

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.9
(321 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
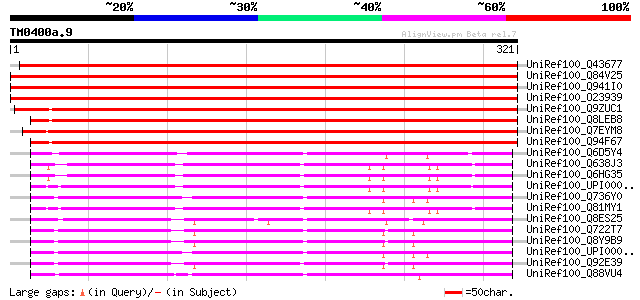
UniRef100_Q43677 Auxin-induced protein [Vigna radiata] 541 e-153
UniRef100_Q84V25 Quinone oxidoreductase [Fragaria ananassa] 480 e-134
UniRef100_Q941I0 Putative quinone oxidoreductase [Fragaria anana... 475 e-133
UniRef100_O23939 Ripening-induced protein [Fragaria vesca] 474 e-132
UniRef100_Q9ZUC1 Quinone oxidoreductase-like protein At1g23740, ... 415 e-115
UniRef100_Q8LEB8 Quinone oxidoreductase-like protein [Arabidopsi... 409 e-113
UniRef100_Q7EYM8 Putative oxidoreductase, zinc-binding [Oryza sa... 406 e-112
UniRef100_Q94F67 Quinone oxidoreductase-like protein [Helianthus... 398 e-109
UniRef100_Q6D5Y4 Probable zinc-binding dehydrogenase [Erwinia ca... 186 6e-46
UniRef100_Q638J3 Quinone oxidoreductase [Bacillus cereus] 181 2e-44
UniRef100_Q6HG35 Quinone oxidoreductase [Bacillus thuringiensis] 181 2e-44
UniRef100_UPI00003CB31E UPI00003CB31E UniRef100 entry 180 4e-44
UniRef100_Q736Y0 Oxidoreductase, zinc-binding [Bacillus cereus] 176 8e-43
UniRef100_Q81MY1 Alcohol dehydrogenase, zinc-containing [Bacillu... 175 2e-42
UniRef100_Q8ES25 Zinc-binding oxidoreductase [Oceanobacillus ihe... 173 5e-42
UniRef100_Q722T7 Alcohol dehydrogenase, zinc-dependent [Listeria... 172 1e-41
UniRef100_Q8Y9B9 Lmo0613 protein [Listeria monocytogenes] 171 2e-41
UniRef100_UPI00003CC0B8 UPI00003CC0B8 UniRef100 entry 170 4e-41
UniRef100_Q92E39 Lin0622 protein [Listeria innocua] 170 5e-41
UniRef100_Q88VU4 Oxidoreductase [Lactobacillus plantarum] 169 7e-41
>UniRef100_Q43677 Auxin-induced protein [Vigna radiata]
Length = 317
Score = 541 bits (1394), Expect = e-153
Identities = 266/315 (84%), Positives = 294/315 (92%)
Query: 7 SNIPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFK 66
++IPS +AW+YS+YG+IEE+LKFDPNVPTPHL +QVLIKV AAA+NPVD KRALGHFK
Sbjct: 2 TSIPSHMRAWIYSEYGNIEEVLKFDPNVPTPHLMENQVLIKVVAAAINPVDYKRALGHFK 61
Query: 67 DIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEE 126
DIDSPLPT PGYDVAGVVVKVG+QVKK +VGDEVYGD+NE+TLDHPKT+GSLAEYTA EE
Sbjct: 62 DIDSPLPTAPGYDVAGVVVKVGNQVKKFQVGDEVYGDINEVTLDHPKTIGSLAEYTAVEE 121
Query: 127 KVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHV 186
KVLAHKPSNLSF+EAASLPLAIITAYQGLETAQFS GKSILVLGGAGGVGSLVIQ+AKH+
Sbjct: 122 KVLAHKPSNLSFVEAASLPLAIITAYQGLETAQFSVGKSILVLGGAGGVGSLVIQIAKHI 181
Query: 187 FEASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKAVKE 246
F ASKIAAT STGKLE LG DLPIDYTKENFEE++EKFDVVYDAVGQS+RALK+VKE
Sbjct: 182 FGASKIAATGSTGKLELFENLGTDLPIDYTKENFEELAEKFDVVYDAVGQSERALKSVKE 241
Query: 247 GGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAFSY 306
GGKVVTI+ P TPPAIPFLLTSDG +LEKL+PYLE+G+VKPILDPKSPFPFSQTVEAFSY
Sbjct: 242 GGKVVTIVAPATPPAIPFLLTSDGDLLEKLRPYLENGQVKPILDPKSPFPFSQTVEAFSY 301
Query: 307 LNTSRATGEVVIYPI 321
L T+RATG+VVIYPI
Sbjct: 302 LKTNRATGKVVIYPI 316
>UniRef100_Q84V25 Quinone oxidoreductase [Fragaria ananassa]
Length = 322
Score = 480 bits (1235), Expect = e-134
Identities = 241/321 (75%), Positives = 278/321 (86%)
Query: 1 MASTPYSNIPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKR 60
MA+ P +IPS KAWVYS+YG ++LKFDP+V P +K DQVLIKV AA+LNPVD KR
Sbjct: 1 MAAAPSESIPSVNKAWVYSEYGKTSDVLKFDPSVAVPEVKEDQVLIKVVAASLNPVDFKR 60
Query: 61 ALGHFKDIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAE 120
ALG+FKD DSPLPTVPGYDVAGVVVKVGSQV K KVGDEVYGD+NE L +P GSLAE
Sbjct: 61 ALGYFKDTDSPLPTVPGYDVAGVVVKVGSQVTKFKVGDEVYGDLNEAALVNPTRFGSLAE 120
Query: 121 YTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVI 180
