

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0387.8
(449 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
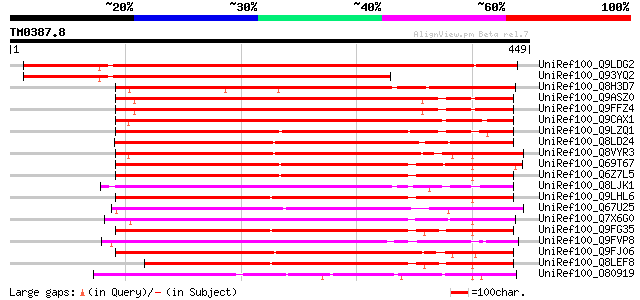
UniRef100_Q9LDG2 Hypothetical protein F24F17.6 [Arabidopsis thal... 607 e-172
UniRef100_Q93YQ2 Hypothetical protein F24F17.6 [Arabidopsis thal... 449 e-124
UniRef100_Q8H3D7 Leaf senescence related protein-like [Oryza sat... 373 e-102
UniRef100_Q9ASZ0 AT5g06230/MBL20_11 [Arabidopsis thaliana] 367 e-100
UniRef100_Q9FFZ4 Gb|AAF00670.1 [Arabidopsis thaliana] 367 e-100
UniRef100_Q9CAX1 Hypothetical protein F24K9.24 [Arabidopsis thal... 364 3e-99
UniRef100_Q9LZQ1 Hypothetical protein T12C14_90 [Arabidopsis tha... 337 5e-91
UniRef100_Q8LD24 Hypothetical protein [Arabidopsis thaliana] 334 3e-90
UniRef100_Q8VYR3 Hypothetical protein At1g60790 [Arabidopsis tha... 327 6e-88
UniRef100_Q69T67 Lustrin A-like [Oryza sativa] 324 3e-87
UniRef100_Q6Z7L5 Hypothetical protein OJ1448_G06.4 [Oryza sativa] 319 9e-86
UniRef100_Q8LJK1 P0018C10.29 protein [Oryza sativa] 314 4e-84
UniRef100_Q9LHL6 Gb|AAF30301.1 [Arabidopsis thaliana] 312 1e-83
UniRef100_Q67U25 Lustrin A-like [Oryza sativa] 311 2e-83
UniRef100_Q7X6G0 OSJNBa0043L24.14 protein [Oryza sativa] 311 3e-83
UniRef100_Q9FG35 Emb|CAB82953.1 [Arabidopsis thaliana] 310 7e-83
UniRef100_Q9FVP8 Hypothetical protein F27K7.9 [Arabidopsis thali... 308 2e-82
UniRef100_Q9FJ06 Emb|CAB82953.1 [Arabidopsis thaliana] 304 4e-81
UniRef100_Q8LEF8 Hypothetical protein [Arabidopsis thaliana] 275 1e-72
UniRef100_O80919 Expressed protein [Arabidopsis thaliana] 256 7e-67
>UniRef100_Q9LDG2 Hypothetical protein F24F17.6 [Arabidopsis thaliana]
Length = 469
Score = 607 bits (1566), Expect = e-172
Identities = 281/439 (64%), Positives = 339/439 (77%), Gaps = 17/439 (3%)
Query: 13 VFEGFRRVKRFRL-FEPSLGVLGFVVVTAIVGCCFFYLGNRDVAARFGFLDQPQSFPWLR 71
+ E RR KR RL FEPSLGVLGF +V + C FF+ R VA +G D+ + F WL+
Sbjct: 16 ICEALRRFKRSRLVFEPSLGVLGFFLVGVCLVCSFFFFDYRSVAKSYGLSDKSERFVWLK 75
Query: 72 LKEQS-----------RVEFLGENGVVVGGGCDLFDGEWVWDESYPLYQSKDCMFLDEGF 120
S RV FL E+G GCD+FDG+WVWDESYPLYQSKDC FLDEGF
Sbjct: 76 FDNISSSSSSSSNSSKRVGFLEESG----SGCDVFDGDWVWDESYPLYQSKDCRFLDEGF 131
Query: 121 RCSENGRRDLFYTQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCML 180
RCS+ GR DLFYTQWRWQP+ CN+PRF+A MLE+LR+KR+VF GDSIGRNQWESLLC+L
Sbjct: 132 RCSDFGRSDLFYTQWRWQPRHCNLPRFDAKLMLEKLRDKRLVFVGDSIGRNQWESLLCLL 191
Query: 181 SSGVSNKESIYEVNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRT 240
SS V N+ IYE+NG+PITKHKGFLVFKF ++NCTVEYYR+PFLV QSRPP G GK++T
Sbjct: 192 SSAVKNESLIYEINGSPITKHKGFLVFKFEEYNCTVEYYRSPFLVPQSRPPIGSPGKVKT 251
Query: 241 TLKVDKMDWNSLKWSNADILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSM 300
+LK+D MDW S KW +AD+L+LNTGHWWN KT R GCYFQEG EVKLK++V+DAYKR++
Sbjct: 252 SLKLDTMDWTSSKWRDADVLVLNTGHWWNEGKTTRTGCYFQEGEEVKLKMNVDDAYKRAL 311
Query: 301 ETVLNWMQSSVNPSKTQVFFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVPNDNWSRF 360
TV+ W+ + ++ +KTQVFFRT APVHFRGGDW+ GG CH ETLP++G+SL ++ W +
Sbjct: 312 NTVVKWIHTELDSNKTQVFFRTFAPVHFRGGDWKTGGTCHMETLPEIGTSLASSETWEQL 371
Query: 361 KAANAVLSSHTNASEIMKIKVLNITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCL 420
K VLS ++N SE +K+K+LNIT M AQRKDGH S+YYLG GPAPLHRQDCSHWCL
Sbjct: 372 KILRDVLSHNSNRSETVKVKLLNITAMAAQRKDGHPSLYYLG-PHGPAPLHRQDCSHWCL 430
Query: 421 PGVPDTWNELLYAMLLKHE 439
PGVPDTWNEL YA+ +K E
Sbjct: 431 PGVPDTWNELFYALFMKQE 449
>UniRef100_Q93YQ2 Hypothetical protein F24F17.6 [Arabidopsis thaliana]
Length = 346
Score = 449 bits (1154), Expect = e-124
Identities = 210/329 (63%), Positives = 253/329 (76%), Gaps = 16/329 (4%)
