

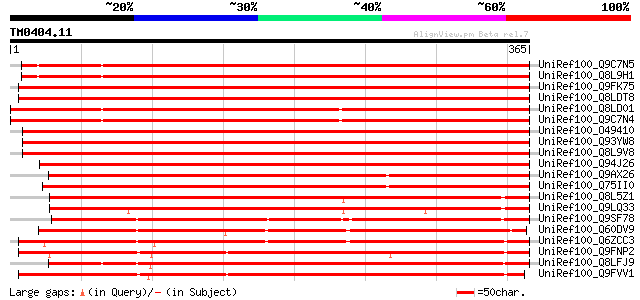
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0404.11
(365 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C7N5 Lipase/hydrolase, putative; 114382-116051 [Arab... 518 e-145
UniRef100_Q8L9H1 Lipase/hydrolase, putative [Arabidopsis thaliana] 513 e-144
UniRef100_Q9FK75 GDSL-motif lipase/hydrolase-like protein [Arabi... 511 e-144
UniRef100_Q8LDT8 GDSL-motif lipase/hydrolase-like protein [Arabi... 509 e-143
UniRef100_Q8LD01 Lipase/hydrolase, putative [Arabidopsis thaliana] 508 e-142
UniRef100_Q9C7N4 Lipase/hydrolase, putative; 118270-120144 [Arab... 507 e-142
UniRef100_O49410 Hypothetical protein AT4g18970 [Arabidopsis tha... 504 e-141
UniRef100_Q93YW8 Hypothetical protein At4g18970 [Arabidopsis tha... 504 e-141
UniRef100_Q8L9V8 GDSL-motif lipase/hydrolase-like protein [Arabi... 501 e-140
UniRef100_Q94J26 Putative GDSL-motif lipase/hydrolase-like prote... 495 e-139
UniRef100_Q9AX26 GDSL-motif lipase/hydrolase-like protein [Oryza... 491 e-137
UniRef100_Q75II0 Putative GDSL-like lipase/hydrolase [Oryza sativa] 484 e-135
UniRef100_Q8L5Z1 Hypothetical protein At1g33810 [Arabidopsis tha... 376 e-103
UniRef100_Q9LQ33 F14M2.7 protein [Arabidopsis thaliana] 358 1e-97
UniRef100_Q9SF78 Putative GDSL-motif lipase/hydrolase; 24593-266... 353 5e-96
UniRef100_Q60DV9 Hypothetical protein P0426G01.13 [Oryza sativa] 344 3e-93
UniRef100_Q6ZCC3 Putative GDSL-motif lipase/hydrolase protein [O... 332 1e-89
UniRef100_Q9FNP2 GDSL-motif lipase/acylhydrolase-like protein [A... 327 2e-88
UniRef100_Q8LFJ9 Hypothetical protein [Arabidopsis thaliana] 327 3e-88
UniRef100_Q9FVV1 Putative GDSL-motif lipase/acylhydrolase; 82739... 322 8e-87
>UniRef100_Q9C7N5 Lipase/hydrolase, putative; 114382-116051 [Arabidopsis thaliana]
Length = 364
Score = 518 bits (1334), Expect = e-145
Identities = 251/357 (70%), Positives = 294/357 (82%), Gaps = 2/357 (0%)
Query: 9 VVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGP 68
+V++ V++LGL G V A PQVPCYFIFGDSLVDNGNNN LRS+ARADY PYGIDF GGP
Sbjct: 10 LVSVWVLLLGL-GFKVKAEPQVPCYFIFGDSLVDNGNNNRLRSIARADYFPYGIDF-GGP 67
Query: 69 SGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRI 128
+GRFSNG+TTVDV+ ELLGFD++IP Y + SG +IL+GVN+ASAAAGIREETG QLG RI
Sbjct: 68 TGRFSNGRTTVDVLTELLGFDNYIPAYSTVSGQEILQGVNYASAAAGIREETGAQLGQRI 127
Query: 129 SFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQY 188
+FSGQV+NY++TV+QVV ILG+E AA+YL +CIYS+G+GSNDYLNNYFMPQFYSTSRQY
Sbjct: 128 TFSGQVENYKNTVAQVVEILGDEYTAADYLKRCIYSVGMGSNDYLNNYFMPQFYSTSRQY 187
Query: 189 TTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDA 248
T +QYAD LI Y +QL LYN+GARK L GIG IGCSPN LAQ SQDG TCV IN A
Sbjct: 188 TPEQYADDLISRYRDQLNALYNYGARKFALVGIGAIGCSPNALAQGSQDGTTCVERINSA 247
Query: 249 NQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNG 308
N+IFNN+L S+V Q NN DA YINAYG FQDIIA+P+ YGF+NTN CCG+GRN G
Sbjct: 248 NRIFNNRLISMVQQLNNAHSDASFTYINAYGAFQDIIANPSAYGFTNTNTACCGIGRNGG 307
Query: 309 QITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
Q+TCLP + PC NR EY+FWDAFHPS A N IA+R+Y+AQ SD YPIDI +LAQ+
Sbjct: 308 QLTCLPGEPPCLNRDEYVFWDAFHPSAAANTAIAKRSYNAQRSSDVYPIDISQLAQL 364
>UniRef100_Q8L9H1 Lipase/hydrolase, putative [Arabidopsis thaliana]
Length = 364
Score = 513 bits (1320), Expect = e-144
Identities = 248/357 (69%), Positives = 292/357 (81%), Gaps = 2/357 (0%)
Query: 9 VVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGP 68
+V++ V++LGL G V A PQVPCYFIFGDSLVDNGNNN LRS+ARADY PYGIDF GGP
Sbjct: 10 LVSVWVLLLGL-GFKVKAEPQVPCYFIFGDSLVDNGNNNRLRSIARADYFPYGIDF-GGP 67
Query: 69 SGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRI 128
+GRFSNG+TTVDV+ ELLGFD++IP Y + SG +IL+GVN+ASAAAGIREETG QLG RI
Sbjct: 68 TGRFSNGRTTVDVLTELLGFDNYIPAYSTVSGQEILQGVNYASAAAGIREETGAQLGQRI 127
Query: 129 SFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQY 188
+FSGQV+NY++TV+QVV ILG+E AA+YL +CIYS+G+GSNDYLNNYFMPQ YSTSRQY
