

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.9
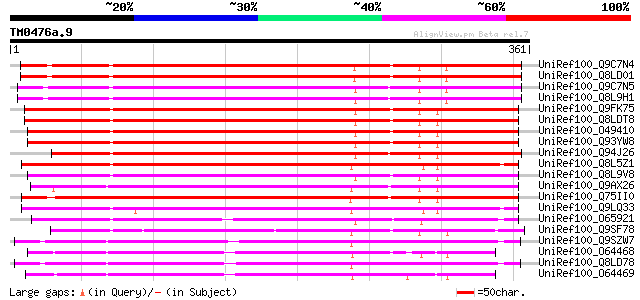
(361 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C7N4 Lipase/hydrolase, putative; 118270-120144 [Arab... 301 2e-80
UniRef100_Q8LD01 Lipase/hydrolase, putative [Arabidopsis thaliana] 301 2e-80
UniRef100_Q9C7N5 Lipase/hydrolase, putative; 114382-116051 [Arab... 291 1e-77
UniRef100_Q8L9H1 Lipase/hydrolase, putative [Arabidopsis thaliana] 291 1e-77
UniRef100_Q9FK75 GDSL-motif lipase/hydrolase-like protein [Arabi... 282 1e-74
UniRef100_Q8LDT8 GDSL-motif lipase/hydrolase-like protein [Arabi... 282 1e-74
UniRef100_O49410 Hypothetical protein AT4g18970 [Arabidopsis tha... 281 3e-74
UniRef100_Q93YW8 Hypothetical protein At4g18970 [Arabidopsis tha... 281 3e-74
UniRef100_Q94J26 Putative GDSL-motif lipase/hydrolase-like prote... 279 8e-74
UniRef100_Q8L5Z1 Hypothetical protein At1g33810 [Arabidopsis tha... 279 1e-73
UniRef100_Q8L9V8 GDSL-motif lipase/hydrolase-like protein [Arabi... 277 3e-73
UniRef100_Q9AX26 GDSL-motif lipase/hydrolase-like protein [Oryza... 276 8e-73
UniRef100_Q75II0 Putative GDSL-like lipase/hydrolase [Oryza sativa] 275 1e-72
UniRef100_Q9LQ33 F14M2.7 protein [Arabidopsis thaliana] 266 9e-70
UniRef100_O65921 Putative GDSL-motif lipase/hydrolase [Arabidops... 260 5e-68
UniRef100_Q9SF78 Putative GDSL-motif lipase/hydrolase; 24593-266... 242 1e-62
UniRef100_Q9SZW7 Hypothetical protein F6G3.170 [Arabidopsis thal... 241 3e-62
UniRef100_O64468 Putative GDSL-motif lipase/hydrolase [Arabidops... 239 1e-61
UniRef100_Q8LD78 Putative GDSL-motif lipase/hydrolase [Arabidops... 238 3e-61
UniRef100_O64469 Putative GDSL-motif lipase/hydrolase [Arabidops... 232 1e-59
>UniRef100_Q9C7N4 Lipase/hydrolase, putative; 118270-120144 [Arabidopsis thaliana]
Length = 363
Score = 301 bits (771), Expect = 2e-80
Identities = 155/361 (42%), Positives = 227/361 (61%), Gaps = 18/361 (4%)
Query: 8 WLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDF 67
W V+ +L+ S+++ ++QVPC FVFGDSL D+GNNN L + A++N+ PYGIDF
Sbjct: 8 WCVVLVLLCFGFSVVKAQA---QAQVPCFFVFGDSLVDNGNNNGLISIARSNYFPYGIDF 64
Query: 68 PATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQ 127
PTGR+SNG+ +D IA++LGF +IP + ++G IL GVNYAS +AGIR+E+G Q
Sbjct: 65 GG--PTGRFSNGKTTVDVIAELLGFNGYIPAYNTVSGRQILSGVNYASAAAGIREETGRQ 122
Query: 128 LGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFN 187
LG ++ Q+ +++ VS++ LG +A YL +C+Y V +G+ND+ NYF+P ++
Sbjct: 123 LGQRISFSGQVRNYQTTVSQVVQLLGDETRAADYLKRCIYSVGLGSNDYLNNYFMPTFYS 182
Query: 188 TSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNG----SCVEE 243
+SR +TP+QYA LI + S L L+++GARK L G+ +GC P L +CV+
Sbjct: 183 SSRQFTPEQYANDLISRYSTQLNALYNYGARKFALSGIGAVGCSPNALAGSPDGRTCVDR 242
Query: 244 KNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCCPT 298
N A +FN++L+SLVD+LN D+KFI+IN+ I D GF VT+ GCC
Sbjct: 243 INSANQIFNNKLRSLVDQLNNN-HPDAKFIYINAYGIFQDMITNPARFGFRVTNAGCCGI 301
Query: 299 NEKG---KCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLI 355
C+ PC++R+ YVFWD H TEAAN++ A SYN+ + + AYP +I L
Sbjct: 302 GRNAGQITCLPGQRPCRDRNAYVFWDAFHPTEAANVIIARRSYNAQSASDAYPMDISRLA 361
Query: 356 Q 356
Q
Sbjct: 362 Q 362
>UniRef100_Q8LD01 Lipase/hydrolase, putative [Arabidopsis thaliana]
Length = 363
Score = 301 bits (771), Expect = 2e-80
Identities = 155/361 (42%), Positives = 227/361 (61%), Gaps = 18/361 (4%)
Query: 8 WLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDF 67
W V+ +L+ S+++ ++QVPC FVFGDSL D+GNNN L + A++N+ PYGIDF
Sbjct: 8 WCVVLVLLCFGFSVVKAQA---QAQVPCFFVFGDSLVDNGNNNGLISIARSNYFPYGIDF 64
Query: 68 PATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQ 127
PTGR+SNG+ +D IA++LGF +IP + ++G IL GVNYAS +AGIR+E+G Q
Sbjct: 65 GG--PTGRFSNGKTTVDVIAELLGFNGYIPAYNTVSGRQILSGVNYASAAAGIREETGRQ 122
Query: 128 LGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFN 187
LG ++ Q+ +++ VS++ LG +A YL +C+Y V +G+ND+ NYF+P ++
