: 181 SSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDF 240
S++K +A ++ FLGM F +G F P HI EL KFPD L++KQIQQFLGI+NYIRDF
Sbjct: 1574 SAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYIRDF 1633
Query: 241 IPHVSKYTSKLSKMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIPGDGKRILQTDASDH 300
IP V+++ S LS ML+K P WGK Q A+K LK+ AQ L IP +GK+ILQTDASD
Sbjct: 1634 IPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQVKSLHIPSEGKKILQTDASDQ 1693
Query: 301 YWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQ 360
YW+AVL+EE +G++ CG ASG+FK +E HYH+TFKE LAVK GIK+F+F L +F V+
Sbjct: 1694 YWSAVLLEEHNGKRKICGFASGKFKVSEQHYHSTFKEILAVKNGIKKFNFFLIHTNFLVE 1753
Query: 361 IDNSSFPKMLEFKNKTPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSRPKQ 415
+D +FPKM+ K PN Q+LR +WFS Y F VKH++G N++ D+LSRP +
Sbjct: 1754 MDMRAFPKMIRLNPKIVPNSQLLRWAQWFSPYQFEVKHLKGKDNILADFLSRPHE 1808
>UniRef100_Q91DM0 ORF I polyprotein [Petunia vein clearing virus]
Length = 2179
Score = 504 bits (1298), Expect = e-141
Identities = 241/415 (58%), Positives = 309/415 (74%)
Query: 1 MTPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFI 60
M P L+ A +EC +L + LIEP+ S+WAC AFYVNKRSEQVR K RLV++Y+PLN F+
Sbjct: 1393 MNPEHLQLAIKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFL 1452
Query: 61 KDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLP 120
+DDKFP+P +L+S++ A +FSKFD+KSGFWQLG+ P +RPKT FCIP+ H+QW V+P
Sbjct: 1453 QDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWKVMP 1512
Query: 121 FGLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVML 180
FGLK APSLFQKAM KIF+PIL ++LVYIDD+LLFS E H K L QF + K GVML
Sbjct: 1513 FGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFGVML 1572
Query: 181 SSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDF 240
S++K +A ++ FLGM F +G F P HI EL KFPD L++KQIQQFLGI+NYIRDF
Sbjct: 1573 SAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYIRDF 1632
Query: 241 IPHVSKYTSKLSKMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIPGDGKRILQTDASDH 300
IP V+++ S LS ML+K P WGK Q A+K LK+ AQ L IP +GK+ILQTDASD
Sbjct: 1633 IPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQVKSLHIPSEGKKILQTDASDQ 1692
Query: 301 YWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQ 360
YW+AVL+EE +G++ CG ASG+FK +E HYH+TFKE LAVK GIK+F+F L +F V+
Sbjct: 1693 YWSAVLLEEHNGKRKICGFASGKFKVSEQHYHSTFKEILAVKNGIKKFNFFLIHTNFLVE 1752
Query: 361 IDNSSFPKMLEFKNKTPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSRPKQ 415
+D +FPKM+ K PN Q+LR +WFS Y F VKH++G N++ D+LSRP +
Sbjct: 1753 MDMRAFPKMIRLNPKIVPNSQLLRWAQWFSPYQFEVKHLKGKDNILADFLSRPHE 1807
>UniRef100_P03554 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 679
Score = 318 bits (814), Expect = 2e-85
Identities = 170/418 (40%), Positives = 249/418 (58%), Gaps = 7/418 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ +I+P+KS AF VN +E+ R KKR+VV+Y+ +N+
Sbjct: 255 SPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATV 314
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + LP L + I+ IFS FD KSGFWQ+ LD + RP TAF P HY+W V+PF
Sbjct: 315 GDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPF 374
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + F VY+DD+L+FS++EE H + C++HG++LS
Sbjct: 375 GLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILS 434
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ K+++FLG+ EG PQ HI+E + KFPD KQ+Q+FLGIL Y D+I
Sbjct: 435 KKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYI 494
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L++N P W KE T ++ +KK Q PPL P + K I++TDASD
Sbjct: 495 PKLAQIRKPLQAKLKENVPWRWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASD 554
Query: 300 HYWA----AVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGH 355
YW A+ I E +L C +ASG FK AE +YH+ KETLAV IK+F +L