Sbjct: 128 TFSGQVENYKNTVAQVVEILGDEYTAADYLKRCIYSVGMGSNDYLNNYFMPQXYSTSRQY 187
Query: 189 TTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDA 248
T +QYAD LI Y +QL LYN+GARK L GIG IGCSPN LAQ S+DG TCV IN A
Sbjct: 188 TPEQYADDLISRYRDQLNALYNYGARKFALVGIGAIGCSPNALAQGSEDGTTCVERINSA 247
Query: 249 NQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNG 308
N+IFNN+L S+V Q NN DA YINAYG FQDII +P+ YGF+NTN CCG+GRN G
Sbjct: 248 NRIFNNRLISMVQQLNNAHSDASFTYINAYGAFQDIITNPSAYGFTNTNTACCGIGRNGG 307
Query: 309 QITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
Q+TCLP + PC NR EY+FWDAFHPS A N IA+R+Y+AQ SD YPIDI +LAQ+
Sbjct: 308 QLTCLPGEPPCLNRDEYVFWDAFHPSAAANTAIAKRSYNAQRSSDVYPIDISQLAQL 364
>UniRef100_Q9FK75 GDSL-motif lipase/hydrolase-like protein [Arabidopsis thaliana]
Length = 362
Score = 511 bits (1317), Expect = e-144
Identities = 243/359 (67%), Positives = 289/359 (79%)
Query: 7 MMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
M ++ M++MV + P PCYFIFGDSLVDNGNNN L+SLARA+Y PYGIDF
Sbjct: 4 MSLMIMMIMVAVTMINIAKSDPIAPCYFIFGDSLVDNGNNNQLQSLARANYFPYGIDFAA 63
Query: 67 GPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGG 126
GP+GRFSNG TTVDVIA+LLGF+D+I PY S G DILRGVN+ASAAAGIR+ETG+QLGG
Sbjct: 64 GPTGRFSNGLTTVDVIAQLLGFEDYITPYASARGQDILRGVNYASAAAGIRDETGRQLGG 123
Query: 127 RISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSR 186
RI+F+GQV N+ +TVSQVVNILG++++A+NYLSKCIYSIGLGSNDYLNNYFMP FYST
Sbjct: 124 RIAFAGQVANHVNTVSQVVNILGDQNEASNYLSKCIYSIGLGSNDYLNNYFMPTFYSTGN 183
Query: 187 QYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDIN 246
Q++ + YAD L+ YTEQL+ LY GARK L G+G IGCSPNELAQ S+DGRTC IN
Sbjct: 184 QFSPESYADDLVARYTEQLRVLYTNGARKFALIGVGAIGCSPNELAQNSRDGRTCDERIN 243
Query: 247 DANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRN 306
AN+IFN+KL S+VD FN PDA+ YINAYGIFQDII +PA YGF TN GCCGVGRN
Sbjct: 244 SANRIFNSKLISIVDAFNQNTPDAKFTYINAYGIFQDIITNPARYGFRVTNAGCCGVGRN 303
Query: 307 NGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
NGQITCLP Q PC NR EY+FWDAFHP EA N+VI +R++ ++ SDA+P DI++LA +
Sbjct: 304 NGQITCLPGQAPCLNRNEYVFWDAFHPGEAANIVIGRRSFKREAASDAHPYDIQQLASL 362
>UniRef100_Q8LDT8 GDSL-motif lipase/hydrolase-like protein [Arabidopsis thaliana]
Length = 362
Score = 509 bits (1312), Expect = e-143
Identities = 242/359 (67%), Positives = 289/359 (80%)
Query: 7 MMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
M ++ M++MV + P PCYFIFGDSLVDNGNNN L+SLARA+Y PYGIDF
Sbjct: 4 MSLMIMMIMVAVTMINIAKSDPIAPCYFIFGDSLVDNGNNNQLQSLARANYFPYGIDFAA 63
Query: 67 GPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGG 126
GP+GRFSNG TTVDVIA+LLGF+D+I PY S G DILRGVN+ASAAAGIR+ETG+QLGG
Sbjct: 64 GPTGRFSNGLTTVDVIAQLLGFEDYITPYASARGQDILRGVNYASAAAGIRDETGRQLGG 123
Query: 127 RISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSR 186
RI+F+GQV N+ +TVSQVVNILG++++A+NYLSKCIYSIGLGSNDYLNNYFMP FYST
Sbjct: 124 RIAFAGQVANHVNTVSQVVNILGDQNEASNYLSKCIYSIGLGSNDYLNNYFMPTFYSTGN 183
Query: 187 QYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDIN 246
Q++ + YAD L+ YTEQL+ LY GARK L G+G IGCSPNELAQ S+DGRTC IN
Sbjct: 184 QFSPESYADDLVARYTEQLRVLYTNGARKFALIGVGAIGCSPNELAQNSRDGRTCDERIN 243
Query: 247 DANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRN 306
AN+IFN+KL S+VD FN PDA+ YINAYGIFQDII +PA YGF TN GCCGVGRN
Sbjct: 244 SANRIFNSKLISIVDAFNQNTPDAKFTYINAYGIFQDIITNPARYGFRVTNAGCCGVGRN 303
Query: 307 NGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
NGQITCLP Q PC NR EY+FWDAFHP EA N+VI +R++ ++ S+A+P DI++LA +
Sbjct: 304 NGQITCLPGQAPCLNRNEYVFWDAFHPGEAANIVIGRRSFKREAASNAHPYDIQQLASL 362
>UniRef100_Q8LD01 Lipase/hydrolase, putative [Arabidopsis thaliana]
Length = 363
Score = 508 bits (1307), Expect = e-142
Identities = 247/365 (67%), Positives = 295/365 (80%), Gaps = 2/365 (0%)
Query: 1 MDLNVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPY 60
M+ + V ++++ G A QVPC+F+FGDSLVDNGNNN L S+AR++Y PY
Sbjct: 1 MESYLTKWCVVLVLLCFGFSVVKAQAQAQVPCFFVFGDSLVDNGNNNGLISIARSNYFPY 60