Sbjct: 123 LGQRISFSGQVRNYQTTVSQVVQLLGDETRAADYLKRCIYSVGLGSNDYLNNYFMPTFYS 182
Query: 188 TSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNG----SCVEE 243
+SR +TP+QYA LI + S L L+++GARK L G+ +GC P L +CV+
Sbjct: 183 SSRQFTPEQYANDLISRYSTQLNALYNYGARKFALSGIGSVGCSPNALAGSPDGRTCVDR 242
Query: 244 KNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCCPT 298
N A +FN++L+SLVD+LN D+KFI+IN+ I D GF VT+ GCC
Sbjct: 243 INSANQIFNNKLRSLVDQLNNN-HPDAKFIYINAYGIFQDMITNPARFGFRVTNAGCCGI 301
Query: 299 NEKG---KCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLI 355
C+ PC++R+ YVFWD H TEAAN++ A SYN+ + + AYP +I L
Sbjct: 302 GRNAGQITCLPGQRPCRDRNAYVFWDAFHPTEAANVIIARRSYNAQSASDAYPMDISRLA 361
Query: 356 Q 356
Q
Sbjct: 362 Q 362
>UniRef100_Q9C7N5 Lipase/hydrolase, putative; 114382-116051 [Arabidopsis thaliana]
Length = 364
Score = 291 bits (746), Expect = 1e-77
Identities = 152/364 (41%), Positives = 220/364 (59%), Gaps = 19/364 (5%)
Query: 6 KTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGI 65
+ W ++S+ V+++ + V E QVPC F+FGDSL D+GNNN L + A+A++ PYGI
Sbjct: 6 RKWCLVSVWVLLLGLGFK---VKAEPQVPCYFIFGDSLVDNGNNNRLRSIARADYFPYGI 62
Query: 66 DFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESG 125
DF PTGR+SNGR +D + ++LGF +IP ++ ++G +IL+GVNYAS +AGIR+E+G
Sbjct: 63 DFGG--PTGRFSNGRTTVDVLTELLGFDNYIPAYSTVSGQEILQGVNYASAAAGIREETG 120
Query: 126 SQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNV 185
+QLG + Q+ +++ V+++ LG A YL +C+Y V +G+ND+ NYF+P
Sbjct: 121 AQLGQRITFSGQVENYKNTVAQVVEILGDEYTAADYLKRCIYSVGMGSNDYLNNYFMPQF 180
Query: 186 FNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----C 240
++TSR YTP+QYA LI + L L+++GARK LVG+ +GC P L GS C
Sbjct: 181 YSTSRQYTPEQYADDLISRYRDQLNALYNYGARKFALVGIGAIGCSPNALAQGSQDGTTC 240
Query: 241 VEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGC 295
VE N A +FN++L S+V +LN D+ F +IN+ D + GFT T+ C
Sbjct: 241 VERINSANRIFNNRLISMVQQLN-NAHSDASFTYINAYGAFQDIIANPSAYGFTNTNTAC 299
Query: 296 CPTNEKG---KCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIK 352
C G C+ PC NR EYVFWD H + AAN A SYN+ + YP +I
Sbjct: 300 CGIGRNGGQLTCLPGEPPCLNRDEYVFWDAFHPSAAANTAIAKRSYNAQRSSDVYPIDIS 359
Query: 353 HLIQ 356
L Q
Sbjct: 360 QLAQ 363
>UniRef100_Q8L9H1 Lipase/hydrolase, putative [Arabidopsis thaliana]
Length = 364
Score = 291 bits (746), Expect = 1e-77
Identities = 152/364 (41%), Positives = 220/364 (59%), Gaps = 19/364 (5%)
Query: 6 KTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGI 65
+ W ++S+ V+++ + V E QVPC F+FGDSL D+GNNN L + A+A++ PYGI
Sbjct: 6 RKWCLVSVWVLLLGLGFK---VKAEPQVPCYFIFGDSLVDNGNNNRLRSIARADYFPYGI 62
Query: 66 DFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESG 125
DF PTGR+SNGR +D + ++LGF +IP ++ ++G +IL+GVNYAS +AGIR+E+G
Sbjct: 63 DFGG--PTGRFSNGRTTVDVLTELLGFDNYIPAYSTVSGQEILQGVNYASAAAGIREETG 120
Query: 126 SQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNV 185
+QLG + Q+ +++ V+++ LG A YL +C+Y V +G+ND+ NYF+P
Sbjct: 121 AQLGQRITFSGQVENYKNTVAQVVEILGDEYTAADYLKRCIYSVGMGSNDYLNNYFMPQX 180
Query: 186 FNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----C 240
++TSR YTP+QYA LI + L L+++GARK LVG+ +GC P L GS C
Sbjct: 181 YSTSRQYTPEQYADDLISRYRDQLNALYNYGARKFALVGIGAIGCSPNALAQGSEDGTTC 240
Query: 241 VEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGC 295
VE N A +FN++L S+V +LN D+ F +IN+ D + GFT T+ C
Sbjct: 241 VERINSANRIFNNRLISMVQQLN-NAHSDASFTYINAYGAFQDIITNPSAYGFTNTNTAC 299
Query: 296 CPTNEKG---KCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIK 352
C G C+ PC NR EYVFWD H + AAN A SYN+ + YP +I
Sbjct: 300 CGIGRNGGQLTCLPGEPPCLNRDEYVFWDAFHPSAAANTAIAKRSYNAQRSSDVYPIDIS 359
Query: 353 HLIQ 356
L Q
Sbjct: 360 QLAQ 363
>UniRef100_Q9FK75 GDSL-motif lipase/hydrolase-like protein [Arabidopsis thaliana]
Length = 362
Score = 282 bits (721), Expect = 1e-74
Identities = 147/358 (41%), Positives = 219/358 (61%), Gaps = 16/358 (4%)