Sbjct: 555 DYWGGMLKAIKINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPV 614
Query: 356 HFEVQIDNSSFPKMLEFKNKTPPN-PQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
HF ++ DN+ F + K + +R + W S Y F V+H++G+ N D+LSR
Sbjct: 615 HFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSR 672
>UniRef100_Q02964 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 679
Score = 318 bits (814), Expect = 2e-85
Identities = 170/418 (40%), Positives = 249/418 (58%), Gaps = 7/418 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ +I+P+KS AF VN +E+ R KKR+VV+Y+ +N+
Sbjct: 255 SPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATI 314
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + LP L + I+ IFS FD KSGFWQ+ LD + RP TAF P HY+W V+PF
Sbjct: 315 GDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPF 374
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + F VY+DD+L+FS++EE H + C++HG++LS
Sbjct: 375 GLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILS 434
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ K+++FLG+ EG PQ HI+E + KFPD KQ+Q+FLGIL Y D+I
Sbjct: 435 KKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYI 494
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L++N P W KE T ++ +KK Q PPL P + K I++TDASD
Sbjct: 495 PKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASD 554
Query: 300 HYWA----AVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGH 355
YW A+ I E +L C +ASG FK AE +YH+ KETLAV IK+F +L
Sbjct: 555 DYWGGMLKAIKINEGTNTELICRYASGSFKAAERNYHSNDKETLAVINTIKKFSIYLTPV 614
Query: 356 HFEVQIDNSSFPKMLEFKNKTPPN-PQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
HF ++ DN+ F + K + +R + W S Y F V+H++G+ N D+LSR
Sbjct: 615 HFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSR 672
>UniRef100_P03555 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 679
Score = 318 bits (814), Expect = 2e-85
Identities = 170/418 (40%), Positives = 249/418 (58%), Gaps = 7/418 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ +I+P+KS AF VN +E+ R KKR+VV+Y+ +N+
Sbjct: 255 SPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATI 314
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + LP L + I+ IFS FD KSGFWQ+ LD + RP TAF P HY+W V+PF
Sbjct: 315 GDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPF 374
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + F VY+DD+L+FS++EE H + C++HG++LS
Sbjct: 375 GLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILS 434
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ K+++FLG+ EG PQ HI+E + KFPD KQ+Q+FLGIL Y D+I
Sbjct: 435 KKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYI 494
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L++N P W KE T ++ +KK Q PPL P + K I++TDASD
Sbjct: 495 PKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASD 554
Query: 300 HYWA----AVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGH 355
YW A+ I E +L C +ASG FK AE +YH+ KETLAV IK+F +L
Sbjct: 555 DYWGGMLKAIKINEGTNTELICRYASGSFKAAERNYHSNDKETLAVINTIKKFSIYLTPV 614
Query: 356 HFEVQIDNSSFPKMLEFKNKTPPN-PQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
HF ++ DN+ F + K + +R + W S Y F V+H++G+ N D+LSR
Sbjct: 615 HFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSR 672
>UniRef100_Q66162 ORF V [Cauliflower mosaic virus]
Length = 680
Score = 317 bits (813), Expect = 3e-85
Identities = 170/418 (40%), Positives = 249/418 (58%), Gaps = 7/418 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ +I+P+KS AF VN +E+ R KKR+VV+Y+ +N+
Sbjct: 256 SPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATI 315
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + LP L + I+ IFS FD KSGFWQ+ LD + RP TAF P HY+W V+PF
Sbjct: 316 GDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPF 375