Query: 61 GIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREET 120
GIDF GGP+GRFSNGKTTVDVIAELLGF+ +IP Y + SG IL GVN+ASAAAGIREET
Sbjct: 61 GIDF-GGPTGRFSNGKTTVDVIAELLGFNGYIPAYNTVSGRQILSGVNYASAAAGIREET 119
Query: 121 GQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQ 180
G+QLG RISFSGQV+NYQ+TVSQVV +LG+E +AA+YL +CIYS+GLGSNDYLNNYFMP
Sbjct: 120 GRQLGQRISFSGQVRNYQTTVSQVVQLLGDETRAADYLKRCIYSVGLGSNDYLNNYFMPT 179
Query: 181 FYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRT 240
FYS+SRQ+T +QYA+ LI Y+ QL LYN+GARK L GIG +GCSPN LA S DGRT
Sbjct: 180 FYSSSRQFTPEQYANDLISRYSTQLNALYNYGARKFALSGIGSVGCSPNALA-GSPDGRT 238
Query: 241 CVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGC 300
CV IN ANQIFNNKL+S+VDQ NN PDA+ IYINAYGIFQD+I +PA +GF TN GC
Sbjct: 239 CVDRINSANQIFNNKLRSLVDQLNNNHPDAKFIYINAYGIFQDMITNPARFGFRVTNAGC 298
Query: 301 CGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIK 360
CG+GRN GQITCLP Q PC +R Y+FWDAFHP+EA NV+IA+R+Y+AQS SDAYP+DI
Sbjct: 299 CGIGRNAGQITCLPGQRPCRDRNAYVFWDAFHPTEAANVIIARRSYNAQSASDAYPMDIS 358
Query: 361 RLAQI 365
RLAQ+
Sbjct: 359 RLAQL 363
>UniRef100_Q9C7N4 Lipase/hydrolase, putative; 118270-120144 [Arabidopsis thaliana]
Length = 363
Score = 507 bits (1306), Expect = e-142
Identities = 247/365 (67%), Positives = 295/365 (80%), Gaps = 2/365 (0%)
Query: 1 MDLNVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPY 60
M+ + V ++++ G A QVPC+F+FGDSLVDNGNNN L S+AR++Y PY
Sbjct: 1 MESYLTKWCVVLVLLCFGFSVVKAQAQAQVPCFFVFGDSLVDNGNNNGLISIARSNYFPY 60
Query: 61 GIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREET 120
GIDF GGP+GRFSNGKTTVDVIAELLGF+ +IP Y + SG IL GVN+ASAAAGIREET
Sbjct: 61 GIDF-GGPTGRFSNGKTTVDVIAELLGFNGYIPAYNTVSGRQILSGVNYASAAAGIREET 119
Query: 121 GQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQ 180
G+QLG RISFSGQV+NYQ+TVSQVV +LG+E +AA+YL +CIYS+GLGSNDYLNNYFMP
Sbjct: 120 GRQLGQRISFSGQVRNYQTTVSQVVQLLGDETRAADYLKRCIYSVGLGSNDYLNNYFMPT 179
Query: 181 FYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRT 240
FYS+SRQ+T +QYA+ LI Y+ QL LYN+GARK L GIG +GCSPN LA S DGRT
Sbjct: 180 FYSSSRQFTPEQYANDLISRYSTQLNALYNYGARKFALSGIGAVGCSPNALA-GSPDGRT 238
Query: 241 CVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGC 300
CV IN ANQIFNNKL+S+VDQ NN PDA+ IYINAYGIFQD+I +PA +GF TN GC
Sbjct: 239 CVDRINSANQIFNNKLRSLVDQLNNNHPDAKFIYINAYGIFQDMITNPARFGFRVTNAGC 298
Query: 301 CGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIK 360
CG+GRN GQITCLP Q PC +R Y+FWDAFHP+EA NV+IA+R+Y+AQS SDAYP+DI
Sbjct: 299 CGIGRNAGQITCLPGQRPCRDRNAYVFWDAFHPTEAANVIIARRSYNAQSASDAYPMDIS 358
Query: 361 RLAQI 365
RLAQ+
Sbjct: 359 RLAQL 363
>UniRef100_O49410 Hypothetical protein AT4g18970 [Arabidopsis thaliana]
Length = 626
Score = 504 bits (1299), Expect = e-141
Identities = 244/356 (68%), Positives = 284/356 (79%)
Query: 10 VAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPS 69
V M+ M + + P PCYFIFGDSLVD+GNNN L SLARA+Y PYGIDF GP+
Sbjct: 271 VMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQYGPT 330
Query: 70 GRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRIS 129
GRFSNGKTTVDVI ELLGFDD+I PY G+DILRGVN+ASAAAGIREETG+QLG RI+
Sbjct: 331 GRFSNGKTTVDVITELLGFDDYITPYSEARGEDILRGVNYASAAAGIREETGRQLGARIT 390
Query: 130 FSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYT 189
F+GQV N+ +TVSQVVNILG+E++AANYLSKCIYSIGLGSNDYLNNYFMP +YST QY+
Sbjct: 391 FAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYSTGSQYS 450
Query: 190 TDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDAN 249
D YA+ LI YTEQL+ +YN GARK L GIG IGCSPNELAQ S+DG TC IN AN
Sbjct: 451 PDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGCSPNELAQNSRDGVTCDERINSAN 510
Query: 250 QIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQ 309
+IFN+KL S+VD FN P A+ YINAYGIFQD++A+P+ YGF TN GCCGVGRNNGQ
Sbjct: 511 RIFNSKLVSLVDHFNQNTPGAKFTYINAYGIFQDMVANPSRYGFRVTNAGCCGVGRNNGQ 570