YTAA+E+VLAHKP +LSFIEAASLPLAI TAY+GLE A+ SAGKSILVLGGAGGVG+ +I
Sbjct: 121 YTAADERVLAHKPKDLSFIEAASLPLAIETAYEGLERAELSAGKSILVLGGAGGVGTHII 180
Query: 181 QLAKHVFEASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRA 240
QLAKHVF ASK+AATAST KL+FLR LG DL IDYTKEN E++ EKFDVVYDAVG++D+A
Sbjct: 181 QLAKHVFGASKVAATASTKKLDFLRTLGVDLAIDYTKENIEDLPEKFDVVYDAVGETDKA 240
Query: 241 LKAVKEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQT 300
+KAVKEGGKVVTI+ P TPPAI F+LTS G+VLEKL+PYLESGKVKP+LDP SP+PF++
Sbjct: 241 VKAVKEGGKVVTIVGPATPPAIHFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTKL 300
Query: 301 VEAFSYLNTSRATGEVVIYPI 321
VEAF YL +SRATG+VV+YPI
Sbjct: 301 VEAFGYLESSRATGKVVVYPI 321
>UniRef100_Q941I0 Putative quinone oxidoreductase [Fragaria ananassa]
Length = 339
Score = 475 bits (1222), Expect = e-133
Identities = 240/322 (74%), Positives = 278/322 (85%), Gaps = 1/322 (0%)
Query: 1 MASTPYSNIPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKR 60
MA+ P +IPS KAWVYS+YG ++LKFDP+V P +K DQVLIKV AA+LNPVD KR
Sbjct: 17 MAAAPSESIPSVNKAWVYSEYGKTSDVLKFDPSVAVPEIKEDQVLIKVVAASLNPVDFKR 76
Query: 61 ALGHFKDIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAE 120
ALG+FKD DSPLPT+PGYDVAGVVVKVGSQV K KVGDEVYGD+NE L +P GSLAE
Sbjct: 77 ALGYFKDTDSPLPTIPGYDVAGVVVKVGSQVTKFKVGDEVYGDLNETALVNPTRFGSLAE 136
Query: 121 YTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVI 180
YTAA E+VLAHKP NLSFIEAASLPLAI TA++GLE A+ SAGKS+LVLGGAGGVG+ +I
Sbjct: 137 YTAAYERVLAHKPKNLSFIEAASLPLAIETAHEGLERAELSAGKSVLVLGGAGGVGTHII 196
Query: 181 QLAKHVFEASKIAATASTGKLEFLRKLG-ADLPIDYTKENFEEVSEKFDVVYDAVGQSDR 239
QLAKHVF ASK+AATAST KL+ LR LG ADL IDYTKENFE++ EKFDVVYDAVG++D+
Sbjct: 197 QLAKHVFGASKVAATASTKKLDLLRTLGAADLAIDYTKENFEDLPEKFDVVYDAVGETDK 256
Query: 240 ALKAVKEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQ 299
A+KAVKEGGKVVTI+ P TPPAI F+LTS G+VLEKL+PYLESGKVKP+LDP SP+PF++
Sbjct: 257 AVKAVKEGGKVVTIVGPATPPAILFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTK 316
Query: 300 TVEAFSYLNTSRATGEVVIYPI 321
VEAF YL +SRATG+VV+YPI
Sbjct: 317 VVEAFGYLESSRATGKVVVYPI 338
>UniRef100_O23939 Ripening-induced protein [Fragaria vesca]
Length = 337
Score = 474 bits (1221), Expect = e-132
Identities = 238/321 (74%), Positives = 277/321 (86%)
Query: 1 MASTPYSNIPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKR 60
MA+ P +IPS KAWV S+YG ++LKFDP+V P +K DQVLIKV AA+LNPVD KR
Sbjct: 16 MAAAPSESIPSVNKAWVXSEYGKTSDVLKFDPSVAVPEIKEDQVLIKVVAASLNPVDFKR 75
Query: 61 ALGHFKDIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAE 120
ALG+FKD DSPLPT+PGY VAGVVVKVGSQV K KVGDEVYGD+NE L +P GSLAE
Sbjct: 76 ALGYFKDTDSPLPTIPGYYVAGVVVKVGSQVTKFKVGDEVYGDLNETALVNPTRFGSLAE 135
Query: 121 YTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVI 180
YTAA+E+VLAHKP NLSFIEAASLPLAI TA++GLE A+ SAGKS+LVLGGAGGVG+ +I
Sbjct: 136 YTAADERVLAHKPKNLSFIEAASLPLAIETAHEGLERAELSAGKSVLVLGGAGGVGTHII 195
Query: 181 QLAKHVFEASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRA 240
QLAKHVF ASK+AATAST KL+ LR LGADL IDYTKENFE++ EKFDVVYDAVG++D+A
Sbjct: 196 QLAKHVFGASKVAATASTKKLDLLRTLGADLAIDYTKENFEDLPEKFDVVYDAVGETDKA 255
Query: 241 LKAVKEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQT 300
+KAVKEGGKVVTI+ P TPPAI F+LTS G+VLEKL+PYLESGKVKP+LDP SP+PF++
Sbjct: 256 VKAVKEGGKVVTIVGPATPPAILFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTKV 315
Query: 301 VEAFSYLNTSRATGEVVIYPI 321
VEAF YL +SRATG+VV+YPI
Sbjct: 316 VEAFGYLESSRATGKVVVYPI 336
>UniRef100_Q9ZUC1 Quinone oxidoreductase-like protein At1g23740, chloroplast
precursor [Arabidopsis thaliana]
Length = 386
Score = 415 bits (1067), Expect = e-115