Query: 11 LSLLVVMV-ASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPA 69
+SL+++M+ ++ + + PC F+FGDSL D+GNNN L + A+AN+ PYGIDF A
Sbjct: 4 MSLMIMMIMVAVTMINIAKSDPIAPCYFIFGDSLVDNGNNNQLQSLARANYFPYGIDFAA 63
Query: 70 TFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLG 129
PTGR+SNG +D IAQ+LGF+++I P+A+ G DIL+GVNYAS +AGIR E+G QLG
Sbjct: 64 G-PTGRFSNGLTTVDVIAQLLGFEDYITPYASARGQDILRGVNYASAAAGIRDETGRQLG 122
Query: 130 HNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTS 189
+ Q+ +H VS++ + LG ++A+ YL++C+Y + +G+ND+ NYF+P ++T
Sbjct: 123 GRIAFAGQVANHVNTVSQVVNILGDQNEASNYLSKCIYSIGLGSNDYLNNYFMPTFYSTG 182
Query: 190 RMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----CVEEK 244
++P+ YA L+ + + L+ L+ GARK L+G+ +GC P L S C E
Sbjct: 183 NQFSPESYADDLVARYTEQLRVLYTNGARKFALIGVGAIGCSPNELAQNSRDGRTCDERI 242
Query: 245 NVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC--- 296
N A +FN +L S+VD N+ D+KF +IN+ I D GF VT+ GCC
Sbjct: 243 NSANRIFNSKLISIVDAFNQN-TPDAKFTYINAYGIFQDIITNPARYGFRVTNAGCCGVG 301
Query: 297 PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHL 354
N + C+ PC NR+EYVFWD H EAAN+V S+ + A+P +I+ L
Sbjct: 302 RNNGQITCLPGQAPCLNRNEYVFWDAFHPGEAANIVIGRRSFKREAASDAHPYDIQQL 359
>UniRef100_Q8LDT8 GDSL-motif lipase/hydrolase-like protein [Arabidopsis thaliana]
Length = 362
Score = 282 bits (721), Expect = 1e-74
Identities = 147/358 (41%), Positives = 219/358 (61%), Gaps = 16/358 (4%)
Query: 11 LSLLVVMV-ASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPA 69
+SL+++M+ ++ + + PC F+FGDSL D+GNNN L + A+AN+ PYGIDF A
Sbjct: 4 MSLMIMMIMVAVTMINIAKSDPIAPCYFIFGDSLVDNGNNNQLQSLARANYFPYGIDFAA 63
Query: 70 TFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLG 129
PTGR+SNG +D IAQ+LGF+++I P+A+ G DIL+GVNYAS +AGIR E+G QLG
Sbjct: 64 G-PTGRFSNGLTTVDVIAQLLGFEDYITPYASARGQDILRGVNYASAAAGIRDETGRQLG 122
Query: 130 HNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTS 189
+ Q+ +H VS++ + LG ++A+ YL++C+Y + +G+ND+ NYF+P ++T
Sbjct: 123 GRIAFAGQVANHVNTVSQVVNILGDQNEASNYLSKCIYSIGLGSNDYLNNYFMPTFYSTG 182
Query: 190 RMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----CVEEK 244
++P+ YA L+ + + L+ L+ GARK L+G+ +GC P L S C E
Sbjct: 183 NQFSPESYADDLVARYTEQLRVLYTNGARKFALIGVGAIGCSPNELAQNSRDGRTCDERI 242
Query: 245 NVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC--- 296
N A +FN +L S+VD N+ D+KF +IN+ I D GF VT+ GCC
Sbjct: 243 NSANRIFNSKLISIVDAFNQN-TPDAKFTYINAYGIFQDIITNPARYGFRVTNAGCCGVG 301
Query: 297 PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHL 354
N + C+ PC NR+EYVFWD H EAAN+V S+ + A+P +I+ L
Sbjct: 302 RNNGQITCLPGQAPCLNRNEYVFWDAFHPGEAANIVIGRRSFKREAASNAHPYDIQQL 359
>UniRef100_O49410 Hypothetical protein AT4g18970 [Arabidopsis thaliana]
Length = 626
Score = 281 bits (718), Expect = 3e-74
Identities = 146/355 (41%), Positives = 214/355 (60%), Gaps = 15/355 (4%)
Query: 13 LLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFP 72
++++ +A M + +G+ PC F+FGDSL DSGNNN L + A+AN+ PYGIDF P
Sbjct: 271 VMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQYG-P 329
Query: 73 TGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
TGR+SNG+ +D I ++LGF ++I P++ G DIL+GVNYAS +AGIR+E+G QLG +
Sbjct: 330 TGRFSNGKTTVDVITELLGFDDYITPYSEARGEDILRGVNYASAAAGIREETGRQLGARI 389
Query: 133 NLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMY 192
Q+ +H VS++ + LG ++A YL++C+Y + +G+ND+ NYF+P ++T Y
Sbjct: 390 TFAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYSTGSQY 449
Query: 193 TPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----CVEEKNVA 247
+P YA LI + + L+ +++ GARK LVG+ +GC P L S C E N A
Sbjct: 450 SPDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGCSPNELAQNSRDGVTCDERINSA 509
Query: 248 TFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC---PTN 299
+FN +L SLVD N+ +KF +IN+ I D GF VT+ GCC N
Sbjct: 510 NRIFNSKLVSLVDHFNQN-TPGAKFTYINAYGIFQDMVANPSRYGFRVTNAGCCGVGRNN 568