Query: 310 ITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
ITCLP Q PC NR EY+FWDAFHP EA NVVI R++ +S SDA+P DI++LA++
Sbjct: 571 ITCLPGQAPCLNRDEYVFWDAFHPGEAANVVIGSRSFQRESASDAHPYDIQQLARL 626
>UniRef100_Q93YW8 Hypothetical protein At4g18970 [Arabidopsis thaliana]
Length = 361
Score = 504 bits (1299), Expect = e-141
Identities = 244/356 (68%), Positives = 284/356 (79%)
Query: 10 VAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPS 69
V M+ M + + P PCYFIFGDSLVD+GNNN L SLARA+Y PYGIDF GP+
Sbjct: 6 VMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQYGPT 65
Query: 70 GRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRIS 129
GRFSNGKTTVDVI ELLGFDD+I PY G+DILRGVN+ASAAAGIREETG+QLG RI+
Sbjct: 66 GRFSNGKTTVDVITELLGFDDYITPYSEARGEDILRGVNYASAAAGIREETGRQLGARIT 125
Query: 130 FSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYT 189
F+GQV N+ +TVSQVVNILG+E++AANYLSKCIYSIGLGSNDYLNNYFMP +YST QY+
Sbjct: 126 FAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYSTGSQYS 185
Query: 190 TDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDAN 249
D YA+ LI YTEQL+ +YN GARK L GIG IGCSPNELAQ S+DG TC IN AN
Sbjct: 186 PDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGCSPNELAQNSRDGVTCDERINSAN 245
Query: 250 QIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQ 309
+IFN+KL S+VD FN P A+ YINAYGIFQD++A+P+ YGF TN GCCGVGRNNGQ
Sbjct: 246 RIFNSKLVSLVDHFNQNTPGAKFTYINAYGIFQDMVANPSRYGFRVTNAGCCGVGRNNGQ 305
Query: 310 ITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
ITCLP Q PC NR EY+FWDAFHP EA NVVI R++ +S SDA+P DI++LA++
Sbjct: 306 ITCLPGQAPCLNRDEYVFWDAFHPGEAANVVIGSRSFQRESASDAHPYDIQQLARL 361
>UniRef100_Q8L9V8 GDSL-motif lipase/hydrolase-like protein [Arabidopsis thaliana]
Length = 361
Score = 501 bits (1290), Expect = e-140
Identities = 243/356 (68%), Positives = 283/356 (79%)
Query: 10 VAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPS 69
V M+ M + + P PCYFIFGDSLVD+GNNN L SLARA+Y PYGIDF GP+
Sbjct: 6 VMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQYGPT 65
Query: 70 GRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRIS 129
GRFSNGKTTVDVI ELLGFDD+I PY G+DILRGVN+ASAAAGIREETG+QLG RI+
Sbjct: 66 GRFSNGKTTVDVITELLGFDDYITPYSEARGEDILRGVNYASAAAGIREETGRQLGARIT 125
Query: 130 FSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYT 189
F+GQV N+ +TVSQVVNILG+E++AANYLSKCIYSIGLGSNDYLNNYFMP +YST QY+
Sbjct: 126 FAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYSTGSQYS 185
Query: 190 TDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDAN 249
D YA+ LI YTEQL+ +YN GARK L GIG IGCSPNELAQ S+DG TC IN AN
Sbjct: 186 PDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGCSPNELAQNSRDGVTCDERINSAN 245
Query: 250 QIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQ 309
+IFN+KL S+VD FN P A+ YINAYGIFQD++A+P+ YGF TN GCCGVGRNNGQ
Sbjct: 246 RIFNSKLVSLVDHFNQNTPGAKFTYINAYGIFQDMVANPSRYGFRVTNAGCCGVGRNNGQ 305
Query: 310 ITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
ITCLP Q PC NR EY+FWDAF P EA NVVI R++ +S SDA+P DI++LA++
Sbjct: 306 ITCLPGQAPCLNRDEYVFWDAFXPGEAANVVIGSRSFQRESASDAHPYDIQQLARL 361
>UniRef100_Q94J26 Putative GDSL-motif lipase/hydrolase-like protein [Oryza sativa]
Length = 363
Score = 495 bits (1274), Expect = e-139
Identities = 225/344 (65%), Positives = 277/344 (80%)
Query: 22 GWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDV 81
G GA PQVPCYF+FGDSLVDNGNNN + S+ARA+Y PYG+DFPGG +GRFSNG TT D
Sbjct: 20 GLAGAEPQVPCYFVFGDSLVDNGNNNNIASMARANYPPYGVDFPGGATGRFSNGLTTADA 79
Query: 82 IAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTV 141
I+ LLGFDD+IPPY + + +L GVNFASAAAGIR++TGQQLG RISFS Q+QNYQ+ V
Sbjct: 80 ISRLLGFDDYIPPYAGATSEQLLTGVNFASAAAGIRDDTGQQLGERISFSAQLQNYQAAV 139
Query: 142 SQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAY 201
Q+V+ILG ED AAN LS+CI+++G+GSNDYLNNYFMP FY TSRQYT +QYADVLI Y
Sbjct: 140 RQLVSILGGEDAAANRLSQCIFTVGMGSNDYLNNYFMPAFYPTSRQYTPEQYADVLINQY 199
Query: 202 TEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVD 261