Identities = 210/318 (66%), Positives = 253/318 (79%), Gaps = 1/318 (0%)
Query: 4 TPYSNIPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALG 63
T ++IP + KAWVYS YG ++ +LK + N+ P +K DQVLIKV AAALNPVD+KR G
Sbjct: 69 TADASIPKEMKAWVYSDYGGVD-VLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQG 127
Query: 64 HFKDIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTA 123
FK DSPLPTVPGYDVAGVVVKVGS VK LK GDEVY +V+E L+ PK GSLAEYTA
Sbjct: 128 KFKATDSPLPTVPGYDVAGVVVKVGSAVKDLKEGDEVYANVSEKALEGPKQFGSLAEYTA 187
Query: 124 AEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLA 183
EEK+LA KP N+ F +AA LPLAI TA +GL +FSAGKSILVL GAGGVGSLVIQLA
Sbjct: 188 VEEKLLALKPKNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLVIQLA 247
Query: 184 KHVFEASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKA 243
KHV+ ASK+AATAST KLE +R LGADL IDYTKEN E++ +K+DVV+DA+G D+A+K
Sbjct: 248 KHVYGASKVAATASTEKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKV 307
Query: 244 VKEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEA 303
+KEGGKVV + TPP F++TS+G VL+KL PY+ESGKVKP++DPK PFPFS+ +A
Sbjct: 308 IKEGGKVVALTGAVTPPGFRFVVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADA 367
Query: 304 FSYLNTSRATGEVVIYPI 321
FSYL T+ ATG+VV+YPI
Sbjct: 368 FSYLETNHATGKVVVYPI 385
>UniRef100_Q8LEB8 Quinone oxidoreductase-like protein [Arabidopsis thaliana]
Length = 309
Score = 409 bits (1050), Expect = e-113
Identities = 205/308 (66%), Positives = 246/308 (79%), Gaps = 1/308 (0%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLP 73
KAWVYS YG ++ +LK + N+ P +K DQVLIKV AA LNPVD+KR G FK DSPLP
Sbjct: 2 KAWVYSDYGGVD-VLKLESNIAVPEIKEDQVLIKVVAAGLNPVDAKRRQGKFKATDSPLP 60
Query: 74 TVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKP 133
TVPGYDVAGVVVKVGS VK K GDEVY +V+E L+ PK GSLAEYTA EEK+LA KP
Sbjct: 61 TVPGYDVAGVVVKVGSAVKDFKEGDEVYANVSEKALEGPKQFGSLAEYTAVEEKLLALKP 120
Query: 134 SNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIA 193
N+ F +AA LPLAI TA +GL +FSAGKSILVL GAGGVGSL+IQLAKHV+ ASK+A
Sbjct: 121 KNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLMIQLAKHVYGASKVA 180
Query: 194 ATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKAVKEGGKVVTI 253
ATASTGKLE +R LGADL IDYTKEN E++ +K+DVV+DA+G D+A+K +KEGGKVV +
Sbjct: 181 ATASTGKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEGGKVVAL 240
Query: 254 LPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAFSYLNTSRAT 313
TPP F++TS+G VL+KL PY+ESGKVKP++DPK PFPFS+ +AFSYL T+ AT
Sbjct: 241 TGAVTPPGFRFVVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYLETNHAT 300
Query: 314 GEVVIYPI 321
G+VV+YPI
Sbjct: 301 GKVVVYPI 308
>UniRef100_Q7EYM8 Putative oxidoreductase, zinc-binding [Oryza sativa]
Length = 390
Score = 406 bits (1044), Expect = e-112
Identities = 208/313 (66%), Positives = 245/313 (77%), Gaps = 1/313 (0%)
Query: 9 IPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI 68
+P+ KAW Y YGD +LK + P + DQVL++V AAALNPVD+KR G FK
Sbjct: 78 VPATMKAWAYDDYGD-GSVLKLNDAAAVPDIADDQVLVRVAAAALNPVDAKRRAGKFKAT 136
Query: 69 DSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKV 128
DSPLPTVPGYDVAGVVVK G +VK LK GDEVYG+++E L+ PK GSLAEYTA EEK+
Sbjct: 137 DSPLPTVPGYDVAGVVVKAGRKVKGLKEGDEVYGNISEKALEGPKQSGSLAEYTAVEEKL 196
Query: 129 LAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFE 188
LA KP +L F +AA LPLAI TA++GLE A FSAGKSIL+LGGAGGVGSL IQLAKHV+
Sbjct: 197 LALKPKSLGFAQAAGLPLAIETAHEGLERAGFSAGKSILILGGAGGVGSLAIQLAKHVYG 256
Query: 189 ASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKAVKEGG 248
ASK+AATAST KLE L+ LGAD+ IDYTKENFE++ +K+DVV DAVGQ ++A+K VKEGG
Sbjct: 257 ASKVAATASTPKLELLKSLGADVAIDYTKENFEDLPDKYDVVLDAVGQGEKAVKVVKEGG 316