Query: 300 EKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHL 354
+ C+ PC NR EYVFWD H EAAN+V S+ + + A+P +I+ L
Sbjct: 569 GQITCLPGQAPCLNRDEYVFWDAFHPGEAANVVIGSRSFQRESASDAHPYDIQQL 623
>UniRef100_Q93YW8 Hypothetical protein At4g18970 [Arabidopsis thaliana]
Length = 361
Score = 281 bits (718), Expect = 3e-74
Identities = 146/355 (41%), Positives = 214/355 (60%), Gaps = 15/355 (4%)
Query: 13 LLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFP 72
++++ +A M + +G+ PC F+FGDSL DSGNNN L + A+AN+ PYGIDF P
Sbjct: 6 VMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQYG-P 64
Query: 73 TGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
TGR+SNG+ +D I ++LGF ++I P++ G DIL+GVNYAS +AGIR+E+G QLG +
Sbjct: 65 TGRFSNGKTTVDVITELLGFDDYITPYSEARGEDILRGVNYASAAAGIREETGRQLGARI 124
Query: 133 NLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMY 192
Q+ +H VS++ + LG ++A YL++C+Y + +G+ND+ NYF+P ++T Y
Sbjct: 125 TFAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYSTGSQY 184
Query: 193 TPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----CVEEKNVA 247
+P YA LI + + L+ +++ GARK LVG+ +GC P L S C E N A
Sbjct: 185 SPDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGCSPNELAQNSRDGVTCDERINSA 244
Query: 248 TFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC---PTN 299
+FN +L SLVD N+ +KF +IN+ I D GF VT+ GCC N
Sbjct: 245 NRIFNSKLVSLVDHFNQN-TPGAKFTYINAYGIFQDMVANPSRYGFRVTNAGCCGVGRNN 303
Query: 300 EKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHL 354
+ C+ PC NR EYVFWD H EAAN+V S+ + + A+P +I+ L
Sbjct: 304 GQITCLPGQAPCLNRDEYVFWDAFHPGEAANVVIGSRSFQRESASDAHPYDIQQL 358
>UniRef100_Q94J26 Putative GDSL-motif lipase/hydrolase-like protein [Oryza sativa]
Length = 363
Score = 279 bits (714), Expect = 8e-74
Identities = 143/340 (42%), Positives = 208/340 (61%), Gaps = 15/340 (4%)
Query: 30 ESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQM 89
E QVPC FVFGDSL D+GNNNN+ + A+AN+ PYG+DFP TGR+SNG D I+++
Sbjct: 25 EPQVPCYFVFGDSLVDNGNNNNIASMARANYPPYGVDFPGG-ATGRFSNGLTTADAISRL 83
Query: 90 LGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIA 149
LGF ++IPP+A +L GVN+AS +AGIR ++G QLG ++ QL +++A V ++
Sbjct: 84 LGFDDYIPPYAGATSEQLLTGVNFASAAAGIRDDTGQQLGERISFSAQLQNYQAAVRQLV 143
Query: 150 HRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYL 209
LGG D A L+QC++ V +G+ND+ NYF+P + TSR YTP+QYA LI Q + L
Sbjct: 144 SILGGEDAAANRLSQCIFTVGMGSNDYLNNYFMPAFYPTSRQYTPEQYADVLINQYAQQL 203
Query: 210 QTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----CVEEKNVATFLFNDQLKSLVDRLNK 264
+TL+++GARK + G+ ++GC P L S C+E N A +FN ++ LV++ N
Sbjct: 204 RTLYNYGARKVAVFGVGQVGCSPNELAQNSRNGVTCIERINSAVRMFNRRVVVLVNQFN- 262
Query: 265 KILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC---PTNEKGKCIRDGIPCQNRHE 316
++L + F +IN I G VT+ GCC N + C+ PC NR E
Sbjct: 263 RLLPGALFTYINCYGIFESIMRTPVEHGLAVTNRGCCGVGRNNGQVTCLPYQAPCANRDE 322
Query: 317 YVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLIQ 356
Y+FWD H TEAAN+ +Y+++ + YP ++ L Q
Sbjct: 323 YLFWDAFHPTEAANIFVGRRAYSAAMRSDVYPVDLSTLAQ 362
>UniRef100_Q8L5Z1 Hypothetical protein At1g33810 [Arabidopsis thaliana]
Length = 370
Score = 279 bits (713), Expect = 1e-73
Identities = 147/363 (40%), Positives = 227/363 (62%), Gaps = 20/363 (5%)
Query: 9 LVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFP 68
L++SL +V+ S ++QVPC+F+FGDSL D+GNNN L + A+AN+ PYGIDFP
Sbjct: 8 LLISLNLVLFGFKTTVSQPQQQAQVPCLFIFGDSLVDNGNNNRLLSLARANYRPYGIDFP 67
Query: 69 ATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQL 128
TGR++NGR +D +AQ+LGF+ +IPP++ + G IL+G N+ASG+AGIR E+G L
Sbjct: 68 QG-TTGRFTNGRTYVDALAQILGFRNYIPPYSRIRGQAILRGANFASGAAGIRDETGDNL 126
Query: 129 GHNVNLGLQL-LHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFN 187
G + ++ Q+ L+ A+ + + G ++ +YL++C++Y +G+ND+ NYF+P+ ++
Sbjct: 127 GAHTSMNQQVELYTTAVQQMLRYFRGDTNELQRYLSRCIFYSGMGSNDYLNNYFMPDFYS 186
Query: 188 TSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV--------NGS 239