Query: 13 VFEGFRRVKRFRL-FEPSLGVLGFVVVTAIVGCCFFYLGNRDVAARFGFLDQPQSFPWLR 71
+ E RR KR RL FEPSLGVLGF +V + C FF+ R VA +G D+ + F WL+
Sbjct: 16 ICEALRRFKRSRLVFEPSLGVLGFFLVGVCLVCSFFFFDYRSVAKSYGLSDKSERFVWLK 75
Query: 72 LKEQS-----------RVEFLGENGVVVGGGCDLFDGEWVWDESYPLYQSKDCMFLDEGF 120
S RV FL E+G GCD+FDG+WVWDESYPLYQSKDC FLDEGF
Sbjct: 76 FDNISSSSSSSSNSSKRVGFLEESG----SGCDVFDGDWVWDESYPLYQSKDCRFLDEGF 131
Query: 121 RCSENGRRDLFYTQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCML 180
RCS+ GR DLFYTQWRWQP+ CN+PRF+A MLE+LR+KR+VF GDSIGRNQWESLLC+L
Sbjct: 132 RCSDFGRSDLFYTQWRWQPRHCNLPRFDAKLMLEKLRDKRLVFVGDSIGRNQWESLLCLL 191
Query: 181 SSGVSNKESIYEVNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRT 240
SS V N+ IYE+NG+PITKHKGFLVFKF ++NCTVEYYR+PFLV QSRPP G GK++T
Sbjct: 192 SSAVKNESLIYEINGSPITKHKGFLVFKFEEYNCTVEYYRSPFLVPQSRPPIGSPGKVKT 251
Query: 241 TLKVDKMDWNSLKWSNADILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSM 300
+LK+D MDW S KW +AD+L+LNTGHWWN KT R GCYFQEG EVKLK++V+DAYKR++
Sbjct: 252 SLKLDTMDWTSSKWRDADVLVLNTGHWWNEGKTTRTGCYFQEGEEVKLKMNVDDAYKRAL 311
Query: 301 ETVLNWMQSSVNPSKTQVFFRTLAPVHFR 329
TV+ W+ + ++ +KTQVFFRT APVHFR
Sbjct: 312 NTVVKWIHTELDSNKTQVFFRTFAPVHFR 340
>UniRef100_Q8H3D7 Leaf senescence related protein-like [Oryza sativa]
Length = 441
Score = 373 bits (957), Expect = e-102
Identities = 177/359 (49%), Positives = 238/359 (65%), Gaps = 18/359 (5%)
Query: 92 CDLFDGEWVWD---ESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFN 148
CD+ DGEWV D E PLY+ + C F+DEGFRC ENGR D + +WRWQP+ C +PRF+
Sbjct: 85 CDVVDGEWVRDDDDERRPLYEPRRCPFVDEGFRCRENGRPDDAFAKWRWQPRHCTLPRFD 144
Query: 149 ATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVS--------NKESIYEVNGNPITK 200
A +LE LRN+R+VF GDSIGRNQWES+LCML++ V+ SIYEVNG+PITK
Sbjct: 145 AKNLLETLRNRRLVFVGDSIGRNQWESMLCMLATAVAFAGDGDGDKAASIYEVNGSPITK 204
Query: 201 HKGFLVFKFRDFNCTVEYYRAPFLVLQSRPP--TGVSGKIRTTLKVDKMDWNSLKWSNAD 258
H+G L F+FRD+NCTVE+YR+P+LV + RPP G +TL++D MD + +W +AD
Sbjct: 205 HEGALSFRFRDYNCTVEHYRSPYLVRRGRPPPRRAHRGAACSTLQLDAMDARAHRWKDAD 264
Query: 259 ILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQV 318
+++ NTGHWW+ E+ ++ C FQ G ++ L + +E AY+R+M T+ +W+ VNP K+ V
Sbjct: 265 VVVFNTGHWWSRERLLQLRCNFQVGKKLILNMSIEAAYQRAMNTLTSWVHREVNPHKSLV 324
Query: 319 FFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMK 378
FRT +P H R + G C ET P+L SS + W N + + K
Sbjct: 325 IFRTYSPAHTRAS---SNGGCAKETTPELNSSRISLHRWPGM--VNPAFEPSRSGTAAAK 379
Query: 379 IKVLNITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLLK 437
++VLN+T MTAQR+DGH S++ + A R DCSHWCLPGVPD WNELLYAM+LK
Sbjct: 380 LRVLNVTLMTAQRRDGHPSVFNVAAAARSPARQRADCSHWCLPGVPDAWNELLYAMILK 438
>UniRef100_Q9ASZ0 AT5g06230/MBL20_11 [Arabidopsis thaliana]
Length = 372
Score = 367 bits (942), Expect = e-100
Identities = 175/354 (49%), Positives = 233/354 (65%), Gaps = 18/354 (5%)
Query: 92 CDLFDGEWVWDESYP------LYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMP 145
CD G+WV S L+ ++C FLD GFRC ++GR+D Y WRWQP C++P
Sbjct: 21 CDYSKGKWVRRASSSSSSVNGLFYGEECRFLDSGFRCHKHGRKDSGYLDWRWQPHGCDLP 80
Query: 146 RFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFL 205
RFNA+ +LER RN R+VF GDSIGRNQWESL+CMLS + NK IYEVNGNPITKHKGFL
Sbjct: 81 RFNASDLLERSRNGRIVFVGDSIGRNQWESLMCMLSQAIPNKSEIYEVNGNPITKHKGFL 140
Query: 206 VFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTG 265
+F N TVEY+R+PFLV+ RPP +I+TT++VD+ +W S +W +D+L+ N+G
Sbjct: 141 SMRFPRENLTVEYHRSPFLVVIGRPPDKSPKEIKTTVRVDEFNWQSKRWVGSDVLVFNSG 200
Query: 266 HWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAP 325
HWWN +KT+ GCYF+EG +V + V +A+ +S++T +W+ ++P K+ VFFR+ +P
Sbjct: 201 HWWNEDKTVLTGCYFEEGRKVNKTMGVMEAFGKSLKTWKSWVLEKLDPDKSYVFFRSYSP 260