Query: 249 KVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAFSYLN 308
VV + PP F++TSDG+VLEKL PYLESGKVKP++DPK PF FSQ VEAFSYL
Sbjct: 317 SVVVLTGAVVPPGFRFVVTSDGSVLEKLNPYLESGKVKPLVDPKGPFAFSQVVEAFSYLE 376
Query: 309 TSRATGEVVIYPI 321
T RATG+VVI PI
Sbjct: 377 TGRATGKVVISPI 389
>UniRef100_Q94F67 Quinone oxidoreductase-like protein [Helianthus annuus]
Length = 309
Score = 398 bits (1022), Expect = e-109
Identities = 199/308 (64%), Positives = 246/308 (79%), Gaps = 1/308 (0%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLP 73
KAW Y +YG ++ +LK +V P +K DQVL+KV AAA+NPVD KR LG+FK IDSPLP
Sbjct: 2 KAWKYDEYGSVD-VLKLATDVAVPEIKDDQVLVKVVAAAVNPVDYKRRLGYFKAIDSPLP 60
Query: 74 TVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKP 133
+PG+DV GVV+KVGSQVK LK GDEVYGD+N+ L+ P G+LAEYTA EE++LA KP
Sbjct: 61 IIPGFDVLGVVLKVGSQVKDLKEGDEVYGDINDKGLEGPTQFGTLAEYTAVEERLLALKP 120
Query: 134 SNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIA 193
NL FI+AA+LPLAI TAY+GLE A+FS GK+ILVL GAGGVGS +IQLAKHV+ ASK+A
Sbjct: 121 KNLDFIQAAALPLAIETAYEGLERAKFSEGKTILVLNGAGGVGSFIIQLAKHVYGASKVA 180
Query: 194 ATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKAVKEGGKVVTI 253
AT+STGKLE L+ LGAD+ IDYTKENFE++ +K+DVVYDA+GQ ++ALKAV E G V+I
Sbjct: 181 ATSSTGKLELLKSLGADVAIDYTKENFEDLPDKYDVVYDAIGQPEKALKAVNETGVAVSI 240
Query: 254 LPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAFSYLNTSRAT 313
P PP F+LTSDG++L KL PYLESGK+K DPKSPFPF + EA++YL ++ T
Sbjct: 241 TGPIPPPGFSFILTSDGSILTKLNPYLESGKIKSDNDPKSPFPFDKLNEAYAYLESTGPT 300
Query: 314 GEVVIYPI 321
G+VVIYPI
Sbjct: 301 GKVVIYPI 308
>UniRef100_Q6D5Y4 Probable zinc-binding dehydrogenase [Erwinia carotovora]
Length = 331
Score = 186 bits (473), Expect = 6e-46
Identities = 122/341 (35%), Positives = 186/341 (53%), Gaps = 50/341 (14%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI-DSPL 72
KA+ ++Y D +I +P P + + VL++V AA++N +D+K G FK I
Sbjct: 2 KAFFINRYKDTGQI----GQLPDPEVGHNDVLVQVHAASINLLDAKIRKGEFKLILPYRF 57
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHK 132
P V G D+AGVVV+VG V++ K GDEVY E +GS AE A +E+ LA K
Sbjct: 58 PLVLGNDLAGVVVRVGRNVRQFKAGDEVYARPPE------DRIGSFAELIAVKEEALALK 111
Query: 133 PSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASK 191
P+NL +AAS+PL +TA+Q L ETAQ G+ +L+ G+GGVG++ IQLAKH+ +
Sbjct: 112 PTNLDMEQAASIPLVALTAWQVLVETAQLKKGQRVLIHAGSGGVGTIAIQLAKHL--GAF 169
Query: 192 IAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQS--DRALKAVKEGGK 249
+A T ST +E+++ LGAD+ IDY + FE V + +DVV +++G +++L+ +K GG+
Sbjct: 170 VATTTSTRNVEWVKSLGADVVIDYKTQAFESVLKDYDVVLNSLGNDVLEKSLQVLKPGGR 229
Query: 250 VVTILPPGTPPAIP--------------------------------FLLTSDGAVLEKLQ 277
+++I P TP + +DG L +
Sbjct: 230 LISISGPPTPAFAKQQGLSWFLKQVLRLLSRRIRQKADKRGVRYNFVFMRADGDQLRDIT 289
Query: 278 PYLESGKVKPILDPKSPFPFSQTVEAFSYLNTSRATGEVVI 318
+ESG +KP++D FP T EA +Y R+ G+V I
Sbjct: 290 ALIESGVIKPVID--RAFPLEATAEALAYAEQGRSKGKVTI 328
>UniRef100_Q638J3 Quinone oxidoreductase [Bacillus cereus]
Length = 331
Score = 181 bits (460), Expect = 2e-44
Identities = 120/340 (35%), Positives = 185/340 (54%), Gaps = 49/340 (14%)
Query: 14 KAWVYSQYGD-----IEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI 68
KA + +GD +EE+ K P L VLI V+A ++NP+D+K G +
Sbjct: 2 KAQIIHSFGDSSVFQLEEVAK-------PKLLPGHVLIHVKATSVNPIDTKMRSGAVSSV 54
Query: 69 DSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKV 128
P + DVAG+V++VG V K K GDEVYG +T G+LAE+ A+ ++
Sbjct: 55 APEFPAILHGDVAGIVIEVGEGVSKFKTGDEVYGCAG----GFKETGGALAEFMLADARL 110
Query: 129 LAHKPSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVF 187