TS Y K +A++LI+ + L L+ FGARK ++ G+ ++GCIP L G
Sbjct: 187 TSTNYNDKTFAESLIKNYTQQLTRLYQFGARKVIVTGVGQIGCIPYQLARYNNRNNSTGR 246
Query: 240 CVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSV-----GFTVTHHG 294
C E+ N A +FN Q+K LVDRLNK L +KF++++S +D +V GF V G
Sbjct: 247 CNEKINNAIVVFNTQVKKLVDRLNKGQLKGAKFVYLDSYKSTYDLAVNGAAYGFEVVDKG 306
Query: 295 CC---PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNI 351
CC N + C+ PC +R +Y+FWD H TE AN++ A +++ S A YP NI
Sbjct: 307 CCGVGRNNGQITCLPLQTPCPDRTKYLFWDAFHPTETANILLAKSNFYSR--AYTYPINI 364
Query: 352 KHL 354
+ L
Sbjct: 365 QEL 367
>UniRef100_Q8L9V8 GDSL-motif lipase/hydrolase-like protein [Arabidopsis thaliana]
Length = 361
Score = 277 bits (709), Expect = 3e-73
Identities = 145/355 (40%), Positives = 213/355 (59%), Gaps = 15/355 (4%)
Query: 13 LLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFP 72
++++ +A M + +G+ PC F+FGDSL DSGNNN L + A+AN+ PYGIDF P
Sbjct: 6 VMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQYG-P 64
Query: 73 TGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
TGR+SNG+ +D I ++LGF ++I P++ G DIL+GVNYAS +AGIR+E+G QLG +
Sbjct: 65 TGRFSNGKTTVDVITELLGFDDYITPYSEARGEDILRGVNYASAAAGIREETGRQLGARI 124
Query: 133 NLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMY 192
Q+ +H VS++ + LG ++A YL++C+Y + +G+ND+ NYF+P ++T Y
Sbjct: 125 TFAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYSTGSQY 184
Query: 193 TPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS-----CVEEKNVA 247
+P YA LI + + L+ +++ GARK LVG+ +GC P L S C E N A
Sbjct: 185 SPDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGCSPNELAQNSRDGVTCDERINSA 244
Query: 248 TFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC---PTN 299
+FN +L SLVD N+ +KF +IN+ I D GF VT+ GCC N
Sbjct: 245 NRIFNSKLVSLVDHFNQN-TPGAKFTYINAYGIFQDMVANPSRYGFRVTNAGCCGVGRNN 303
Query: 300 EKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHL 354
+ C+ PC NR EYVFWD EAAN+V S+ + + A+P +I+ L
Sbjct: 304 GQITCLPGQAPCLNRDEYVFWDAFXPGEAANVVIGSRSFQRESASDAHPYDIQQL 358
>UniRef100_Q9AX26 GDSL-motif lipase/hydrolase-like protein [Oryza sativa]
Length = 363
Score = 276 bits (705), Expect = 8e-73
Identities = 147/355 (41%), Positives = 214/355 (59%), Gaps = 18/355 (5%)
Query: 15 VVMVASIMQHSTVLG--ESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFP 72
VVM A++ ++ + QVPC F+FGDSL D+GNNN + + A+AN+ PYGIDF A P
Sbjct: 9 VVMAAAVSSALVMVARCDPQVPCYFIFGDSLVDNGNNNYIVSLARANYPPYGIDF-AGGP 67
Query: 73 TGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
+GR++NG +D IAQ+LGF FIPP+A +G IL G N+AS +AGIR E+G QLG +
Sbjct: 68 SGRFTNGLTTVDVIAQLLGFDNFIPPYAATSGDQILNGANFASAAAGIRAETGQQLGGRI 127
Query: 133 NLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMY 192
Q+ +++ V + LG D A+ L++C++ V +G+ND+ NYF+P +NT Y
Sbjct: 128 PFAGQVQNYQTAVQTLISILGDQDTASDRLSKCIFSVGMGSNDYLNNYFMPAFYNTGSQY 187
Query: 193 TPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV-----NGSCVEEKNVA 247
TP+Q+A +LI Y+Q L+++GARK V++G+ ++GC P L +CV + A
Sbjct: 188 TPEQFADSLIADYRRYVQVLYNYGARKVVMIGVGQVGCSPNELARYSADGATCVARIDSA 247
Query: 248 TFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC---PTN 299
+FN +L LVD +N L + F FIN+ I D S GFT T GCC N
Sbjct: 248 IQIFNRRLVGLVDEMN--TLPGAHFTFINAYNIFSDILANAASYGFTETTAGCCGVGRNN 305
Query: 300 EKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHL 354
+ C+ PC NR +++FWD H +EAAN++ SY + +P AYP +I L
Sbjct: 306 GQVTCLPYEAPCSNRDQHIFWDAFHPSEAANIIVGRRSYRAESPNDAYPMDIATL 360
>UniRef100_Q75II0 Putative GDSL-like lipase/hydrolase [Oryza sativa]
Length = 365
Score = 275 bits (703), Expect = 1e-72
Identities = 148/359 (41%), Positives = 220/359 (61%), Gaps = 20/359 (5%)
Query: 9 LVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFP 68
+V + +V+ VA+ +Q + QVPC FVFGDSL D+GNNN++ + A+AN+ PYGIDF
Sbjct: 11 VVAAAVVLAVAAPVQAAP-----QVPCYFVFGDSLVDNGNNNDIVSLARANYPPYGIDFA 65