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + F VY+DD+L+FS++EE H + C++HG++LS
Sbjct: 376 GLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILS 435
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ K+++FLG+ EG PQ HI+E + KFPD KQ+Q+FLGIL Y D+I
Sbjct: 436 KKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYI 495
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L++N P W KE T ++ +KK Q PPL P + K I++TDASD
Sbjct: 496 PKLAQIRKPLQAKLKENVPWRWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASD 555
Query: 300 HYWA----AVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGH 355
YW A+ I E +L C +ASG FK AE +YH+ KETLAV IK+F +L
Sbjct: 556 DYWGGMLKAIKINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPV 615
Query: 356 HFEVQIDNSSFPKMLEFKNKTPPN-PQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
HF ++ DN+ F + K + +R + W S Y F V+H++G+ N D+LSR
Sbjct: 616 HFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSR 673
>UniRef100_P03556 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 674
Score = 315 bits (808), Expect = 1e-84
Identities = 169/418 (40%), Positives = 248/418 (58%), Gaps = 7/418 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ +I+P+KS AF VN +E+ R KKR+VV+Y+ +N+
Sbjct: 250 SPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATV 309
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + P L + I+ IFS FD KSGFWQ+ LD + RP TAF P HY+W V+PF
Sbjct: 310 GDAYNPPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPF 369
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + F VY+DD+L+FS++EE H + C++HG++LS
Sbjct: 370 GLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILS 429
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ K+++FLG+ EG PQ HI+E + KFPD KQ+Q+FLGIL Y D+I
Sbjct: 430 KKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYI 489
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L++N P W KE T ++ +KK Q PPL P + K I++TDASD
Sbjct: 490 PKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASD 549
Query: 300 HYWA----AVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGH 355
YW A+ I E +L C +ASG FK AE +YH+ KETLAV IK+F +L
Sbjct: 550 DYWGGMLKAIKINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPV 609
Query: 356 HFEVQIDNSSFPKMLEFKNKTPPN-PQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
HF ++ DN+ F + K + +R + W S Y F V+H++G+ N D+LSR
Sbjct: 610 HFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSR 667
>UniRef100_Q9WI36 Reverse transcriptase [Cauliflower mosaic virus]
Length = 674
Score = 315 bits (807), Expect = 2e-84
Identities = 169/418 (40%), Positives = 247/418 (58%), Gaps = 7/418 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ +I+P+KS AF VN +E+ R KKR+VV+Y+ +N+
Sbjct: 250 SPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATV 309
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + LP L + I+ IFS FD KSGFWQ+ D + RP TAF P HY+W V+PF
Sbjct: 310 GDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLPDQESRPLTAFTCPQGHYEWNVVPF 369
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + F VY+DD+L+FS+ EE H + C++HG++LS
Sbjct: 370 GLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNTEEDHLLHVAMILQKCNQHGIILS 429
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ K+++FLG+ EG PQ HI+E + KFPD KQ+Q+FLGIL Y D+I
Sbjct: 430 KKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYI 489
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L++N P W KE T ++ +KK Q PPL P + K I++TDASD
Sbjct: 490 PKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHRPLPEEKLIIETDASD 549
Query: 300 HYWA----AVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGH 355
YW A+ I E +L C +ASG FK AE +YH+ KETLAV IK+F +L
Sbjct: 550 DYWGGMLKAIQINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPV 609
Query: 356 HFEVQIDNSSFPKMLEFKNKTPPN-PQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