+AHKP N++ EAA+LPL ITA++ L + A +G+++L+ G GGVG + IQLAK +
Sbjct: 111 IAHKPKNVTMEEAAALPLVAITAWESLFDRANMKSGQNVLIHGATGGVGHVAIQLAK--W 168
Query: 188 EASKIAATAS-TGKLEFLRKLGADLPIDYTKENFEEVSEK------FDVVYDAVG--QSD 238
+K+ TAS K+E +LGAD+ I+Y +E+ +E +K F+V++D VG D
Sbjct: 169 AGAKVFTTASQQNKMEIAHRLGADIAINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLD 228
Query: 239 RALKAVKEGGKVVTILPPGTPPAIPF---------------LLTSD-----GAVLEKLQP 278
+ +A G VVTI T P +L +D G +L K+
Sbjct: 229 NSFEAAAVNGTVVTIAARSTHDLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITQ 288
Query: 279 YLESGKVKPILDPKSPFPFSQTVEAFSYLNTSRATGEVVI 318
+E GK++P+LD KS F F + +A YL +++A G++V+
Sbjct: 289 IVEEGKLRPLLDSKS-FTFDEVAQAHEYLESNKAIGKIVL 327
>UniRef100_Q6HG35 Quinone oxidoreductase [Bacillus thuringiensis]
Length = 331
Score = 181 bits (459), Expect = 2e-44
Identities = 120/340 (35%), Positives = 185/340 (54%), Gaps = 49/340 (14%)
Query: 14 KAWVYSQYGD-----IEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI 68
KA + +GD +EE+ K P L VLI V+A ++NP+D+K G +
Sbjct: 2 KAQIIHSFGDSSVFQLEEVAK-------PKLLPGHVLIHVKATSVNPIDTKMRSGAVSSV 54
Query: 69 DSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKV 128
P + DVAG+V++VG V K K GDEVYG +T G+LAE+ A+ ++
Sbjct: 55 APEFPAILHGDVAGIVIEVGEGVSKFKTGDEVYGCAG----GFKETGGALAEFMLADARL 110
Query: 129 LAHKPSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVF 187
+AHKP N++ EAA+LPL ITA++ L + A +G+++L+ G GGVG + IQLAK +
Sbjct: 111 IAHKPKNITMEEAAALPLVAITAWESLFDRANIKSGQNVLIHGATGGVGHVAIQLAK--W 168
Query: 188 EASKIAATAS-TGKLEFLRKLGADLPIDYTKENFEEVSEK------FDVVYDAVG--QSD 238
+K+ TAS K+E +LGAD+ I+Y +E+ +E +K F+V++D VG D
Sbjct: 169 AGAKVFTTASQQNKMEISHRLGADVAINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLD 228
Query: 239 RALKAVKEGGKVVTILPPGTPPAIPF---------------LLTSD-----GAVLEKLQP 278
+ +A G VVTI T P +L +D G +L K+
Sbjct: 229 NSFEAAAVNGTVVTIAARSTHDLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITQ 288
Query: 279 YLESGKVKPILDPKSPFPFSQTVEAFSYLNTSRATGEVVI 318
+E GK++P+LD KS F F + +A YL +++A G++V+
Sbjct: 289 IVEEGKLRPLLDSKS-FTFDEVAQAHEYLESNKAIGKIVL 327
>UniRef100_UPI00003CB31E UPI00003CB31E UniRef100 entry
Length = 331
Score = 180 bits (457), Expect = 4e-44
Identities = 119/335 (35%), Positives = 183/335 (54%), Gaps = 39/335 (11%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLP 73
KA + +GD + + + V P L VLI V+A ++NP+D+K G + P
Sbjct: 2 KAQIIHSFGD-SSVFQLE-EVSKPKLLPGHVLIHVKATSVNPIDTKMRSGAVSAVAPEFP 59
Query: 74 TVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKP 133
+ DVAG+V++VG V K K GDEVYG +T G+LAE+ A+ +++AHKP
Sbjct: 60 AILHGDVAGIVIEVGEGVSKFKSGDEVYGCAG----GFKETGGALAEFMLADARLIAHKP 115
Query: 134 SNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKI 192
+NL+ EAA+LPL ITA++ L + A G+++L+ G GGVG + IQLAK + +K+
Sbjct: 116 NNLTMEEAAALPLVAITAWESLFDRANIKPGQNVLIHGATGGVGHVAIQLAK--WAGAKV 173
Query: 193 AATAS-TGKLEFLRKLGADLPIDYTKENFEEVSEK------FDVVYDAVG--QSDRALKA 243
TAS K+E LGAD+ I+Y +E+ +E +K F+V++D VG D + +A
Sbjct: 174 FTTASQQSKMEISHHLGADIAINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLDHSFEA 233
Query: 244 VKEGGKVVTILPPGTPPAIPF---------------LLTSD-----GAVLEKLQPYLESG 283
G VVTI T P +L +D G +L K+ +E G
Sbjct: 234 AAVNGTVVTIAARSTHDLSPLHTKGLTLHVTFMALKILHTDKRNHCGEILTKITQIIEEG 293
Query: 284 KVKPILDPKSPFPFSQTVEAFSYLNTSRATGEVVI 318
K++P+LD K PF F + +A YL +++A G++V+
Sbjct: 294 KLRPLLDSK-PFTFDEVAQAHEYLESNKAIGKIVL 327
>UniRef100_Q736Y0 Oxidoreductase, zinc-binding [Bacillus cereus]
Length = 308
Score = 176 bits (446), Expect = 8e-43
Identities = 121/314 (38%), Positives = 178/314 (56%), Gaps = 19/314 (6%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKD-IDSPL 72