+QL+TLYN+GARK+ +FG+GQ+GCSPNELAQ S++G TC+ IN A ++FN ++ +V+
Sbjct: 200 AQQLRTLYNYGARKVAVFGVGQVGCSPNELAQNSRNGVTCIERINSAVRMFNRRVVVLVN 259
Query: 262 QFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCEN 321
QFN LP A YIN YGIF+ I+ +P +G + TN GCCGVGRNNGQ+TCLP Q PC N
Sbjct: 260 QFNRLLPGALFTYINCYGIFESIMRTPVEHGLAVTNRGCCGVGRNNGQVTCLPYQAPCAN 319
Query: 322 RREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
R EYLFWDAFHP+EA N+ + +RAYSA SD YP+D+ LAQ+
Sbjct: 320 RDEYLFWDAFHPTEAANIFVGRRAYSAAMRSDVYPVDLSTLAQL 363
>UniRef100_Q9AX26 GDSL-motif lipase/hydrolase-like protein [Oryza sativa]
Length = 363
Score = 491 bits (1263), Expect = e-137
Identities = 226/338 (66%), Positives = 278/338 (81%), Gaps = 1/338 (0%)
Query: 28 PQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLG 87
PQVPCYFIFGDSLVDNGNNN + SLARA+Y PYGIDF GGPSGRF+NG TTVDVIA+LLG
Sbjct: 27 PQVPCYFIFGDSLVDNGNNNYIVSLARANYPPYGIDFAGGPSGRFTNGLTTVDVIAQLLG 86
Query: 88 FDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNI 147
FD+FIPPY +TSGD IL G NFASAAAGIR ETGQQLGGRI F+GQVQNYQ+ V +++I
Sbjct: 87 FDNFIPPYAATSGDQILNGANFASAAAGIRAETGQQLGGRIPFAGQVQNYQTAVQTLISI 146
Query: 148 LGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQT 207
LG++D A++ LSKCI+S+G+GSNDYLNNYFMP FY+T QYT +Q+AD LI Y +Q
Sbjct: 147 LGDQDTASDRLSKCIFSVGMGSNDYLNNYFMPAFYNTGSQYTPEQFADSLIADYRRYVQV 206
Query: 208 LYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQL 267
LYN+GARK+V+ G+GQ+GCSPNELA+ S DG TCV+ I+ A QIFN +L +VD+ N L
Sbjct: 207 LYNYGARKVVMIGVGQVGCSPNELARYSADGATCVARIDSAIQIFNRRLVGLVDEMNT-L 265
Query: 268 PDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLF 327
P A +INAY IF DI+A+ A+YGF+ T GCCGVGRNNGQ+TCLP + PC NR +++F
Sbjct: 266 PGAHFTFINAYNIFSDILANAASYGFTETTAGCCGVGRNNGQVTCLPYEAPCSNRDQHIF 325
Query: 328 WDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
WDAFHPSEA N+++ +R+Y A+SP+DAYP+DI LA +
Sbjct: 326 WDAFHPSEAANIIVGRRSYRAESPNDAYPMDIATLASV 363
>UniRef100_Q75II0 Putative GDSL-like lipase/hydrolase [Oryza sativa]
Length = 365
Score = 484 bits (1247), Expect = e-135
Identities = 230/343 (67%), Positives = 276/343 (80%), Gaps = 2/343 (0%)
Query: 24 VGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPS-GRFSNGKTTVDVI 82
V AAPQVPCYF+FGDSLVDNGNNN + SLARA+Y PYGIDF GG + GRFSNG TTVDVI
Sbjct: 24 VQAAPQVPCYFVFGDSLVDNGNNNDIVSLARANYPPYGIDFAGGAATGRFSNGLTTVDVI 83
Query: 83 AELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVS 142
++LLGF+DFIPP+ S D +L GVNFASAAAGIREETGQQLG RISFSGQVQNYQS V
Sbjct: 84 SKLLGFEDFIPPFAGASSDQLLTGVNFASAAAGIREETGQQLGARISFSGQVQNYQSAVQ 143
Query: 143 QVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYT 202
Q+V+ILG+ED AA +LS+CI+++G+GSNDYLNNYFMP FY+T QYT +QYAD L Y
Sbjct: 144 QLVSILGDEDTAAAHLSQCIFTVGMGSNDYLNNYFMPAFYNTGSQYTPEQYADDLAARYA 203
Query: 203 EQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQ 262
+ L+ +Y+ GARK+ L G+GQ+GCSPNELAQ+S +G TCV IN A +IFN KL +VDQ
Sbjct: 204 QLLRAMYSNGARKVALVGVGQVGCSPNELAQQSANGVTCVERINSAIRIFNQKLVGLVDQ 263
Query: 263 FNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENR 322
FN LP A YIN YGIF DI+ +P ++G TN+GCCGVGRNNGQ+TCLP QTPC NR
Sbjct: 264 FNT-LPGAHFTYINIYGIFDDILGAPGSHGLKVTNQGCCGVGRNNGQVTCLPFQTPCANR 322
Query: 323 REYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
EY FWDAFHP+EA NV++ QR YSA+ SD +P+D++ LA +
Sbjct: 323 HEYAFWDAFHPTEAANVLVGQRTYSARLQSDVHPVDLRTLASL 365
>UniRef100_Q8L5Z1 Hypothetical protein At1g33810 [Arabidopsis thaliana]
Length = 370
Score = 376 bits (965), Expect = e-103
Identities = 183/342 (53%), Positives = 243/342 (70%), Gaps = 7/342 (2%)
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGF 88
QVPC FIFGDSLVDNGNNN L SLARA+Y PYGIDFP G +GRF+NG+T VD +A++LGF
Sbjct: 31 QVPCLFIFGDSLVDNGNNNRLLSLARANYRPYGIDFPQGTTGRFTNGRTYVDALAQILGF 90
Query: 89 DDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
++IPPY G ILRG NFAS AAGIR+ETG LG S + QV+ Y + V Q++