HF ++ DN+ F + K + +R + W S Y F V+H++G+ N D+LSR
Sbjct: 610 HFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNRFADFLSR 667
>UniRef100_Q83169 Reverse transcriptase [Cauliflower mosaic virus]
Length = 680
Score = 315 bits (807), Expect = 2e-84
Identities = 169/418 (40%), Positives = 248/418 (58%), Gaps = 7/418 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ +I+P+KS AF VN +E+ R KKR+VV+Y+ +N+
Sbjct: 256 SPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATV 315
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + LP L + I+ IFS FD KSGFWQ+ LD + RP TAF P HY+W V+PF
Sbjct: 316 GDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPF 375
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + F VY+DD+L+FS++EE H + C++HG++LS
Sbjct: 376 GLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILS 435
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ K+++FLG+ EG PQ HI+E + KFPD KQ+Q+FLGIL Y D+I
Sbjct: 436 KKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYI 495
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L++N P W KE T ++ +KK Q PPL P + K I++TDASD
Sbjct: 496 PKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASD 555
Query: 300 HYWA----AVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGH 355
YW A+ I E +L C +ASG FK AE +YH+ KETLAV IK+F +L
Sbjct: 556 DYWGGMLKAIKINEGINTELICRYASGSFKAAERNYHSNDKETLAVINTIKKFSIYLTPA 615
Query: 356 HFEVQIDNSSFPKMLEFKNKTPPN-PQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
HF + DN+ F + K + +R + W + Y F V+H++G+ N D+LSR
Sbjct: 616 HFLTRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLNHYSFDVEHIKGTDNHFVDFLSR 673
>UniRef100_Q00962 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 680
Score = 313 bits (803), Expect = 4e-84
Identities = 167/418 (39%), Positives = 247/418 (58%), Gaps = 7/418 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ +I+P+KS AF VN +E R KR+VV+Y+ +N+
Sbjct: 256 SPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAENGRGNKRMVVNYKAMNKATV 315
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + LP L + I+ IFS FD KSGFWQ+ LD + RP TAF P HY+W V+PF
Sbjct: 316 GDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPF 375
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + F VY+DD+++FS++EE H + C++HG++LS
Sbjct: 376 GLKQAPSIFQRHMDEAFRVFRKFCCVYVDDIVVFSNNEEDHLLHVAMILQKCNQHGIILS 435
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ K+++FLG+ EG PQ HI+E + KFPD KQ+Q+FLGIL Y D+I
Sbjct: 436 KKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYI 495
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P++++ L L++N P W KE T ++ +KK Q PPL P + K I++TDASD
Sbjct: 496 PNLAQMRQPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASD 555
Query: 300 HYWA----AVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGH 355
YW A+ I E +L C + SG FK AE +YH+ KETLAV IK+F +L
Sbjct: 556 DYWGGMLKAIKINEGTNTELICRYRSGSFKAAERNYHSNDKETLAVINTIKKFSIYLTPV 615
Query: 356 HFEVQIDNSSFPKMLEFKNKTPPN-PQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
HF ++ DN+ F + K + +R + W S Y F V+H++G+ N D+LSR
Sbjct: 616 HFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSR 673
>UniRef100_Q8JTA2 , complete genome [Mirabilis mosaic virus]
Length = 674
Score = 294 bits (752), Expect = 4e-78
Identities = 157/421 (37%), Positives = 248/421 (58%), Gaps = 5/421 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + + + + ++L+ +I P+KS+ AF V K +E+ R KKR+VV+Y+ LN+
Sbjct: 248 SPEDRKEFEIQIKELLDLKVIIPSKSQHMSPAFLVEKEAEKRRGKKRMVVNYKKLNEVTI 307
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D LP + L + ++ +IFS FD KSGFWQ+ LD + + TAF P HYQW V+PF
Sbjct: 308 GDSHNLPNMQELITLLRGKNIFSSFDCKSGFWQVLLDDESQKLTAFTCPQGHYQWRVVPF 367
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M + SLVY+DD+++FS ++ H+ + + G++LS