KA V QYG +EE+ + V P +K ++VLI++ A ++NPVD K G ++ +
Sbjct: 2 KAIVIDQYGSVEELK--ERQVLKPVVKNNEVLIRILATSINPVDWKIRKGDLQEQLRFSF 59
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHK 132
P + G DVAGV+ +VG +++ K+GD+VY I GS AE+ + ++++
Sbjct: 60 PIILGLDVAGVIEEVGEEIECFKIGDKVYTKPENIGQ------GSYAEFITVKSDLVSYM 113
Query: 133 PSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASK 191
P+NLSF EAAS+PLA +TA+Q L + A+ +L+ GAGGVGSL IQ+AK +
Sbjct: 114 PTNLSFEEAASIPLAGLTAWQSLVDYAKIKEHDRVLIHAGAGGVGSLAIQIAKSF--GAF 171
Query: 192 IAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVG--QSDRALKAVKEGGK 249
+A TAS+ EFL+ LGADL IDY K+ FEE+ FD+V D +G +++ + +K GG
Sbjct: 172 VATTASSKNEEFLKSLGADLVIDYKKDKFEELLSDFDIVLDTIGGEVQEKSFQILKPGGV 231
Query: 250 VVTIL--PPGTPPAIP--FL-LTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAF 304
+V+I+ P I FL L +G LE L + GKVKPI+ PF EA
Sbjct: 232 LVSIVHEPIQKVKEIKSGFLWLKPNGKQLEALSDLIVHGKVKPIISKVMPFSEEGIREAH 291
Query: 305 SYLNTSRATGEVVI 318
G++VI
Sbjct: 292 ILSEGQHVRGKIVI 305
>UniRef100_Q81MY1 Alcohol dehydrogenase, zinc-containing [Bacillus anthracis]
Length = 335
Score = 175 bits (443), Expect = 2e-42
Identities = 116/339 (34%), Positives = 184/339 (54%), Gaps = 43/339 (12%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLP 73
KA + +GD + + + V P L VLI V+A ++NP+D+K G + P
Sbjct: 2 KAQIIHSFGD-SSVFQLE-EVSKPKLLPGHVLIDVKATSVNPIDTKMRSGAVSAVAPEFP 59
Query: 74 TVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKP 133
+ DVAG+V++VG V K K GDEVYG +T G+LAE+ A+ +++AHKP
Sbjct: 60 AILHGDVAGIVIEVGEGVSKFKCGDEVYGCAG----GFKETGGALAEFMLADARLIAHKP 115
Query: 134 SNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKI 192
+N++ EAA+LPL ITA++ L + A +G+++L+ G GGVG + IQLAK + + +
Sbjct: 116 NNITMEEAAALPLVAITAWESLFDRANIKSGQNVLIHGATGGVGHVAIQLAK--WAGANV 173
Query: 193 AATAS-TGKLEFLRKLGADLPIDYTKENFEEVSEK----------FDVVYDAVG--QSDR 239
TAS K+E +LGAD+ I+Y +E+ +E ++ F+V++D VG D
Sbjct: 174 FTTASQQNKMEIAHRLGADVAINYKEESVQESVQEYVQKHTNGNGFEVIFDTVGGKNLDN 233
Query: 240 ALKAVKEGGKVVTILPPGTPPAIPF---------------LLTSD-----GAVLEKLQPY 279
+ +A G VVTI T P +L +D G +L K+
Sbjct: 234 SFEAAAVNGTVVTIAARSTHDLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITKI 293
Query: 280 LESGKVKPILDPKSPFPFSQTVEAFSYLNTSRATGEVVI 318
+E GK++P+LD KS F F + +A YL +++A G++V+
Sbjct: 294 VEEGKLRPLLDSKS-FTFDEVAQAHEYLESNKAIGKIVL 331
>UniRef100_Q8ES25 Zinc-binding oxidoreductase [Oceanobacillus iheyensis]
Length = 311
Score = 173 bits (439), Expect = 5e-42
Identities = 124/323 (38%), Positives = 176/323 (54%), Gaps = 34/323 (10%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKD-IDSPL 72
KA V YG +E+ + D VP P + +Q+LI+ A ++NP+D K G+ KD +
Sbjct: 2 KAIVIENYGHADELHEQD--VPKPTINDNQILIEQYATSINPIDWKLREGYLKDGFNFEF 59
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTV--GSLAEYTAAEEKVLA 130
P + G+D AGVV +VG V + KVGD V+ P T G+ AE+ A EE +A
Sbjct: 60 PIILGWDSAGVVAEVGKNVSQFKVGDRVFS--------RPATTAQGTYAEFVAVEESRVA 111
Query: 131 HKPSNLSFIEAASLPLAIITAYQGLETAQFSA---GKSILVLGGAGGVGSLVIQLAKHVF 187
P N+SF EAAS+PLA +TA+Q L FSA G +L+ G+GGVGS IQ AKH
Sbjct: 112 KIPENVSFEEAASVPLAGLTAWQCL--VDFSAIKKGDKVLIHAGSGGVGSFAIQFAKHF- 168
Query: 188 EASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQS--DRALKAVK 245
+ +A TAS +++LGAD I+Y +ENF EV E +D+V D +G +++ + +K
Sbjct: 169 -GAYVATTASGKNESLVKELGADRFINYKEENFNEVLEDYDIVVDTLGGDILEQSFEVLK 227
Query: 246 EGGKVVTILPPGTPP---------AIPFL-LTSDGAVLEKLQPYLESGKVKPILDPKSPF 295
EGGK+V+I G P FL L +G L ++ LE+GKVK ++ P
Sbjct: 228 EGGKLVSI--AGNPDEEKAKEKGIKAGFLWLEENGKQLSEVADLLENGKVKSVIGHTFPL 285
Query: 296 PFSQTVEAFSYLNTSRATGEVVI 318
