

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
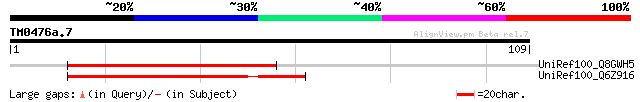
Query= TM0476a.7
(109 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GWH5 Hypothetical protein At1g78895/F9K20.31 [Arabid... 57 8e-08
UniRef100_Q6Z916 Pentatricopeptide (PPR) repeat-containing prote... 56 2e-07
>UniRef100_Q8GWH5 Hypothetical protein At1g78895/F9K20.31 [Arabidopsis thaliana]
Length = 164
Score = 57.4 bits (137), Expect = 8e-08
Identities = 31/44 (70%), Positives = 33/44 (74%)
Query: 13 IYQDTVKAIVACLANTVDSAESVLRVAATGHDKRLYFKVSPSLY 56
I QDT IVA LANTV +AE VLRVAATGHDKRL+ KV LY
Sbjct: 66 ISQDTASNIVARLANTVGAAEGVLRVAATGHDKRLFVKVVICLY 109
>UniRef100_Q6Z916 Pentatricopeptide (PPR) repeat-containing protein-like [Oryza
sativa]
Length = 135
Score = 55.8 bits (133), Expect = 2e-07
Identities = 29/50 (58%), Positives = 38/50 (76%), Gaps = 2/50 (4%)
Query: 13 IYQDTVKAIVACLANTVDSAESVLRVAATGHDKRLYFKVSPSLY*ILTHF 62
I Q+ +IVA LANT+ +AESVLRVAATGHDK+L+FK P L+ ++ F
Sbjct: 84 ISQEMANSIVASLANTIGAAESVLRVAATGHDKKLFFK--PELFDMIAIF 131
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.366 0.161 0.597
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 139,151,768
Number of Sequences: 2790947
Number of extensions: 3704110
Number of successful extensions: 27627
Number of sequences better than 10.0: 2
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 27625
Number of HSP's gapped (non-prelim): 2
length of query: 109
length of database: 848,049,833
effective HSP length: 85
effective length of query: 24
effective length of database: 610,819,338
effective search space: 14659664112
effective search space used: 14659664112
T: 11
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0476a.7