Sbjct: 368 GLKQAPSIFQRHMQDALRGLEEFSLVYVDDIIVFSDNKNDHQDHVMKVLRRIESLGIILS 427
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+ + ++++FLG+ G PQ HI++ + FPD KQ+Q+FLG+L Y +I
Sbjct: 428 KKKANLFKEKINFLGLEIDRGTHTPQNHILDHIHTFPDRIEDKKQLQRFLGVLTYADSYI 487
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L+K+ W + T+ +K +KK N P L +P + I++TDASD
Sbjct: 488 PKLAEKRKPLQVKLKKDQVWIWTQSDTDYVKKIKKGLINFPKLYLPKKEDSLIIETDASD 547
Query: 300 HYWAAVL-IEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFE 358
H+W VL + +G++L C ++SG FK AEL+YH+ KE LAVK I +F +L F
Sbjct: 548 HFWGGVLKAQTTEGEELICRYSSGTFKPAELNYHSNEKELLAVKQVITKFSIYLTPVCFT 607
Query: 359 VQIDNSSFPKMLEFKNKTPPNPQ--ILRLKEWFSLYDFSVKHVQGSKNLIPDYLSRPKQQ 416
V+ DN + K K T + Q ++R + WFS Y F V H++G +N++ DYL+R
Sbjct: 608 VRTDNVNLLKGFMNKKITGDSKQGRLIRWQMWFSHYTFKVDHLKGEQNVLADYLTREFHN 667
Query: 417 G 417
G
Sbjct: 668 G 668
>UniRef100_P09523 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Figwort mosaic virus]
Length = 666
Score = 291 bits (745), Expect = 2e-77
Identities = 154/415 (37%), Positives = 241/415 (57%), Gaps = 4/415 (0%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + ++ ++L+ GLI P+KS+ AF V +E+ R KKR+VV+Y+ +NQ
Sbjct: 248 SPQDREGFAKQIKELLDLGLIIPSKSQHMSPAFLVENEAERRRGKKRMVVNYKAINQATI 307
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D LP + L + ++ IFS FD KSGFWQ+ LD + + TAF P H+QW V+PF
Sbjct: 308 GDSHNLPNMQELLTLLRGKSIFSSFDCKSGFWQVVLDEESQKLTAFTCPQGHFQWKVVPF 367
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M +VY+DD+++FS+ E H + I K+G++LS
Sbjct: 368 GLKQAPSIFQRHMQTALNGADKFCMVYVDDIIVFSNSELDHYNHVYAVLKIVEKYGIILS 427
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+ + ++++FLG+ +G PQ HI+E + KFPD K +Q+FLG+L Y +I
Sbjct: 428 KKKANLFKEKINFLGLEIDKGTHCPQNHILENIHKFPDRLEDKKHLQRFLGVLTYAETYI 487
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P +++ L L+K+ W + ++ +K +KK + P L +P + I++TDASD
Sbjct: 488 PKLAEIRKPLQVKLKKDVTWNWTQSDSDYVKKIKKNLGSFPKLYLPKPEDHLIIETDASD 547
Query: 300 HYWAAVL-IEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFE 358
+W VL +DG +L C ++SG FK AE +YH+ KE LAVK I +F +L F
Sbjct: 548 SFWGGVLKARALDGVELICRYSSGSFKQAEKNYHSNDKELLAVKQVITKFSAYLTPVRFT 607
Query: 359 VQIDNSSFPKMLEFKNK-TPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
V+ DN +F L K +++R + WFS Y F V+H++G KN++ D L+R
Sbjct: 608 VRTDNKNFTYFLRINLKGDSKQGRLVRWQNWFSKYQFDVEHLEGVKNVLADCLTR 662
>UniRef100_Q5W1F1 Polyprotein [Carnation etched ring virus]
Length = 656
Score = 276 bits (706), Expect = 8e-73
Identities = 148/412 (35%), Positives = 235/412 (56%), Gaps = 3/412 (0%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+PS+ ++ ++ + +I+P+KS AF V +E+ R KKR+VV+Y+ +N+ K
Sbjct: 236 SPSDREEFDKQIKEPLDLKVIKPSKSPHMSPAFLVENEAERRRGKKRMVVNYKAMNKATK 295
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D LP L + ++ I+S FD KSGFWQ+ LD + + TAF P HYQW V+PF
Sbjct: 296 GDAHNLPNKDELLALVRGKKIYSSFDCKSGFWQVLLDKESQLLTAFTCPQGHYQWNVVPF 355
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK APS+FQ+ M F VY+DD+L+FS+ EE H + C K G++LS
Sbjct: 356 GLKQAPSIFQRHMQTAFNQHSKYCCVYVDDILVFSNTEEEHYIHVLNILRRCEKLGIILS 415
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K+Q+ ++++FLG+ +G PQ HI+E + KFPD KQ+Q+ LGIL Y D+I
Sbjct: 416 KKKAQLFKEKINFLGLEIDQGTHCPQNHILEHIHKFPDRIEDKKQLQRSLGILTYASDYI 475
Query: 242 PHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
P ++ L L++++ W ++ + +KK ++ P L P + K +++TDAS+
Sbjct: 476 PKLASIRKPLQSKLKEDSTWTWNDTDSQYMAKIKKNLKSFPKLYHPEPNDKLVIETDASE 535
Query: 300 HYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEV 359
+W +L + + C +ASG FK AE +YH+ KE LAV I +F +L F +
Sbjct: 536 EFWGGILKAIHNSHEYICRYASGSFKAAERNYHSNEKELLAVFRVIYKFSIYLTPCRFLI 595
Query: 360 QIDNSSFPKMLEFKNK-TPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYL 410