Query: 326 VHFRGGDWRNGGNCHSETLPDLGSSLVPND---NWSRFKAANAVLSSHTNASEIMKIKVL 382
VH+R G W GG C +E P+ + D N +K + H+ K+K L
Sbjct: 261 VHYRNGTWNTGGLCDAEIEPETDKRKLEPDASHNEYIYKVIEEMRYRHS------KVKFL 314
Query: 383 NITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
NIT++T RKDGH S Y R G + QDCSHWCLPGVPDTWNE+LYA LL
Sbjct: 315 NITYLTEFRKDGHISRY---REQGTSVDVPQDCSHWCLPGVPDTWNEILYAQLL 365
>UniRef100_Q9FFZ4 Gb|AAF00670.1 [Arabidopsis thaliana]
Length = 413
Score = 367 bits (942), Expect = e-100
Identities = 175/354 (49%), Positives = 233/354 (65%), Gaps = 18/354 (5%)
Query: 92 CDLFDGEWVWDESYP------LYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMP 145
CD G+WV S L+ ++C FLD GFRC ++GR+D Y WRWQP C++P
Sbjct: 62 CDYSKGKWVRRASSSSSSVNGLFYGEECRFLDSGFRCHKHGRKDSGYLDWRWQPHGCDLP 121
Query: 146 RFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFL 205
RFNA+ +LER RN R+VF GDSIGRNQWESL+CMLS + NK IYEVNGNPITKHKGFL
Sbjct: 122 RFNASDLLERSRNGRIVFVGDSIGRNQWESLMCMLSQAIPNKSEIYEVNGNPITKHKGFL 181
Query: 206 VFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTG 265
+F N TVEY+R+PFLV+ RPP +I+TT++VD+ +W S +W +D+L+ N+G
Sbjct: 182 SMRFPRENLTVEYHRSPFLVVIGRPPDKSPKEIKTTVRVDEFNWQSKRWVGSDVLVFNSG 241
Query: 266 HWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAP 325
HWWN +KT+ GCYF+EG +V + V +A+ +S++T +W+ ++P K+ VFFR+ +P
Sbjct: 242 HWWNEDKTVLTGCYFEEGRKVNKTMGVMEAFGKSLKTWKSWVLEKLDPDKSYVFFRSYSP 301
Query: 326 VHFRGGDWRNGGNCHSETLPDLGSSLVPND---NWSRFKAANAVLSSHTNASEIMKIKVL 382
VH+R G W GG C +E P+ + D N +K + H+ K+K L
Sbjct: 302 VHYRNGTWNTGGLCDAEIEPETDKRKLEPDASHNEYIYKVIEEMRYRHS------KVKFL 355
Query: 383 NITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
NIT++T RKDGH S Y R G + QDCSHWCLPGVPDTWNE+LYA LL
Sbjct: 356 NITYLTEFRKDGHISRY---REQGTSVDVPQDCSHWCLPGVPDTWNEILYAQLL 406
>UniRef100_Q9CAX1 Hypothetical protein F24K9.24 [Arabidopsis thaliana]
Length = 427
Score = 364 bits (934), Expect = 3e-99
Identities = 173/348 (49%), Positives = 224/348 (63%), Gaps = 9/348 (2%)
Query: 92 CDLFDGEWVW---DESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFN 148
CD G WV D Y ++C FLD GFRC NGR+D + QWRWQP C++PRFN
Sbjct: 79 CDYSYGRWVRRRRDVDETSYYGEECRFLDPGFRCLNNGRKDSGFRQWRWQPHGCDLPRFN 138
Query: 149 ATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFK 208
A+ LER RN R+VF GDSIGRNQWESLLCMLS VSNK IYEVNGNPI+KHKGFL +
Sbjct: 139 ASDFLERSRNGRIVFVGDSIGRNQWESLLCMLSQAVSNKSEIYEVNGNPISKHKGFLSMR 198
Query: 209 FRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWW 268
F + N TVEY+R PFLV+ RPP ++ T++VD+ +W S KW +D+L+ NTGHWW
Sbjct: 199 FPEQNLTVEYHRTPFLVVVGRPPENSPVDVKMTVRVDEFNWQSKKWVGSDVLVFNTGHWW 258
Query: 269 NYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHF 328
N +KT GCYFQEG ++ + V + +++S++T +W+ ++ ++ VFFR+ +PVH+
Sbjct: 259 NEDKTFIAGCYFQEGGKLNKTMGVMEGFEKSLKTWKSWVLERLDSERSHVFFRSFSPVHY 318
Query: 329 RGGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMT 388
R G W GG C ++T P+ + D + A+ S K+K LNIT++T
Sbjct: 319 RNGTWNLGGLCDADTEPETDMKKMEPDPIHNNYISQAIQEMRYEHS---KVKFLNITYLT 375
Query: 389 AQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
RKD H S Y AP QDCSHWCLPGVPDTWNE+LYA LL
Sbjct: 376 EFRKDAHPSRYREPGTPEDAP---QDCSHWCLPGVPDTWNEILYAQLL 420
>UniRef100_Q9LZQ1 Hypothetical protein T12C14_90 [Arabidopsis thaliana]
Length = 475
Score = 337 bits (863), Expect = 5e-91
Identities = 162/350 (46%), Positives = 219/350 (62%), Gaps = 20/350 (5%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
CD+ G+WV+D YPLY + C F+DEGF C NGR DL Y WRW+P+DC+ PRFNATK
Sbjct: 138 CDVTKGKWVYDSDYPLYTNASCPFIDEGFGCQSNGRLDLNYMNWRWEPQDCHAPRFNATK 197
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRD 211