Query: 69 ATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQL 128
TGR+SNG +D I+++LGF++FIPPFA + +L GVN+AS +AGIR+E+G QL
Sbjct: 66 GGAATGRFSNGLTTVDVISKLLGFEDFIPPFAGASSDQLLTGVNFASAAAGIREETGQQL 125
Query: 129 GHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNT 188
G ++ Q+ ++++ V ++ LG D A +L+QC++ V +G+ND+ NYF+P +NT
Sbjct: 126 GARISFSGQVQNYQSAVQQLVSILGDEDTAAAHLSQCIFTVGMGSNDYLNNYFMPAFYNT 185
Query: 189 SRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLL----VNG-SCVEE 243
YTP+QYA L + + L+ ++ GARK LVG+ ++GC P L NG +CVE
Sbjct: 186 GSQYTPEQYADDLAARYAQLLRAMYSNGARKVALVGVGQVGCSPNELAQQSANGVTCVER 245
Query: 244 KNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCC-- 296
N A +FN +L LVD+ N L + F +IN I D S G VT+ GCC
Sbjct: 246 INSAIRIFNQKLVGLVDQFN--TLPGAHFTYINIYGIFDDILGAPGSHGLKVTNQGCCGV 303
Query: 297 -PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHL 354
N + C+ PC NRHEY FWD H TEAAN++ +Y++ + +P +++ L
Sbjct: 304 GRNNGQVTCLPFQTPCANRHEYAFWDAFHPTEAANVLVGQRTYSARLQSDVHPVDLRTL 362
>UniRef100_Q9LQ33 F14M2.7 protein [Arabidopsis thaliana]
Length = 390
Score = 266 bits (679), Expect = 9e-70
Identities = 146/383 (38%), Positives = 227/383 (59%), Gaps = 40/383 (10%)
Query: 9 LVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFP 68
L++SL +V+ S ++QVPC+F+FGDSL D+GNNN L + A+AN+ PYGIDFP
Sbjct: 8 LLISLNLVLFGFKTTVSQPQQQAQVPCLFIFGDSLVDNGNNNRLLSLARANYRPYGIDFP 67
Query: 69 ATFPTGRYSNGRNPIDKI-------------AQMLGFQEFIPPFANLNGSDILKGVNYAS 115
TGR++NGR +D + +Q+LGF+ +IPP++ + G IL+G N+AS
Sbjct: 68 QG-TTGRFTNGRTYVDALGIFVGEFYMYRALSQILGFRNYIPPYSRIRGQAILRGANFAS 126
Query: 116 GSAGIRKESGSQLGHNVNLGLQL-LHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTN 174
G+AGIR E+G LG + ++ Q+ L+ A+ + + G ++ +YL++C++Y +G+N
Sbjct: 127 GAAGIRDETGDNLGAHTSMNQQVELYTTAVQQMLRYFRGDTNELQRYLSRCIFYSGMGSN 186
Query: 175 DFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKL 234
D+ NYF+P+ ++TS Y K +A++LI+ + L L+ FGARK ++ G+ ++GCIP
Sbjct: 187 DYLNNYFMPDFYSTSTNYNDKTFAESLIKNYTQQLTRLYQFGARKVIVTGVGQIGCIPYQ 246
Query: 235 LV--------NGSCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSV 286
L G C E+ N A +FN Q+K LVDRLNK L +KF++++S +D +V
Sbjct: 247 LARYNNRNNSTGRCNEKINNAIVVFNTQVKKLVDRLNKGQLKGAKFVYLDSYKSTYDLAV 306
Query: 287 ------------GFTVTHHGCC---PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANL 331
GF V GCC N + C+ PC +R +Y+FWD H TE AN+
Sbjct: 307 NGAAYVIYIDPTGFEVVDKGCCGVGRNNGQITCLPLQTPCPDRTKYLFWDAFHPTETANI 366
Query: 332 VTAITSYNSSNPAIAYPTNIKHL 354
+ A +++ S A YP NI+ L
Sbjct: 367 LLAKSNFYSR--AYTYPINIQEL 387
>UniRef100_O65921 Putative GDSL-motif lipase/hydrolase [Arabidopsis thaliana]
Length = 344
Score = 260 bits (664), Expect = 5e-68
Identities = 144/347 (41%), Positives = 206/347 (58%), Gaps = 20/347 (5%)
Query: 16 VMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGR 75
++ A I +TV+ Q PC FVFGDS+SD+GNNNNL + AK NF PYG DFP PTGR
Sbjct: 7 LVAAIIFTAATVVYGQQAPCFFVFGDSMSDNGNNNNLKSEAKVNFSPYGNDFPKG-PTGR 65
Query: 76 YSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLG 135
+SNGR D I ++ GF++FIPPFA + G+NYASG +G+R+E+ LG +++
Sbjct: 66 FSNGRTIPDIIGELSGFKDFIPPFAEASPEQAHTGMNYASGGSGLREETSEHLGDRISIR 125
Query: 136 LQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPK 195
QL +H+ +++ + + L QCLY +NIG+ND+ NYF+ +NT R YTPK
Sbjct: 126 KQLQNHKTSITKA-------NVPAERLQQCLYMINIGSNDYINNYFMSKPYNTKRRYTPK 178
Query: 196 QYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNGS----CVEEKNVATFLF 251
QYA +LI +L+ LH GARK + GL ++GC PK++ + S C E N A +F
Sbjct: 179 QYAYSLIIIYRSHLKNLHRLGARKVAVFGLSQIGCTPKIMKSHSDGKICSREVNEAVKIF 238
Query: 252 NDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKS---VGFTVTHHGCCPTNE-KGKCIRD 307
N L LV NKK+ +KF +++ + ++ +GF V CC N + C+ +
Sbjct: 239 NKNLDDLVMDFNKKV-RGAKFTYVDLFSGGDPQAFIFLGFKVGGKSCCTVNPGEELCVPN 297
Query: 308 GIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHL 354
C NR EYVFWD +H+TEA N+V A S++ I+ P +I L