+ DN +F + K +++R + W S YDF V+H+ G+KN+ D+L
Sbjct: 596 RSDNKNFTHFVNINLKGDRKQGRLVRWQMWLSQYDFDVEHIAGTKNVFADFL 647
>UniRef100_P05400 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Carnation etched ring virus]
Length = 659
Score = 267 bits (682), Expect = 5e-70
Identities = 146/414 (35%), Positives = 233/414 (56%), Gaps = 5/414 (1%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+PS+ + ++L++ +I+P+KS AF V +E+ R KKR+VV+Y+ +N+ K
Sbjct: 237 SPSDREEFDRQIKELLELKVIKPSKSTHMSPAFLVENEAERRRGKKRMVVNYKAMNKATK 296
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D LP L + ++ I+S FD KSG WQ+ LD + + TAF P HYQW V+PF
Sbjct: 297 GDAHNLPNKDELLTLVRGKKIYSSFDCKSGLWQVLLDKESQLLTAFTCPQGHYQWNVVPF 356
Query: 122 GLKVAPSLFQKAMTKIF-EPILHTSLVYIDDVLLFSH-DEESHKKFLQQFYDICSKHGVM 179
GLK APS+F K VY+DD+L+FS+ + H + C K G++
Sbjct: 357 GLKQAPSIFPKTYANSHSNQYSKYCCVYVDDILVFSNTGRKEHYIHVLNILRRCEKLGII 416
Query: 180 LSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRD 239
LS +K+Q+ ++++FLG+ +G PQ HI+E + KFPD KQ+Q+FLGIL Y D
Sbjct: 417 LSKKKAQLFKEKINFLGLEIDQGTHCPQNHILEHIHKFPDRIEDKKQLQRFLGILTYASD 476
Query: 240 FIPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDA 297
+IP ++ L L++++ W ++ + +KK ++ P L P + K +++TDA
Sbjct: 477 YIPKLASIRKPLQSKLKEDSTWTWNDTDSQYMAKIKKNLKSFPKLYHPEPNDKLVIETDA 536
Query: 298 SDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHF 357
S+ +W +L + + C +ASG FK AE +YH+ KE LAV IK+F +L F
Sbjct: 537 SEEFWGGILKAIHNSHEYICRYASGSFKAAERNYHSNEKELLAVIRVIKKFSIYLTPSRF 596
Query: 358 EVQIDNSSFPKMLEFKNK-TPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYL 410
++ DN +F + K +++R + W S YDF V+H+ G+KN+ D+L
Sbjct: 597 LIRTDNKNFTHFVNINLKGDRKQGRLVRWQMWLSQYDFDVEHIAGTKNVFADFL 650
>UniRef100_Q88442 ORF V protein [Strawberry vein banding virus]
Length = 708
Score = 253 bits (646), Expect = 7e-66
Identities = 142/418 (33%), Positives = 229/418 (53%), Gaps = 9/418 (2%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
+P + + + ++L+K G+I P+KS + AF V +E R K R+V++Y+ LN K
Sbjct: 287 SPQDREEFKTQIEELLKLGIIRPSKSPHSSPAFMVRNHAEIKRGKARMVINYKKLNDHTK 346
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
D + LP L I +S FD KSGFWQ+ L P+ TAF P HY+W V+PF
Sbjct: 347 GDGYLLPNKEQLLQRIGGKTFYSSFDCKSGFWQVRLAPETIQLTAFSCPQGHYEWLVMPF 406
Query: 122 GLKVAPSLFQKAMTKIFEPIL-HTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVML 180
GLK AP++FQ+ M + + VY+DD+++FS EE H ++ + C G++L
Sbjct: 407 GLKQAPAIFQRHMDESLSNMYPQFCAVYVDDIIVFSKTEEEHLGHVKIVLNRCKALGIVL 466
Query: 181 SSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDF 240
S +K+Q+ ++FLG+ G Q HI L FPD+ +Q+FLG+LNYI +
Sbjct: 467 SKKKAQLCKTTINFLGLVIERGNLKVQSHIGLHLVAFPDQLSDRNALQRFLGLLNYISAY 526
Query: 241 IPHVSKYTSKLSKMLRKNAP-PWGKEQTEAIKFLKKAAQNPPPLKIPG-DGKRILQTDAS 298
P ++ S L L+K W ++ TE ++ +K + P L P + K I++ DAS
Sbjct: 527 FPKIANLRSPLQVKLKKEITWSWTEKDTETVRKIKSLVKTLPDLYNPSPEDKPIIECDAS 586
Query: 299 DHYWAAVLIEEI-DGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHF 357
D +W A+L ++ +G+++ C +ASG FK AE +YH+ KE L++ IK F ++ + F
Sbjct: 587 DDHWGAILKAKLPEGKEVICRYASGTFKPAEKNYHSNEKEILSIIKAIKAFRAYILPYKF 646
Query: 358 EVQIDNSSFPKMLEFKNKTPPNPQILRLKEW---FSLYDFSVKHVQGSKNLIPDYLSR 412
V+ DN++ + + + + RL W Y F ++HV G KN++ D ++R
Sbjct: 647 LVRTDNTNAAYFV--RTNIAGDYKQSRLVRWQMALREYSFDIEHVSGQKNVLADIMTR 702
>UniRef100_Q7TD08 Putative multifunctional pol protein [Cestrum yellow leaf curling
virus]
Length = 643
Score = 238 bits (607), Expect = 2e-61
Identities = 152/429 (35%), Positives = 236/429 (54%), Gaps = 17/429 (3%)
Query: 5 ELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDK 64
++ +EC L+++G+IE +KS + AFYV +E R+K+R+V++Y+ LN+ +
Sbjct: 214 DINEFSQECADLVRKGIIEESKSPHSAPAFYVENHNEIKRKKRRMVINYKALNKATIGNA 273
Query: 65 FPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAF-CIPNAHYQWTVLPFGL 123