EA T A G++VI
Sbjct: 286 TEQGLREAHQLSETHHARGKIVI 308
>UniRef100_Q722T7 Alcohol dehydrogenase, zinc-dependent [Listeria monocytogenes]
Length = 313
Score = 172 bits (435), Expect = 1e-41
Identities = 121/321 (37%), Positives = 177/321 (54%), Gaps = 29/321 (9%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI-DSPL 72
KA V YG EE+ + V P +QV++K A ++NP+D K G+ K + D
Sbjct: 2 KAVVIENYGGKEELK--EKEVAMPKAGKNQVIVKEAATSINPIDWKLREGYLKQMMDWEF 59
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTV--GSLAEYTAAEEKVLA 130
P + G+DVAGV+ +VG V KVGDEV+ P+T G+ AEYTA ++ +LA
Sbjct: 60 PIILGWDVAGVISEVGEGVTDWKVGDEVFA--------RPETTRFGTYAEYTAVDDHLLA 111
Query: 131 HKPSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEA 189
P +SF EAAS+PLA +TA+Q L + A+ G+ +L+ GAGGVG+L IQLAKH
Sbjct: 112 PLPEGISFDEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAKHA--G 169
Query: 190 SKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVG---QSDRALKAVKE 246
+++ TAS E L+ LGAD IDY + NF++V DVV+D +G ++D + +KE
Sbjct: 170 AEVITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETD-SYDVLKE 228
Query: 247 G-GKVVTIL--------PPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPF 297
G G++V+I+ A L +G L++L L + VKPI+ PF
Sbjct: 229 GTGRLVSIVGISNEERAKEKNVTATGIWLQPNGEQLKELGKLLANKTVKPIVGATFPFSE 288
Query: 298 SQTVEAFSYLNTSRATGEVVI 318
+A + T A G++VI
Sbjct: 289 KGVFDAHALSETHHAVGKIVI 309
>UniRef100_Q8Y9B9 Lmo0613 protein [Listeria monocytogenes]
Length = 313
Score = 171 bits (434), Expect = 2e-41
Identities = 120/321 (37%), Positives = 177/321 (54%), Gaps = 29/321 (9%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI-DSPL 72
KA V YG EE+ + V P +QV++K A ++NP+D K G+ K + D
Sbjct: 2 KAVVIENYGGKEELK--EKEVAMPKAGKNQVIVKEAATSINPIDWKLREGYLKQMMDWEF 59
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTV--GSLAEYTAAEEKVLA 130
P + G+DVAGV+ +VG V KVGDEV+ P+T G+ AEYTA ++ +LA
Sbjct: 60 PIILGWDVAGVISEVGEGVTDWKVGDEVFA--------RPETTRFGTYAEYTAVDDHLLA 111
Query: 131 HKPSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEA 189
P +SF EAAS+PLA +TA+Q L + A+ G+ +L+ GAGGVG+L IQLAKH
Sbjct: 112 PLPEGISFDEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAKHA--G 169
Query: 190 SKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVG---QSDRALKAVKE 246
+++ TAS E L+ LGAD IDY + NF++V DVV+D +G ++D + +KE
Sbjct: 170 AEVITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETD-SYDVLKE 228
Query: 247 G-GKVVTIL--------PPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPF 297
G G++V+I+ A L +G L++L L + +KPI+ PF
Sbjct: 229 GTGRLVSIVGISNEDRAKEKNVTATGIWLQPNGEQLKELGKLLANKTIKPIVGATFPFSE 288
Query: 298 SQTVEAFSYLNTSRATGEVVI 318
+A + T A G++VI
Sbjct: 289 KGVFDAHALSETHHAVGKIVI 309
>UniRef100_UPI00003CC0B8 UPI00003CC0B8 UniRef100 entry
Length = 308
Score = 170 bits (431), Expect = 4e-41
Identities = 118/314 (37%), Positives = 175/314 (55%), Gaps = 19/314 (6%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKD-IDSPL 72
KA V QYG +EE+ + V P +K ++VLI++ A ++NP D K G ++ +
Sbjct: 2 KAIVIDQYGSVEELK--ERRVLKPVVKNNEVLIRILATSINPGDWKIRKGDLQEQLGFSF 59
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHK 132
P + G DVAGV+ ++G +V+ K+GD+V+ I GS AE+ + ++++
Sbjct: 60 PIILGLDVAGVIEEIGEEVECFKIGDKVFTKPENIGW------GSYAEFITVKTDLVSYM 113
Query: 133 PSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASK 191
P+NLSF+EAAS+PLA +TA+Q L + A+ +L+ GAGGVGSL IQ+AK +
Sbjct: 114 PTNLSFVEAASIPLAGLTAWQSLVDYAKIKEHDRVLIHAGAGGVGSLAIQIAKSF--GAF 171
Query: 192 IAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVG--QSDRALKAVKEGGK 249
+A TAS+ EFL+ LGADL IDY K+ FEE+ FD+V +G +++ + +K GG