MLE +R KR+VF GDSI RNQWES+LC+L V + + +YE + ITK KG F+F D
Sbjct: 198 MLEMIRGKRLVFVGDSINRNQWESMLCLLFQAVKDPKRVYETHNRRITKEKGNYSFRFVD 257
Query: 212 FNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYE 271
+ CTVE+Y FLV + R G K R TL++D MD S +W A+IL+ NT HWW++
Sbjct: 258 YKCTVEFYVTHFLVREGRARIG--KKRRETLRIDAMDRTSSRWKGANILVFNTAHWWSHY 315
Query: 272 KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGG 331
KT G Y+QEG + KL V A+K++++T +W+ +V+P KT+VFFR+ AP HF GG
Sbjct: 316 KTKSGVNYYQEGDLIHPKLDVSTAFKKALQTWSSWVDKNVDPKKTRVFFRSAAPSHFSGG 375
Query: 332 DWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMTAQR 391
+W +GG+C +P L + P+ + + + + T + +LN++ ++ R
Sbjct: 376 EWNSGGHCREANMP-LNQTFKPSYSSKKSIVEDVLKQMRT------PVTLLNVSGLSQYR 428
Query: 392 KDGHSSIYYLGRVAGPAPLHR-----QDCSHWCLPGVPDTWNELLYAMLL 436
D H SIY G P +R QDCSHWCLPGVPDTWN LY LL
Sbjct: 429 IDAHPSIY------GTKPENRRSRAVQDCSHWCLPGVPDTWNHFLYLHLL 472
>UniRef100_Q8LD24 Hypothetical protein [Arabidopsis thaliana]
Length = 485
Score = 334 bits (857), Expect = 3e-90
Identities = 159/348 (45%), Positives = 218/348 (61%), Gaps = 9/348 (2%)
Query: 91 GCDLFDGEWVWDES-YPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNA 149
GCDL+ G WV D+ YPLYQ C ++D+ F C NGRRD Y WRW+P C++PRFNA
Sbjct: 140 GCDLYRGSWVKDDDEYPLYQPGSCPYVDDAFDCQRNGRRDSDYLNWRWKPDGCDLPRFNA 199
Query: 150 TKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKF 209
T L +LR K ++ GDS+ RNQ+ES+LC+L G+S+K +YEV+G+ ITK +G+ VFKF
Sbjct: 200 TDFLVKLRGKSLMLVGDSMNRNQFESMLCVLREGLSDKSRMYEVHGHNITKGRGYFVFKF 259
Query: 210 RDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWN 269
D+NCTVE+ R+ FLV + G TL +D++D + KW ADIL+ NTGHWW
Sbjct: 260 EDYNCTVEFVRSHFLVREG-VRANAQGNTNPTLSIDRIDKSHAKWKRADILVFNTGHWWV 318
Query: 270 YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFR 329
+ KT RG Y++EG + K +AY+RS++T W+ +VNP K VF+R + HFR
Sbjct: 319 HGKTARGKNYYKEGDYIYPKFDATEAYRRSLKTWAKWIDQNVNPKKQLVFYRGYSSAHFR 378
Query: 330 GGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMTA 389
GG+W +GG+C+ E P S++ + + ++ ++ I +LN+T +T
Sbjct: 379 GGEWDSGGSCNGEVEPVKKGSIIDS-----YPLKMKIVQEAIKEMQVPVI-LLNVTKLTN 432
Query: 390 QRKDGHSSIYYLGRVAG-PAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
RKDGH SIY G RQDCSHWCLPGVPD WN L+YA LL
Sbjct: 433 FRKDGHPSIYGKTNTDGKKVSTRRQDCSHWCLPGVPDVWNHLIYASLL 480
>UniRef100_Q8VYR3 Hypothetical protein At1g60790 [Arabidopsis thaliana]
Length = 541
Score = 327 bits (837), Expect = 6e-88
Identities = 162/360 (45%), Positives = 220/360 (61%), Gaps = 15/360 (4%)
Query: 92 CDLFDGEWVW--DESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNA 149
CD++DG WV DE+ P Y C ++D F C NGR D Y +WRWQP C++PR N
Sbjct: 190 CDIYDGSWVRADDETMPYYPPGSCPYIDRDFNCHANGRPDDAYVKWRWQPNGCDIPRLNG 249
Query: 150 TKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKF 209
T LE+LR K++VF GDSI RN WESL+C+L + +K+ +YE++G K KGF F+F
Sbjct: 250 TDFLEKLRGKKLVFVGDSINRNMWESLICILRHSLKDKKRVYEISGRREFKKKGFYAFRF 309
Query: 210 RDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWN 269
D+NCTV++ +PF V +S GV+G TL++D MD + + +ADILI NTGHWW
Sbjct: 310 EDYNCTVDFVGSPFFVRES-SFKGVNGTTLETLRLDMMDKTTSMYRDADILIFNTGHWWT 368
Query: 270 YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFR 329
++KT G Y+QEG V +L V +AYKR++ T W+ +++ S+T + FR + HFR
Sbjct: 369 HDKTKLGENYYQEGNVVYPRLKVLEAYKRALITWAKWVDKNIDRSQTHIVFRGYSVTHFR 428
Query: 330 GGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKV--LNITHM 387
GG W +GG CH ET P +S + S+ KA +L + MK V +NI+ +
Sbjct: 429 GGPWNSGGQCHKETEPIFNTSYLAKYP-SKMKALEYIL------RDTMKTPVIYMNISRL 481
Query: 388 TAQRKDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLLKHEVDQRW 444
T RKDGH SIY Y + QDCSHWCLPGVPDTWN+LLY LLK + +W