LP+I S+ + +K ++ FS D KSG+WQL L P+ +P TAF C P HYQW VLPFGL
Sbjct: 274 HKLPRIDSILTKVKGSNWFSTLDAKSGYWQLRLHPQSKPLTAFSCPPQKHYQWNVLPFGL 333
Query: 124 KVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSH-DEESHKKFLQQFYDICSKHGVMLSS 182
K AP ++Q M K E + + L YIDD+L+F++ E H L + C + G++LS
Sbjct: 334 KQAPGIYQNFMDKNLEGLENFCLAYIDDILVFTNSSREEHLSKLLVVLERCKEKGLILSK 393
Query: 183 RKSQIATKEVDFLGMHFTE-GKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRD-- 239
+K+ IA + +DFLG+ E G+ QP+++E+L FPD KQ+Q+FLG LNYI D
Sbjct: 394 KKAIIARQTIDFLGLTLQENGEIKLQPNVLEKLELFPDAIEDRKQLQRFLGCLNYIADKG 453
Query: 240 FIPHVSKYTSKL-SKMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDA 297
F+ ++K T L K+ N W ++ + +KK ++ PL P + I++TDA
Sbjct: 454 FLKEIAKETKNLYPKVSITNPWHWSDLDSKLVNQIKKKCKDLSPLYFPKPEDYLIIETDA 513
Query: 298 SDHYWAAVL-IEEI---DGQK-----LYCGHASGQFKDAELHYHTTFKETLAVKYGIKRF 348
S WA L E+ G K C + SG F AE Y KETLA ++++
Sbjct: 514 SGDTWAGCLKAAELLFPKGTKNKVVERLCKYTSGIFSSAEQKYTVHEKETLAALKTMRKW 573
Query: 349 DFHLRGHHFEVQIDNSSFPKMLEFKNKTPPNP-QILRLKEWFSLYDFSVKHVQGSKNLIP 407
L F ++ D+S K N +++R + F Y V++++G KN +
Sbjct: 574 KAELLPKEFTLRTDSSYVTGFARHNLKANYNQGRLVRWQLEFLQYPARVEYIKGEKNSLA 633
Query: 408 DYLSRPKQQ 416
D L+R +Q
Sbjct: 634 DTLTREWKQ 642
>UniRef100_Q89703 ORF 3 [Cassava vein mosaic virus]
Length = 652
Score = 235 bits (599), Expect = 2e-60
Identities = 143/421 (33%), Positives = 231/421 (53%), Gaps = 29/421 (6%)
Query: 14 QQLIKQGLIEPTKS----EWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFPLPK 69
+++IK+G IE + +++ AF VNK SEQ R K R+V+DY+ LN+ K K+P+P
Sbjct: 224 EEMIKEGFIEEKTNFEDKKYSSPAFIVNKHSEQKRGKTRMVIDYKDLNKKAKVVKYPIPN 283
Query: 70 IYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSL 129
+L A +SKFD KSGF+ + L+ + TAF +P +YQW VLPFG +PS+
Sbjct: 284 KDTLIHRSIQARYYSKFDCKSGFYHIKLEEDSKKYTAFTVPQGYYQWKVLPFGYHNSPSI 343
Query: 130 FQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIAT 189
FQ+ M +IF P +VYIDD+L+FS E HK + +F DI +G+++S +K+++
Sbjct: 344 FQQFMDRIFRPYYDFIIVYIDDILVFSKTIEEHKIHIAKFRDITLANGLIISKKKTELCK 403
Query: 190 KEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTS 249
+++DFLG+ +G QPHI+ ++ + + ++Q LG+LN IR FIPH+++
Sbjct: 404 EKIDFLGVQIEQGGIELQPHIINKILEKHTKIKNKTELQSILGLLNQIRHFIPHLAQILL 463
Query: 250 KLSKMLR---KNAPPWGKEQTEAIKFLKKAAQN-PPPLKIP---GDGKRILQTDASDHYW 302
+ K L+ + W KE E IK ++ ++N +K P D I++ DAS++ +
Sbjct: 464 PIQKKLKIKDEEIWTWTKEDEEKIKLIQDYSKNLVIKMKYPINKEDMNWIIEVDASNNAY 523
Query: 303 AAVLIEEIDGQKL--YCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQ 360
+ L + K+ C + SG FK+ E Y KE +AV G++ + + V+
Sbjct: 524 GSCLKYKPKNSKIEYLCRYNSGTFKENEQKYDINRKELIAVYQGLQSYSLFTCEGNKLVR 583
Query: 361 IDNSSF---------PKMLEFKNKTPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYLS 411
DNS K +EF+N I L ++Y+F ++ + G N+I DYLS
Sbjct: 584 TDNSQVYYWIKNDTNKKSIEFRN-------IKYLLAKIAVYNFEIQLIDGKTNIIADYLS 636
Query: 412 R 412
R
Sbjct: 637 R 637
>UniRef100_Q8H6Q8 CTV.20 [Poncirus trifoliata]
Length = 3148
Score = 232 bits (591), Expect = 2e-59
Identities = 121/287 (42%), Positives = 180/287 (62%), Gaps = 10/287 (3%)
Query: 7 RAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFP 66
R +EE L+K+GLI ++S W+C AFYVNK SE R RLV++Y+PLN+ +K ++P
Sbjct: 2620 RHCKEEINDLVKKGLIVKSRSPWSCAAFYVNKNSEIERGTPRLVINYKPLNKALKWIRYP 2679
Query: 67 LPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVA 126
+P L + +A IFSKFDMKSGFWQ+ + PKDR KTAF +P Y+WTV+PFGLK A
Sbjct: 2680 IPNKKDLLQKLHSAFIFSKFDMKSGFWQIQIHPKDRYKTAFTVPFGQYEWTVMPFGLKNA 2739
Query: 127 PSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQ 186
PS FQ+ M I+ P +VYIDDVL+FS+ + H K L+ FY K G+ +S+ K
Sbjct: 2740 PSEFQRIMNDIYNPYSDFCIVYIDDVLIFSNSIDQHFKHLKTFYFATRKAGLAISNSKVS 2799
Query: 187 IATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSK 246