Sbjct: 172 VATTASSKNEEFLKSLGADLVIDYKKDKFEELLSDFDIVLGTIGGEVQEKSFQILKPGGV 231
Query: 250 VVTIL--PPGTPPAIP--FL-LTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAF 304
V+I+ P I FL L G LE L + GKVKPI+ PF EA
Sbjct: 232 FVSIVHEPIQKVKEIKSGFLWLKPSGKQLEALSDLIVHGKVKPIISKVMPFSEEGIREAH 291
Query: 305 SYLNTSRATGEVVI 318
G++VI
Sbjct: 292 ILSEGQHVRGKIVI 305
>UniRef100_Q92E39 Lin0622 protein [Listeria innocua]
Length = 313
Score = 170 bits (430), Expect = 5e-41
Identities = 121/321 (37%), Positives = 177/321 (54%), Gaps = 29/321 (9%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI-DSPL 72
KA V YG EE+ + V P +QV++K A ++NP+D K G+ K + D
Sbjct: 2 KAVVIENYGGKEELK--EKEVAMPKAGKNQVIVKEVATSINPIDWKLREGYLKQMMDWEF 59
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTV--GSLAEYTAAEEKVLA 130
P + G+DVAGV+ +VG V KVGDEV+ P+T G+ AEYTA ++ +LA
Sbjct: 60 PIILGWDVAGVISEVGEGVTDWKVGDEVFA--------RPETTRFGTYAEYTAVDDHLLA 111
Query: 131 HKPSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEA 189
P +SF EAAS+PLA +TA+Q L + A+ G+ +L+ GAGGVG+L IQLAK +
Sbjct: 112 PLPEGISFEEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAK--YAG 169
Query: 190 SKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVG---QSDRALKAVKE 246
+++ TAS E L+ LGAD IDY + NF++V DVV+D +G ++D + +KE
Sbjct: 170 AEVITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETD-SYDVLKE 228
Query: 247 G-GKVVTIL--------PPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPF 297
G G++V+I+ A L +G L+KL L + VKPI+ PF
Sbjct: 229 GTGRLVSIVGISNEDRAKEKNVTANGIWLQPNGEQLKKLGELLANKTVKPIVGATFPFSE 288
Query: 298 SQTVEAFSYLNTSRATGEVVI 318
+A + T A G++VI
Sbjct: 289 KGVFDAHALSETHHAVGKIVI 309
>UniRef100_Q88VU4 Oxidoreductase [Lactobacillus plantarum]
Length = 314
Score = 169 bits (429), Expect = 7e-41
Identities = 114/314 (36%), Positives = 173/314 (54%), Gaps = 16/314 (5%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDI-DSPL 72
KA + +QYG EE+ + P + D+VL+ V+A ++NP+D K G K + D +
Sbjct: 2 KAVIINQYGGPEELQM--ATIERPSIDQDEVLVAVQATSINPIDWKARQGLLKGMFDWQM 59
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHK 132
P V G+DVAG +V+ G+ V KVGDEV+ +I D + G+ AEY A +E LA K
Sbjct: 60 PVVLGWDVAGTIVEAGAAVTNFKVGDEVFARP-DIYADGRR--GTYAEYAAVKEDKLALK 116
Query: 133 PSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASK 191
P++LSF +AA+LPLA +TAYQ + + + AG+ +L+ GAGGVG IQ+AK++ ++
Sbjct: 117 PASLSFEQAAALPLAGLTAYQVIHDQLKIQAGEKVLIQAGAGGVGLNAIQIAKYL--GAE 174
Query: 192 IAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKAVKEGGKVV 251
+A TAS LR++GAD IDY ++V +D V+D + D AL +K G++V
Sbjct: 175 VATTASANHHTLLRQIGADHVIDYHTTAIQDVLTDYDAVFDTIDAIDEALAILKPTGRLV 234
Query: 252 TILPPGT-------PPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAF 304
TI T P + L +G L L G ++ I+D PF + A
Sbjct: 235 TIDGQPTAAQKSQGPTVSSWWLQPNGRELAILGQLAVEGHLQVIIDQVFPFSTAGLQAAH 294
Query: 305 SYLNTSRATGEVVI 318
Y S + G+++I
Sbjct: 295 RYSENSHSAGKLII 308
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 543,850,896
Number of Sequences: 2790947
Number of extensions: 23458437
Number of successful extensions: 67875
Number of sequences better than 10.0: 3953
Number of HSP's better than 10.0 without gapping: 1497
Number of HSP's successfully gapped in prelim test: 2457
Number of HSP's that attempted gapping in prelim test: 61412
Number of HSP's gapped (non-prelim): 4773
length of query: 321
length of database: 848,049,833
effective HSP length: 127
effective length of query: 194
effective length of database: 493,599,564
effective search space: 95758315416
effective search space used: 95758315416
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0400a.9