Sbjct: 482 TDFRKDGHPSIYRMVYRTEKEKREAVSHQDCSHWCLPGVPDTWNQLLYVSLLKAGLASKW 541
>UniRef100_Q69T67 Lustrin A-like [Oryza sativa]
Length = 847
Score = 324 bits (831), Expect = 3e-87
Identities = 165/358 (46%), Positives = 223/358 (62%), Gaps = 14/358 (3%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
CD+F G WV DESYPLY C +DE F C NGR D Y + RWQP CN+PR N T
Sbjct: 498 CDMFHGNWVRDESYPLYPEGSCPHIDEPFDCYLNGRPDRAYQKLRWQPSSCNIPRLNPTD 557
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRD 211
MLERLR KR+VF GDS+ RN WESL+C+L + V +K ++E +G K +G F F D
Sbjct: 558 MLERLRGKRLVFVGDSLNRNMWESLVCILRNSVKDKRKVFEASGRHEFKTEGSYSFLFTD 617
Query: 212 FNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYE 271
+NC+VE++R+PFLV + +GK + TL++D ++ +S K+ +AD LI NTGHWW +E
Sbjct: 618 YNCSVEFFRSPFLVQEWEMKVS-NGKKKETLRLDIVEQSSPKYKDADFLIFNTGHWWTHE 676
Query: 272 KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGG 331
KT G Y+QEG V +L+V DA+ +++ T W+ ++VNP KT V FR + HF GG
Sbjct: 677 KTSLGKDYYQEGNHVYSELNVVDAFHKALVTWSRWIDANVNPKKTTVLFRGYSASHFSGG 736
Query: 332 DWRNGGNCHSETLPDLGSSLVPNDNW-SRFKAANAVLSSHTNASEIMKIKVLNITHMTAQ 390
W +GG+C ET P + N+ + S + ++L + + + LNIT MT
Sbjct: 737 QWNSGGSCDKETEP------IRNEQYLSTYPPKMSILEDVIHKMK-TPVVYLNITRMTDY 789
Query: 391 RKDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL--KHEVDQR 443
RKD H SIY L +P QDCSHWCLPGVPD+WNELLYA LL +H++ Q+
Sbjct: 790 RKDAHPSIYRKRNLTEDERRSPERYQDCSHWCLPGVPDSWNELLYAQLLIKQHQMLQQ 847
>UniRef100_Q6Z7L5 Hypothetical protein OJ1448_G06.4 [Oryza sativa]
Length = 700
Score = 319 bits (818), Expect = 9e-86
Identities = 155/348 (44%), Positives = 215/348 (61%), Gaps = 10/348 (2%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
CD+F G WV D+SYPLY C +DE F C NGR D Y + RWQP C++PR N +
Sbjct: 351 CDMFYGNWVRDDSYPLYPEGSCPHIDESFNCPLNGRPDNAYQRLRWQPSGCSIPRLNPSD 410
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRD 211
MLERLR KR+VF GDS+ RN WESL+C+L + V +K ++EV+G + +G F F+D
Sbjct: 411 MLERLRGKRLVFVGDSLNRNMWESLVCILRNSVKDKRKVFEVSGRQQFRAEGSYSFLFQD 470
Query: 212 FNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYE 271
+NC+VE++R+PFLV + P G + TL++D + + ++ +ADI+I NTGHWW +E
Sbjct: 471 YNCSVEFFRSPFLVQEWEFPVR-KGLTKETLRLDMISNSFPRYKDADIIIFNTGHWWTHE 529
Query: 272 KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGG 331
KT G Y+QEG V +L+V+DA+++++ T W+ SSVNP KT VFFR + HF GG
Sbjct: 530 KTSLGKDYYQEGNRVYSELNVDDAFQKALITWAKWVDSSVNPKKTTVFFRGYSSSHFSGG 589
Query: 332 DWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMTAQR 391
W +GG+C ET P + N+ + + + + LNIT MT R
Sbjct: 590 QWNSGGSCDKETEP------ITNEKFLTPYPRKMSILEDVLSGMKTPVVYLNITRMTDYR 643
Query: 392 KDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
K+ H S+Y L +P QDCSHWCLPGVPD+WNELLYA ++
Sbjct: 644 KEAHPSVYRKQKLTEEEKKSPQIYQDCSHWCLPGVPDSWNELLYAQIM 691
>UniRef100_Q8LJK1 P0018C10.29 protein [Oryza sativa]
Length = 434
Score = 314 bits (804), Expect = 4e-84
Identities = 163/360 (45%), Positives = 214/360 (59%), Gaps = 25/360 (6%)
Query: 79 EFLGENGVVVGGGCDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQ 138
EF G+N CD+FDG WV D YPLY S DC F++ GF C NGR+D Y +WRW+
Sbjct: 81 EFAGDNLE----SCDVFDGSWVPDRRYPLYNSSDCPFVERGFNCLANGRKDTGYLKWRWK 136
Query: 139 PKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPI 198
P+ C++PRF+A +LERLR KRVVF GDS+ R QWES +CML +GV N +++YEVNGN I
Sbjct: 137 PRGCDLPRFSARDVLERLRGKRVVFVGDSMSRTQWESFICMLMAGVENPKTVYEVNGNQI 196
Query: 199 TKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNAD 258
+K FL +F FN VE++R+ FLV QS P ++R LK+DKMD S KW NAD
Sbjct: 197 SKTIRFLGVRFASFNLNVEFFRSVFLVQQSPAPRSSPKRVRAILKLDKMDNISRKWENAD 256