+ ++ FLG H ++G P + KFPD+ L Q+Q+FLG LNY+ DF P++S+
Sbjct: 2800 LFQTKIRFLGHHISKGTITPIERSLAFADKFPDKILDKTQLQRFLGSLNYVLDFCPNISR 2859
Query: 247 YT-SKLSKMLRKNAPPWGKEQ-------TEAIKFLKKAAQNPPPLKI 285
++ S++S ++ PP +++ T+++K K++ P K+
Sbjct: 2860 FSNSRISS--KQIMPPKRRDKGKGIAKDTDSLKPSKESQSTPSKEKL 2904
Score = 229 bits (584), Expect = 1e-58
Identities = 120/289 (41%), Positives = 176/289 (60%), Gaps = 10/289 (3%)
Query: 7 RAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFP 66
R +EE L+K+GLI ++S W+C AFYVNK SE R RLV++Y+PLN+ +K ++P
Sbjct: 1155 RHCKEEINDLVKKGLIVKSRSPWSCAAFYVNKNSEIERGTPRLVINYKPLNKALKWIRYP 1214
Query: 67 LPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVA 126
+P L + +A IFSKFDMKS FWQ+ + PK+R KTAF +P Y+WTV+PFGLK A
Sbjct: 1215 IPNKKDLLQKLHSAFIFSKFDMKSVFWQIQIHPKERYKTAFTVPFGQYEWTVMPFGLKNA 1274
Query: 127 PSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQ 186
PS FQ+ M I+ P +VYIDDVL+FS+ + H K L+ FY K G+ +S+ K
Sbjct: 1275 PSEFQRIMNDIYNPYSDFCIVYIDDVLIFSNSIDQHFKHLKTFYFATRKAGLAISNSKVS 1334
Query: 187 IATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSK 246
+ ++ FLG H ++G P + KFPD+ L Q+Q+FLG LNY+ DF P++S+
Sbjct: 1335 LFQTKIRFLGHHISKGTITPIERSLAFADKFPDKILDKTQLQRFLGSLNYVLDFCPNISR 1394
Query: 247 YTSKLSKMLRKN---APPWGKEQ-------TEAIKFLKKAAQNPPPLKI 285
+ L L+K PP +++ T+++K K++ P K+
Sbjct: 1395 LSKPLHDRLKKKPAAMPPKRRDKGKGIAKDTDSLKPSKESQSTPSKEKL 1443
>UniRef100_Q7XRB3 OSJNBa0006B20.7 protein [Oryza sativa]
Length = 685
Score = 228 bits (581), Expect = 2e-58
Identities = 136/421 (32%), Positives = 225/421 (53%), Gaps = 12/421 (2%)
Query: 2 TPSELRAAQEECQQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIK 61
TP ++ + ++L+K G I ++S AF V +E+VR K R+V++Y+ LN
Sbjct: 189 TPKDIEEFKMHIEELLKLGAIRESRSPHRSAAFIVRNHAEEVRGKSRMVINYKRLNNNTI 248
Query: 62 DDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPF 121
+D + +P + I+ + FSKFD+K+GFWQ+ + + TAF P HY+W V+P
Sbjct: 249 EDAYNIPNKQEWINRIQGSKYFSKFDLKAGFWQVKMAEESIEWTAFTCPQGHYEWLVMPL 308
Query: 122 GLKVAPSLFQKAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLS 181
GLK AP++FQ+ M IF LVYIDD+L+FS + H L+ F ++G++LS
Sbjct: 309 GLKNAPAIFQRKMQNIFNENQEFILVYIDDLLVFSRTYKEHIAHLEIFIRKVEQNGLILS 368
Query: 182 SRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFI 241
+K +I ++++FLG EGK Q HI +++ +FPD K++QQFLG++NY R+ I
Sbjct: 369 KKKMEICKEKINFLGHEIGEGKIHLQEHIAKKILQFPDAMNDKKKLQQFLGLVNYARNHI 428
Query: 242 PHVSKYTSKLSKMLRKNAPPW-GKEQTEAIKFLKKAAQNPPPLKIP-GDGKRILQTDASD 299
+++K L LRKN + E + ++ +K+ + PL++P + I++TDAS
Sbjct: 429 NNLAKLAGPLYAKLRKNGQRYFNSEDIKLVRLIKERVKELKPLELPLEESYFIIETDASQ 488
Query: 300 HYWAAVLIE-----EIDGQKLYCGHASGQFKDAELHYHTTFKETLAVKYGIKRFDFHLRG 354
H W A+L + ++ C +ASG +K + + T +E +A+ I F +L
Sbjct: 489 HGWGAILKQRPTKFSAKSEEKICRYASGSYKLKTV--NNTDREIMAIINAINAFRLYLGF 546
Query: 355 HHFEVQIDNSSFPKMLEFKNKTPPNPQILRLKEWF---SLYDFSVKHVQGSKNLIPDYLS 411
F V D + + N + + L E + Y +H++G N +PD LS
Sbjct: 547 KEFTVITDCEAICRYYNQLNSKKSSTRRWVLFEDIITGNGYKVIFEHIKGKDNNLPDMLS 606
Query: 412 R 412
R
Sbjct: 607 R 607
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 745,849,344
Number of Sequences: 2790947
Number of extensions: 32579591
Number of successful extensions: 132126
Number of sequences better than 10.0: 29036
Number of HSP's better than 10.0 without gapping: 27847
Number of HSP's successfully gapped in prelim test: 1189
Number of HSP's that attempted gapping in prelim test: 72961
Number of HSP's gapped (non-prelim): 29736
length of query: 418
length of database: 848,049,833
effective HSP length: 130
effective length of query: 288
effective length of database: 485,226,723
effective search space: 139745296224
effective search space used: 139745296224
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0490.11