Sbjct: 91 RNYIPPYSRIRGQAILRGANFASGAAGIRDETGDNLGAHTSMNQQVELYTTAVQQMLRYF 150
Query: 149 -GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQT 207
G+ ++ YLS+CI+ G+GSNDYLNNYFMP FYSTS Y +A+ LI+ YT+QL
Sbjct: 151 RGDTNELQRYLSRCIFYSGMGSNDYLNNYFMPDFYSTSTNYNDKTFAESLIKNYTQQLTR 210
Query: 208 LYNFGARKMVLFGIGQIGCSPNELAQ---RSQDGRTCVSDINDANQIFNNKLKSVVDQFN 264
LY FGARK+++ G+GQIGC P +LA+ R+ C IN+A +FN ++K +VD+ N
Sbjct: 211 LYQFGARKVIVTGVGQIGCIPYQLARYNNRNNSTGRCNEKINNAIVVFNTQVKKLVDRLN 270
Query: 265 -NQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRR 323
QL A+ +Y+++Y D+ + A YGF ++GCCGVGRNNGQITCLP+QTPC +R
Sbjct: 271 KGQLKGAKFVYLDSYKSTYDLAVNGAAYGFEVVDKGCCGVGRNNGQITCLPLQTPCPDRT 330
Query: 324 EYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
+YLFWDAFHP+E N+++A+ + S + YPI+I+ LA +
Sbjct: 331 KYLFWDAFHPTETANILLAKSNF--YSRAYTYPINIQELANL 370
>UniRef100_Q9LQ33 F14M2.7 protein [Arabidopsis thaliana]
Length = 390
Score = 358 bits (920), Expect = 1e-97
Identities = 182/362 (50%), Positives = 243/362 (66%), Gaps = 27/362 (7%)
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVI------ 82
QVPC FIFGDSLVDNGNNN L SLARA+Y PYGIDFP G +GRF+NG+T VD +
Sbjct: 31 QVPCLFIFGDSLVDNGNNNRLLSLARANYRPYGIDFPQGTTGRFTNGRTYVDALGIFVGE 90
Query: 83 -------AELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQ 135
+++LGF ++IPPY G ILRG NFAS AAGIR+ETG LG S + QV+
Sbjct: 91 FYMYRALSQILGFRNYIPPYSRIRGQAILRGANFASGAAGIRDETGDNLGAHTSMNQQVE 150
Query: 136 NYQSTVSQVVNIL-GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYA 194
Y + V Q++ G+ ++ YLS+CI+ G+GSNDYLNNYFMP FYSTS Y +A
Sbjct: 151 LYTTAVQQMLRYFRGDTNELQRYLSRCIFYSGMGSNDYLNNYFMPDFYSTSTNYNDKTFA 210
Query: 195 DVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQ---RSQDGRTCVSDINDANQI 251
+ LI+ YT+QL LY FGARK+++ G+GQIGC P +LA+ R+ C IN+A +
Sbjct: 211 ESLIKNYTQQLTRLYQFGARKVIVTGVGQIGCIPYQLARYNNRNNSTGRCNEKINNAIVV 270
Query: 252 FNNKLKSVVDQFN-NQLPDARVIYINAYGIFQDIIASPATY-------GFSNTNEGCCGV 303
FN ++K +VD+ N QL A+ +Y+++Y D+ + A Y GF ++GCCGV
Sbjct: 271 FNTQVKKLVDRLNKGQLKGAKFVYLDSYKSTYDLAVNGAAYVIYIDPTGFEVVDKGCCGV 330
Query: 304 GRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLA 363
GRNNGQITCLP+QTPC +R +YLFWDAFHP+E N+++A+ + S + YPI+I+ LA
Sbjct: 331 GRNNGQITCLPLQTPCPDRTKYLFWDAFHPTETANILLAKSNF--YSRAYTYPINIQELA 388
Query: 364 QI 365
+
Sbjct: 389 NL 390
>UniRef100_Q9SF78 Putative GDSL-motif lipase/hydrolase; 24593-26678 [Arabidopsis
thaliana]
Length = 384
Score = 353 bits (905), Expect = 5e-96
Identities = 169/336 (50%), Positives = 237/336 (70%), Gaps = 5/336 (1%)
Query: 30 VPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFD 89
VP F+FGDSL+DNGNNN + S A+A+Y PYGIDF GGP+GRF NG T VD IA+LLG
Sbjct: 53 VPALFVFGDSLIDNGNNNNIPSFAKANYFPYGIDFNGGPTGRFCNGLTMVDGIAQLLGLP 112
Query: 90 DFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILG 149
IP Y +GD +LRGVN+ASAAAGI +TG GRI F Q+ N+++T+ QV + G
Sbjct: 113 -LIPAYSEATGDQVLRGVNYASAAAGILPDTGGNFVGRIPFDQQIHNFETTLDQVASKSG 171
Query: 150 NEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLY 209
A+ +++ ++ IG+GSNDYLNNY MP F T QY + Q+ D+L+Q YT+QL LY
Sbjct: 172 GAVAIADSVTRSLFFIGMGSNDYLNNYLMPNF-PTRNQYNSQQFGDLLVQHYTDQLTRLY 230
Query: 210 NFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPD 269
N G RK V+ G+G++GC P+ LAQ DG+ C ++N FN +K+++ N LPD
Sbjct: 231 NLGGRKFVVAGLGRMGCIPSILAQ-GNDGK-CSEEVNQLVLPFNTNVKTMISNLNQNLPD 288
Query: 270 ARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWD 329
A+ IY++ +F+DI+A+ A YG + ++GCCG+G+N GQITCLP +TPC NR +Y+FWD
Sbjct: 289 AKFIYLDIAHMFEDIVANQAAYGLTTMDKGCCGIGKNRGQITCLPFETPCPNRDQYVFWD 348
Query: 330 AFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
AFHP+E N+++A++A+ A + AYPI+I++LA +
Sbjct: 349 AFHPTEKVNLIMAKKAF-AGDRTVAYPINIQQLASL 383
>UniRef100_Q60DV9 Hypothetical protein P0426G01.13 [Oryza sativa]
Length = 393
Score = 344 bits (882), Expect = 3e-93
Identities = 168/345 (48%), Positives = 235/345 (67%), Gaps = 7/345 (2%)