Query: 259 ILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQV 318
+LI N+GHWW K GCYF+ G +KL + A+K ++ET +W++ V+ +T V
Sbjct: 257 VLIFNSGHWWTPSKLFDMGCYFEAGGLLKLGTSINSAFKMALETWASWVKEKVDLKRTHV 316
Query: 319 FFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVPNDNWSRFK--AANAVLSSHTNASEI 376
FFRT P H+ G N C P ++ D+ S F A V++ A+
Sbjct: 317 FFRTYEPSHWSGS---NQKVCEVTEFP---TAEAKGDDRSEFGDILAGVVVNMSVPAT-- 368
Query: 377 MKIKVLNITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
+LN+T M A R D H I+ P DCSHWCLPGVPD WNEL+ + LL
Sbjct: 369 ----ILNVTLMGAFRSDAHIGIW-------SHPSTILDCSHWCLPGVPDAWNELVISHLL 417
>UniRef100_Q9LHL6 Gb|AAF30301.1 [Arabidopsis thaliana]
Length = 556
Score = 312 bits (799), Expect = 1e-83
Identities = 148/348 (42%), Positives = 213/348 (60%), Gaps = 10/348 (2%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
C+ F+G+WV D+SYPLY+ C +DE F C NGR D+ + + +W+PK C++PR N K
Sbjct: 196 CEFFEGDWVKDDSYPLYKPGSCNLIDEQFNCISNGRPDVDFQKLKWKPKQCSLPRLNGGK 255
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRD 211
+LE +R +R+VF GDS+ RN WESL+C+L V ++ ++E +G + + F F+D
Sbjct: 256 LLEMIRGRRLVFVGDSLNRNMWESLVCILKGSVKDESQVFEAHGRHQFRWEAEYSFVFKD 315
Query: 212 FNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYE 271
+NCTVE++ +PFLV Q T +G + TL++D + +S ++ ADIL+ NTGHWW +E
Sbjct: 316 YNCTVEFFASPFLV-QEWEVTEKNGTKKETLRLDLVGKSSEQYKGADILVFNTGHWWTHE 374
Query: 272 KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGG 331
KT +G Y+QEG+ V KL V++A+++++ T W+ +VNP K+ VFFR +P HF GG
Sbjct: 375 KTSKGEDYYQEGSTVHPKLDVDEAFRKALTTWGRWVDKNVNPKKSLVFFRGYSPSHFSGG 434
Query: 332 DWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMTAQR 391
W GG C ET P + N+ + + + LNIT +T R
Sbjct: 435 QWNAGGACDDETEP------IKNETYLTPYMLKMEILERVLRGMKTPVTYLNITRLTDYR 488
Query: 392 KDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
KD H SIY L +PL QDCSHWCLPGVPD+WNE+ YA LL
Sbjct: 489 KDAHPSIYRKQKLSAEESKSPLLYQDCSHWCLPGVPDSWNEIFYAELL 536
>UniRef100_Q67U25 Lustrin A-like [Oryza sativa]
Length = 463
Score = 311 bits (797), Expect = 2e-83
Identities = 148/368 (40%), Positives = 220/368 (59%), Gaps = 28/368 (7%)
Query: 89 GGG--CDLFDGEWVWDESY-PLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMP 145
GGG CDL+DGEW DE+ PLY C ++DE + C+ NGR D YT+WRW P+ C +P
Sbjct: 109 GGGTTCDLYDGEWARDEAARPLYAPGTCPYVDEAYACASNGRPDAAYTRWRWAPRRCRLP 168
Query: 146 RFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFL 205
RFNAT L LR KR++ GDS+ RNQ+ESLLC+L + +K ++E +G I+K +G+
Sbjct: 169 RFNATDFLATLRGKRLMLVGDSMNRNQFESLLCILREAIPDKTRMFETHGYRISKGRGYF 228
Query: 206 VFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTG 265
VFKF D++CTVE+ R+ FLV + L++D++D ++ +W D+L+ NTG
Sbjct: 229 VFKFVDYDCTVEFVRSHFLVREGVRYNRQKNS-NPILQIDRIDKSASRWRKVDVLVFNTG 287
Query: 266 HWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAP 325
HWW + KT RG Y++EG + + +AY+R+++T W+ +++P+K+ VF+R +
Sbjct: 288 HWWTHGKTARGINYYKEGDTLYPQFDSTEAYRRALKTWARWIDKNMDPAKSVVFYRGYST 347
Query: 326 VHFRGGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMK------- 378
HFRGG+W +GG+C E P L AV+ S+ I++
Sbjct: 348 AHFRGGEWDSGGSCSGEREPTL---------------RGAVIGSYPRKMRIVEEVIGRMR 392
Query: 379 --IKVLNITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
+++LN+T +T R+DGH S+Y +QDCSHWCLPGVPD WNEL+YA L+
Sbjct: 393 FPVRLLNVTKLTNFRRDGHPSVYGKAAAGKKVSRRKQDCSHWCLPGVPDAWNELIYASLV 452
Query: 437 KHEVDQRW 444
+ W
Sbjct: 453 LEPKPRSW 460
>UniRef100_Q7X6G0 OSJNBa0043L24.14 protein [Oryza sativa]
Length = 711
Score = 311 bits (796), Expect = 3e-83
Identities = 152/363 (41%), Positives = 215/363 (58%), Gaps = 14/363 (3%)
Query: 83 ENGVVVGGGCDLFDGEWVWDE-----SYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRW 137