Sbjct: 298 QPVCANRTEYVFWDDLHSTEATNMVVAKGSFDG---IISKPYSIAQL 341
>UniRef100_Q9SF78 Putative GDSL-motif lipase/hydrolase; 24593-26678 [Arabidopsis
thaliana]
Length = 384
Score = 242 bits (618), Expect = 1e-62
Identities = 130/341 (38%), Positives = 195/341 (57%), Gaps = 16/341 (4%)
Query: 29 GESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQ 88
G+ VP +FVFGDSL D+GNNNN+P+ AKAN+ PYGIDF PTGR+ NG +D IAQ
Sbjct: 49 GDGIVPALFVFGDSLIDNGNNNNIPSFAKANYFPYGIDFNGG-PTGRFCNGLTMVDGIAQ 107
Query: 89 MLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRI 148
+LG IP ++ G +L+GVNYAS +AGI ++G + Q+ + + ++
Sbjct: 108 LLGL-PLIPAYSEATGDQVLRGVNYASAAAGILPDTGGNFVGRIPFDQQIHNFETTLDQV 166
Query: 149 AHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHY 208
A + GG + + L+++ +G+ND+ NY +PN F T Y +Q+ L+Q +
Sbjct: 167 ASKSGGAVAIADSVTRSLFFIGMGSNDYLNNYLMPN-FPTRNQYNSQQFGDLLVQHYTDQ 225
Query: 209 LQTLHHFGARKSVLVGLDRLGCIPKLLV---NGSCVEEKNVATFLFNDQLKSLVDRLNKK 265
L L++ G RK V+ GL R+GCIP +L +G C EE N FN +K+++ LN+
Sbjct: 226 LTRLYNLGGRKFVVAGLGRMGCIPSILAQGNDGKCSEEVNQLVLPFNTNVKTMISNLNQN 285
Query: 266 ILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCCPTNE-KGK--CIRDGIPCQNRHEY 317
L D+KFI+++ + D + G T GCC + +G+ C+ PC NR +Y
Sbjct: 286 -LPDAKFIYLDIAHMFEDIVANQAAYGLTTMDKGCCGIGKNRGQITCLPFETPCPNRDQY 344
Query: 318 VFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLIQSN 358
VFWD H TE NL+ A ++ + + +AYP NI+ L N
Sbjct: 345 VFWDAFHPTEKVNLIMAKKAF-AGDRTVAYPINIQQLASLN 384
>UniRef100_Q9SZW7 Hypothetical protein F6G3.170 [Arabidopsis thaliana]
Length = 348
Score = 241 bits (614), Expect = 3e-62
Identities = 139/357 (38%), Positives = 198/357 (54%), Gaps = 19/357 (5%)
Query: 4 ESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPY 63
ESK ++ V VA++ + + Q PC FVFGDS+ D+GNNN L T AK N+LPY
Sbjct: 5 ESKALWIILATVFAVAAV---APAVHGQQTPCYFVFGDSVFDNGNNNALNTKAKVNYLPY 61
Query: 64 GIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKE 123
GID+ PTGR+SNGRN D IA++ GF IPPFA + + G+NYASG+ GIR+E
Sbjct: 62 GIDY-FQGPTGRFSNGRNIPDVIAELAGFNNPIPPFAGASQAQANIGLNYASGAGGIREE 120
Query: 124 SGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLP 183
+ +G ++L Q+ +H + + A L L QCLY +NIG+ND+ NYFL
Sbjct: 121 TSENMGERISLRQQVNNHFSAIITAAVPL-------SRLRQCLYTINIGSNDYLNNYFLS 173
Query: 184 NVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV----NGS 239
R++ P QYA++LI YL L+ GAR L G+ ++GC P+++
Sbjct: 174 PPTLARRLFNPDQYARSLISLYRIYLTQLYVLGARNVALFGIGKIGCTPRIVATLGGGTG 233
Query: 240 CVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSVGFTVTHHGCCPTN 299
C EE N A +FN +LK+LV N K ++ + S ++G TV CC N
Sbjct: 234 CAEEVNQAVIIFNTKLKALVTDFNNKPGAMFTYVDLFSGNAEDFAALGITVGDRSCCTVN 293
Query: 300 E-KGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLI 355
+ C +G C +R++++FWD +HTTE N V A ++N IA P NI L+
Sbjct: 294 PGEELCAANGPVCPDRNKFIFWDNVHTTEVINTVVANAAFNG---PIASPFNISQLV 347
>UniRef100_O64468 Putative GDSL-motif lipase/hydrolase [Arabidopsis thaliana]
Length = 349
Score = 239 bits (609), Expect = 1e-61
Identities = 135/338 (39%), Positives = 199/338 (57%), Gaps = 26/338 (7%)
Query: 13 LLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFP 72
LLV+ + LG+ +VPC FVFGDS+ D+GNNN L TSAK N+ PYGIDF A P
Sbjct: 10 LLVIATTAFATTEAALGQ-RVPCYFVFGDSVFDNGNNNVLNTSAKVNYSPYGIDF-ARGP 67
Query: 73 TGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
TGR+SNGRN D IA+++ F ++IPPF + G+NYASG GIR+E+ LG +
Sbjct: 68 TGRFSNGRNIPDIIAELMRFSDYIPPFTGASPEQAHIGINYASGGGGIREETSQHLGEII 127
Query: 133 NLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMY 192
+ Q+ +HR+++ + LN+CLY +NIG+ND+ NYF+P + T++ +
Sbjct: 128 SFKKQIKNHRSMIMTAK-------VPEEKLNKCLYTINIGSNDYLNNYFMPAPYMTNKKF 180
Query: 193 TPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVN----GSCVEEKNVAT 248
+ +YA +LI+ YL++L+ GARK + G+ +LGC P+++ + C E N A
Sbjct: 181 SFDEYADSLIRSYRSYLKSLYVLGARKVAVFGVSKLGCTPRMIASHGGGNGCAAEVNKAV 240