Query: 21 GGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVD 80
GG AA VP F+FGDSL DNGNNN + SLA+A+Y+PYGIDF GGP+GRFSNG T VD
Sbjct: 50 GGGAAAAASVPAMFVFGDSLTDNGNNNDMTSLAKANYLPYGIDFAGGPTGRFSNGYTMVD 109
Query: 81 VIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQST 140
IAELLG +P + +GD L GVN+ASAAAGI + TGQ GR F+ Q++N+++T
Sbjct: 110 EIAELLGLP-LLPSHNDATGDAALHGVNYASAAAGILDNTGQNFVGRSPFNQQIKNFEAT 168
Query: 141 VSQVVNILGN--EDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLI 198
+ Q+ LG + A L++ I+ +G+GSNDYLNNY MP Y+T +Y DQY+ +L+
Sbjct: 169 LQQISGKLGGGAAGKLAPSLARSIFYVGMGSNDYLNNYLMPN-YNTRNEYNGDQYSTLLV 227
Query: 199 QAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKS 258
Q YT+QL LYN GAR+ V+ G+G + C PN A+ + C D++D FN+K+KS
Sbjct: 228 QQYTKQLTRLYNLGARRFVIAGVGSMACIPNMRARNPAN--MCSPDVDDLIIPFNSKVKS 285
Query: 259 VVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTP 318
+V+ N LP A+ I+++ Y + +++ +P +YGFS + GCCG+GRN G ITCLP Q P
Sbjct: 286 MVNTLNVNLPRAKFIFVDTYAMISEVLRNPWSYGFSVVDRGCCGIGRNRGMITCLPFQRP 345
Query: 319 CENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLA 363
C NR Y+FWDAFHP+E N+++ + AYS + +P++I++LA
Sbjct: 346 CLNRNTYIFWDAFHPTERVNILLGKAAYSGGADL-VHPMNIQQLA 389
>UniRef100_Q6ZCC3 Putative GDSL-motif lipase/hydrolase protein [Oryza sativa]
Length = 381
Score = 332 bits (850), Expect = 1e-89
Identities = 169/368 (45%), Positives = 240/368 (64%), Gaps = 14/368 (3%)
Query: 7 MMVVAMLVMVLGLWGGW------VGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPY 60
++VV + +V+ GG G A VP F+FGDSL+DNGNNN L S A+A+Y PY
Sbjct: 19 VVVVGAVFVVVAEGGGSEEAAASTGKAAMVPALFVFGDSLIDNGNNNNLASFAKANYYPY 78
Query: 61 GIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSG--DDILRGVNFASAAAGIRE 118
GIDF GP+GRF NG T VD +AELLG +PPY SG +L+GVNFASAAAGI +
Sbjct: 79 GIDFAAGPTGRFCNGYTIVDELAELLGLP-LVPPYSQASGHVQQLLQGVNFASAAAGILD 137
Query: 119 ETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFM 178
E+G GRI F+ Q+ N+++TV Q+ +G ++ AA+ +++ I +GLGSNDYLNNY M
Sbjct: 138 ESGGNFVGRIPFNQQIDNFEATVEQIAGAVGGKEAAASMVARSILFVGLGSNDYLNNYLM 197
Query: 179 PQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDG 238
P Y+T R+YT Q+AD+L Y QL LY GARK V+ G+G +GC PN LAQ +
Sbjct: 198 PN-YNTRRRYTPRQFADLLADRYAAQLTRLYKAGARKFVVAGVGSMGCIPNVLAQSVES- 255
Query: 239 RTCVSDINDANQIFNNKLKSVVDQFNNQ-LPDARVIYINAYGIFQDIIASPATYGFSNTN 297
C +++ FN +++++ + + LP A +++++ YG+F+ I+ PA +GF+ +
Sbjct: 256 -RCSPEVDALVVPFNANVRAMLGRLDGGGLPGASLVFLDNYGVFKAILGDPAAHGFAVVD 314
Query: 298 EGCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPI 357
GCCG+GRN GQ+TCLP PC+ R Y+FWDAFHP+ A NV+IA+ A+ PI
Sbjct: 315 RGCCGIGRNAGQVTCLPFMPPCDGRDRYVFWDAFHPTAAVNVLIAREAFYG-GADVVSPI 373
Query: 358 DIKRLAQI 365
+++RLA +
Sbjct: 374 NVRRLAAL 381
>UniRef100_Q9FNP2 GDSL-motif lipase/acylhydrolase-like protein [Arabidopsis thaliana]
Length = 385
Score = 327 bits (839), Expect = 2e-88
Identities = 168/367 (45%), Positives = 240/367 (64%), Gaps = 11/367 (2%)
Query: 7 MMVVAMLVMVLGLWGGWVGA--APQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDF 64
+++V ++V L GG + P F+FGDSLVDNGNNN L SLAR++Y+PYGIDF
Sbjct: 22 LVLVPWFLVVFVLAGGEDSSETTAMFPAMFVFGDSLVDNGNNNHLNSLARSNYLPYGIDF 81
Query: 65 PGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVST--SGDDILRGVNFASAAAGIREETG 121
G P+GRFSNGKT VD I ELLG + IP ++ T G DIL GVN+ASAA GI EETG
Sbjct: 82 AGNQPTGRFSNGKTIVDFIGELLGLPE-IPAFMDTVDGGVDILHGVNYASAAGGILEETG 140
Query: 122 QQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQF 181
+ LG R S QV+N++ T+ ++ + E Y++K + + LG+NDY+NNY P+
Sbjct: 141 RHLGERFSMGRQVENFEKTLMEISRSMRKES-VKEYMAKSLVVVSLGNNDYINNYLKPRL 199
Query: 182 YSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTC 241
+ +S Y +AD+L+ +T L LY G RK V+ G+G +GC P++LA ++ C
Sbjct: 200 FLSSSIYDPTSFADLLLSNFTTHLLELYGKGFRKFVIAGVGPLGCIPDQLAAQAALPGEC 259
Query: 242 VSDINDANQIFNNKLKSVVDQFNNQ---LPDARVIYINAYGIFQDIIASPATYGFSNTNE 298
V +N+ ++FNN+L S+VD+ N+ +A +Y N YG DI+ +P YGF T+