++G+V CD+F G WV D+ +YP Y C +D+ F C +NGR D + +WRW
Sbjct: 347 QSGLVSFAKCDVFSGRWVRDDDEGGGAYPFYPPGSCPHIDDDFNCHKNGRADTGFLRWRW 406
Query: 138 QPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNP 197
QP C++PR N LERLR +R++F GDS+ RN WESL+C+L GV +K +YE +G
Sbjct: 407 QPHGCDIPRLNPIDFLERLRGQRIIFVGDSLNRNMWESLVCILRHGVRDKRRMYEASGRN 466
Query: 198 ITKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNA 257
K +G+ F+FRD+NC+V++ R+ FLV + T + L++D++D + + A
Sbjct: 467 QFKTRGYYSFRFRDYNCSVDFIRSIFLVKEMINETKGGAVVDAKLRLDELDETTPAYRTA 526
Query: 258 DILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQ 317
DI++ NTGHWW + KT RG Y+QEG V L V DAYKR++ T W+ +++ ++TQ
Sbjct: 527 DIVVFNTGHWWTHWKTSRGLNYYQEGNYVHPSLEVMDAYKRALTTWARWVDKNIDSTRTQ 586
Query: 318 VFFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIM 377
V FR + HFRGG W +GG CH ET P + + + + +L
Sbjct: 587 VVFRGYSLTHFRGGQWNSGGRCHRETEP-----IFNRTHLAEYPEKMRILEQVLGRMRTP 641
Query: 378 KIKVLNITHMTAQRKDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAM 434
I LNI+ MT RKD H S+Y Y A + +QDCSHWCLPGVPD+WNELLYA
Sbjct: 642 VI-YLNISAMTDYRKDAHPSVYRVRYETEEERMAAVAKQDCSHWCLPGVPDSWNELLYAS 700
Query: 435 LLK 437
LL+
Sbjct: 701 LLQ 703
>UniRef100_Q9FG35 Emb|CAB82953.1 [Arabidopsis thaliana]
Length = 608
Score = 310 bits (793), Expect = 7e-83
Identities = 151/353 (42%), Positives = 216/353 (60%), Gaps = 20/353 (5%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
C+ FDGEW+ D+SYPLY+ C +DE F C NGR D + + +W+PK C++PR N
Sbjct: 255 CEFFDGEWIKDDSYPLYKPGSCNLIDEQFNCITNGRPDKDFQKLKWKPKKCSLPRLNGAI 314
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRD 211
+LE LR +R+VF GDS+ RN WESL+C+L V ++ +YE G + + F F+D
Sbjct: 315 LLEMLRGRRLVFVGDSLNRNMWESLVCILKGSVKDETKVYEARGRHHFRGEAEYSFVFQD 374
Query: 212 FNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYE 271
+NCTVE++ +PFLV Q G + TL++D + +S ++ AD+++ NTGHWW +E
Sbjct: 375 YNCTVEFFVSPFLV-QEWEIVDKKGTKKETLRLDLVGKSSEQYKGADVIVFNTGHWWTHE 433
Query: 272 KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGG 331
KT +G Y+QEG+ V +L V +A+++++ T W++ +VNP+K+ VFFR + HF GG
Sbjct: 434 KTSKGEDYYQEGSNVYHELAVLEAFRKALTTWGRWVEKNVNPAKSLVFFRGYSASHFSGG 493
Query: 332 DWRNGGNCHSETLPDLGSSLVPNDNW-----SRFKAANAVLSSHTNASEIMKIKVLNITH 386
W +GG C SET P + ND + S+ K VL + LNIT
Sbjct: 494 QWNSGGACDSETEP------IKNDTYLTPYPSKMKVLEKVLRGMKT-----PVTYLNITR 542
Query: 387 MTAQRKDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
+T RKDGH S+Y L +PL QDCSHWCLPGVPD+WNE+LYA L+
Sbjct: 543 LTDYRKDGHPSVYRKQSLSEKEKKSPLLYQDCSHWCLPGVPDSWNEILYAELI 595
>UniRef100_Q9FVP8 Hypothetical protein F27K7.9 [Arabidopsis thaliana]
Length = 485
Score = 308 bits (789), Expect = 2e-82
Identities = 157/368 (42%), Positives = 221/368 (59%), Gaps = 26/368 (7%)
Query: 80 FLGENGV------VVGGG-CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFY 132
FLG NG VVG CD+FDG WV D++YPLY + +C F+++GF C NGR Y
Sbjct: 96 FLGYNGALEFNSSVVGDTECDIFDGNWVVDDNYPLYNASECPFVEKGFNCLGNGRGHDEY 155
Query: 133 TQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYE 192
+WRW+PK C +PRF +L+RLR KR+VF GDS+ R QWESL+CML +G+ +K S+YE
Sbjct: 156 LKWRWKPKHCTVPRFEVRDVLKRLRGKRIVFVGDSMSRTQWESLICMLMTGLEDKRSVYE 215
Query: 193 VNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSL 252
VNGN ITK FL +F +N TVE+YR+ FLV R ++++TLK+D +D +
Sbjct: 216 VNGNNITKRIRFLGVRFSSYNFTVEFYRSVFLVQPGRLRWHAPKRVKSTLKLDVLDVINH 275
Query: 253 KWSNADILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVN 312
+WS+AD LI NTG WW K GCYFQ G ++L + + AY+ ++ET +W++S+V+
Sbjct: 276 EWSSADFLIFNTGQWWVPGKLFETGCYFQVGNSLRLGMSIPAAYRVALETWASWIESTVD 335
Query: 313 PSKTQVFFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTN 372