Query: 249 FLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKS------VGFTVTHHGCCPTNEKG 302
FN LK+LV N+ D+KF F++ I +S +GF VT CC T + G
Sbjct: 241 EPFNKNLKALVYEFNRD-FADAKFTFVD---IFSGQSPFAFFMLGFRVTDKSCC-TVKPG 295
Query: 303 K--CIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSY 338
+ C + C + YV+WD +H+TEAAN+V A +Y
Sbjct: 296 EELCATNEPVCPVQRRYVYWDNVHSTEAANMVVAKAAY 333
>UniRef100_Q8LD78 Putative GDSL-motif lipase/hydrolase [Arabidopsis thaliana]
Length = 348
Score = 238 bits (606), Expect = 3e-61
Identities = 138/358 (38%), Positives = 198/358 (54%), Gaps = 21/358 (5%)
Query: 4 ESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPY 63
ESK ++ V VA++ + + Q PC FVFGDS+ D+GNNN L T AK N+LPY
Sbjct: 5 ESKALWIILATVFAVAAV---APAVHGQQTPCYFVFGDSVFDNGNNNALNTKAKVNYLPY 61
Query: 64 GIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKE 123
GID+ PTGR+SNG N D IA++ GF IPPFA + + G+NYASG+ GIR+E
Sbjct: 62 GIDY-FQGPTGRFSNGPNIPDVIAELAGFNNPIPPFAGASQAQANIGLNYASGAGGIREE 120
Query: 124 SGSQLGHNVNLGLQLLHH-RAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFL 182
+ +G ++L Q+ +H AI++ + L QCLY +NIG+ND+ NYFL
Sbjct: 121 TSENMGERISLRQQVNNHFSAIITAVV--------PLSRLRQCLYTINIGSNDYLNNYFL 172
Query: 183 PNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV----NG 238
R++ P QYA++LI YL L+ GAR L G+ ++GC P+++
Sbjct: 173 SPPTLARRLFNPDQYARSLISLYRIYLTQLYVLGARNVALFGIGKIGCTPRIVATLGGGT 232
Query: 239 SCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSVGFTVTHHGCCPT 298
C EE N A +FN +LK+LV N K ++ + S ++G TV CC
Sbjct: 233 GCAEEVNQAVIIFNTKLKALVTDFNNKPGAMFTYVDLFSGNAEDFAALGITVGDRSCCTV 292
Query: 299 NE-KGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLI 355
N + C +G C +R++++FWD +HTTE N V A ++N IA P NI L+
Sbjct: 293 NPGEELCAANGPVCPDRNKFIFWDNVHTTEVINTVVANAAFNG---PIASPFNISQLV 347
>UniRef100_O64469 Putative GDSL-motif lipase/hydrolase [Arabidopsis thaliana]
Length = 349
Score = 232 bits (591), Expect = 1e-59
Identities = 133/337 (39%), Positives = 191/337 (56%), Gaps = 20/337 (5%)
Query: 12 SLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATF 71
+LL +++ V G+ VPC FVFGDS+ D+GNNN L T AK N+ PYGIDF A
Sbjct: 8 ALLWAFATAVVMAEAVRGQL-VPCYFVFGDSVFDNGNNNELDTLAKVNYSPYGIDF-ARG 65
Query: 72 PTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHN 131
PTGR+SNGRN D IA+ L IPPF + G+NYASG AG+ +E+ LG
Sbjct: 66 PTGRFSNGRNIPDFIAEELRISYDIPPFTRASTEQAHTGINYASGGAGLLEETSQHLGER 125
Query: 132 VNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRM 191
++ Q+ +HR ++ + L +CLY +NIG+ND+ NYF+P + T+
Sbjct: 126 ISFEKQITNHRKMIMTAG-------VPPEKLKKCLYTINIGSNDYLNNYFMPAPYTTNEN 178
Query: 192 YTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVN----GSCVEEKNVA 247
++ +YA LIQ YL++L+ GARK + G+ +LGC P+++ + C E N A
Sbjct: 179 FSFDEYADFLIQSYRSYLKSLYVLGARKVAVFGVSKLGCTPRMIASHGGGKGCATEVNKA 238
Query: 248 TFLFNDQLKSLVDRLNKKILMD-SKFIFI---NSTAIIHDKSVGFTVTHHGCCPTNEKGK 303
FN +LK L+ N+ ++D +KF F+ +S I +GFTVT CC T E G+
Sbjct: 239 VEPFNKKLKDLISEFNRISVVDHAKFTFVDLFSSQNPIEYFILGFTVTDKSCC-TVESGQ 297
Query: 304 --CIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSY 338
C + C NR YV+WD +H+TEAAN V ++
Sbjct: 298 ELCAANKPVCPNRERYVYWDNVHSTEAANKVVVKAAF 334
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 604,543,653
Number of Sequences: 2790947
Number of extensions: 25990202
Number of successful extensions: 57718
Number of sequences better than 10.0: 374
Number of HSP's better than 10.0 without gapping: 190
Number of HSP's successfully gapped in prelim test: 184
Number of HSP's that attempted gapping in prelim test: 56470
Number of HSP's gapped (non-prelim): 442
length of query: 361
length of database: 848,049,833
effective HSP length: 128
effective length of query: 233
effective length of database: 490,808,617
effective search space: 114358407761
effective search space used: 114358407761
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0476a.9