Sbjct: 260 VEAVNEMAELFNNRLVSLVDRLNSDNKTASEAIFVYGNTYGAAVDILTNPFNYGFEVTDR 319
Query: 299 GCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPID 358
GCCGVGRN G+ITCLP+ PC R ++FWDAFHP++A N++IA RA++ S SD YPI+
Sbjct: 320 GCCGVGRNRGEITCLPLAVPCAFRDRHVFWDAFHPTQAFNLIIALRAFNG-SKSDCYPIN 378
Query: 359 IKRLAQI 365
+ +L+++
Sbjct: 379 LSQLSRL 385
>UniRef100_Q8LFJ9 Hypothetical protein [Arabidopsis thaliana]
Length = 364
Score = 327 bits (838), Expect = 3e-88
Identities = 160/342 (46%), Positives = 231/342 (66%), Gaps = 7/342 (2%)
Query: 28 PQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLG 87
P P +F+FGDSLVD+GNNN + +LARA+Y PYGIDF G P+GRF NG+T VD A LG
Sbjct: 26 PLAPAFFVFGDSLVDSGNNNYIPTLARANYFPYGIDF-GFPTGRFCNGRTVVDYGATYLG 84
Query: 88 FDDFIPPYVS--TSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVS-QV 144
+PPY+S + G + LRGVN+ASAAAGI +ETG+ G R +F+GQ+ ++ T+ ++
Sbjct: 85 LP-LVPPYLSPLSIGQNALRGVNYASAAAGILDETGRHYGARTTFNGQISQFEITIELRL 143
Query: 145 VNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQ 204
N YL+K I I +GSNDY+NNY MP+ YSTS+ Y+ + YAD+LI+ + Q
Sbjct: 144 RRFFQNPADLRKYLAKSIIGINIGSNDYINNYLMPERYSTSQTYSGEDYADLLIKTLSAQ 203
Query: 205 LQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRT-CVSDINDANQIFNNKLKSVVDQF 263
+ LYN GARKMVL G G +GC P++L+ + + + CV+ IN+ +FN++LK + +
Sbjct: 204 ISRLYNLGARKMVLAGSGPLGCIPSQLSMVTGNNTSGCVTKINNMVSMFNSRLKDLANTL 263
Query: 264 NNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRR 323
N LP + +Y N + +F D++ +P+ YG +NE CCG GR G +TCLP+Q PC +R
Sbjct: 264 NTTLPGSFFVYQNVFDLFHDMVVNPSRYGLVVSNEACCGNGRYGGALTCLPLQQPCLDRN 323
Query: 324 EYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
+Y+FWDAFHP+E N +IA +S +S + +YPI + LA++
Sbjct: 324 QYVFWDAFHPTETANKIIAHNTFS-KSANYSYPISVYELAKL 364
>UniRef100_Q9FVV1 Putative GDSL-motif lipase/acylhydrolase; 82739-81282 [Arabidopsis
thaliana]
Length = 374
Score = 322 bits (826), Expect = 8e-87
Identities = 158/358 (44%), Positives = 236/358 (65%), Gaps = 5/358 (1%)
Query: 7 MMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
++++A+ V V+ V +VP F+ GDSLVD GNNN L+++ARA+++PYGID
Sbjct: 16 VLILALTVSVILQQPELVTGQARVPAMFVLGDSLVDAGNNNFLQTVARANFLPYGIDMNY 75
Query: 67 GPSGRFSNGKTTVDVIAELLGFDDFIPPYV--STSGDDILRGVNFASAAAGIREETGQQL 124
P+GRFSNG T +D++A LL PP+ +TSG+ IL+GVN+ASAAAGI + +G
Sbjct: 76 QPTGRFSNGLTFIDLLARLLEIPS-PPPFADPTTSGNRILQGVNYASAAAGILDVSGYNY 134
Query: 125 GGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYST 184
GGR S + Q+ N ++T+SQ+ ++ ++ +YL++ + + GSNDY+NNY MP Y +
Sbjct: 135 GGRFSLNQQMVNLETTLSQLRTMMSPQN-FTDYLARSLVVLVFGSNDYINNYLMPNLYDS 193
Query: 185 SRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSD 244
S ++ +A++L+ Y QL TLY+ G RK+ + G+ +GC PN+ A+ CV
Sbjct: 194 SIRFRPPDFANLLLSQYARQLLTLYSLGLRKIFIPGVAPLGCIPNQRARGISPPDRCVDS 253
Query: 245 INDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVG 304
+N FN LKS+VDQ N + P A +Y N Y DI+ +PA YGFS + CCG+G
Sbjct: 254 VNQILGTFNQGLKSLVDQLNQRSPGAIYVYGNTYSAIGDILNNPAAYGFSVVDRACCGIG 313
Query: 305 RNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRL 362
RN GQITCLP+QTPC NR +Y+FWDAFHP++ N ++A+RA+ PSDAYP++++++
Sbjct: 314 RNQGQITCLPLQTPCPNRNQYVFWDAFHPTQTANSILARRAFYG-PPSDAYPVNVQQM 370
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 628,731,290
Number of Sequences: 2790947
Number of extensions: 28247442
Number of successful extensions: 71717
Number of sequences better than 10.0: 414
Number of HSP's better than 10.0 without gapping: 294
Number of HSP's successfully gapped in prelim test: 121
Number of HSP's that attempted gapping in prelim test: 70154
Number of HSP's gapped (non-prelim): 481
length of query: 365
length of database: 848,049,833
effective HSP length: 129
effective length of query: 236
effective length of database: 488,017,670
effective search space: 115172170120
effective search space used: 115172170120
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0404.11