P+KT+V FRT P H W + +C+ P D R K+ + +
Sbjct: 336 PNKTRVLFRTFEPSH-----WSDHRSCNVTKYP-------APDTEGRDKSIFSEMIKEVV 383
Query: 373 ASEIMKIKVLNITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLY 432
+ + + +L++T M+A R DGH ++ PL DCSHWCLPGVPD WNE+L
Sbjct: 384 KNMTIPVSILDVTSMSAFRSDGHVGLW------SDNPL-VPDCSHWCLPGVPDIWNEILL 436
Query: 433 AMLLKHEV 440
L + V
Sbjct: 437 FFLFRQPV 444
>UniRef100_Q9FJ06 Emb|CAB82953.1 [Arabidopsis thaliana]
Length = 457
Score = 304 bits (778), Expect = 4e-81
Identities = 156/352 (44%), Positives = 217/352 (61%), Gaps = 16/352 (4%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
CD+FDG WV+D+S P+Y C F+++ F C +NGR D + + RWQP C++PRF+ K
Sbjct: 100 CDIFDGTWVFDDSEPVYLPGYCPFVEDKFNCFKNGRPDSGFLRHRWQPHGCSIPRFDGKK 159
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPIT-KHKGFLVFKFR 210
ML+ LR KRVVF GDS+ RN WESL+C L S + +K + ++ G ++GF F+F
Sbjct: 160 MLKMLRGKRVVFVGDSLNRNMWESLVCSLRSTLEDKNRVSKIIGKQSNLPNEGFYGFRFN 219
Query: 211 DFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLK-WSNADILILNTGHWWN 269
DF C++++ ++PFLV +S V GK R TL++D + + K + NADI+I NTGHWW
Sbjct: 220 DFECSIDFIKSPFLVQESE-VVDVYGKRRETLRLDMIQRSMTKIYKNADIVIFNTGHWWT 278
Query: 270 YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFR 329
++KT G Y+QEG V +L V++AY +++ T +W+ S++N +KT+VFF + HFR
Sbjct: 279 HQKTYEGKGYYQEGNRVYERLEVKEAYTKAIHTWADWVDSNINSTKTRVFFVGYSSSHFR 338
Query: 330 GGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKV--LNITHM 387
G W +GG C ET P + W K +V+S MK V +NIT M
Sbjct: 339 KGAWNSGGQCDGETRPIQNETYTGVYPW-MMKVVESVISE-------MKTPVFYMNITKM 390
Query: 388 TAQRKDGHSSIYYL---GRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
T R DGH S+Y R PA QDCSHWCLPGVPD+WN+LLYA LL
Sbjct: 391 TWYRTDGHPSVYRQPADPRGTSPAAGMYQDCSHWCLPGVPDSWNQLLYATLL 442
>UniRef100_Q8LEF8 Hypothetical protein [Arabidopsis thaliana]
Length = 329
Score = 275 bits (704), Expect = 1e-72
Identities = 138/328 (42%), Positives = 198/328 (60%), Gaps = 20/328 (6%)
Query: 117 DEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESL 176
DE F C NGR D + + +W+PK C++PR N +LE LR +R+VF GDS+ RN WESL
Sbjct: 1 DEQFNCITNGRPDKDFQKLKWKPKKCSLPRLNGAILLEMLRGRRLVFVGDSLNRNMWESL 60
Query: 177 LCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTGVSG 236
+C+L V ++ +YE G + + F F+D+NCTVE++ +PFLV Q G
Sbjct: 61 VCILKGSVKDETKVYEARGRHHFRGEAEYSFVFQDYNCTVEFFVSPFLV-QEWEIVXKKG 119
Query: 237 KIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAY 296
+ TL++D + +S ++ AD+++ NTGHWW +EKT +G Y+QEG+ V +L V +A+
Sbjct: 120 XKKETLRLDLVGKSSEQYKGADVIVFNTGHWWTHEKTSKGEDYYQEGSNVYHELAVLEAF 179
Query: 297 KRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVPNDN 356
++++ T W++ +VNP+K+ VFFR + HF GG W +GG C SET P + ND
Sbjct: 180 RKALTTWGRWVEKNVNPAKSLVFFRGYSASHFSGGQWNSGGACDSETEP------IKNDT 233
Query: 357 W-----SRFKAANAVLSSHTNASEIMKIKVLNITHMTAQRKDGHSSIY---YLGRVAGPA 408
+ S+ K VL + LNIT +T RKDGH S+Y L +
Sbjct: 234 YLTPYPSKMKVLEKVLRGMKT-----PVTYLNITRLTDYRKDGHPSVYRKQSLSEKEKKS 288
Query: 409 PLHRQDCSHWCLPGVPDTWNELLYAMLL 436
PL QDCSHWCLPGVPD+WNE+LYA L+
Sbjct: 289 PLLYQDCSHWCLPGVPDSWNEILYAELI 316
>UniRef100_O80919 Expressed protein [Arabidopsis thaliana]
Length = 410
Score = 256 bits (655), Expect = 7e-67
Identities = 140/380 (36%), Positives = 220/380 (57%), Gaps = 26/380 (6%)
Query: 73 KEQSRVEFLGENGVVVGGGCDLFDGEWVWDE-SYPLYQSKDCMFLDEGFRCSENGRRDLF 131
KE + + + G G C+LF+G+WV+D SYPLY+ +DC F+ + C + GR+DL
Sbjct: 40 KENPQSHGVTDRGGDSGRECNLFEGKWVFDNVSYPLYKEEDCKFMSDQLACEKFGRKDLS 99
Query: 132 YTQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIY 191
Y WRWQP C++PRFN TK+LERLRNKR+V+ GDS+ R QW S++CM+SS ++N +++Y
Sbjct: 100 YKFWRWQPHTCDLPRFNGTKLLERLRNKRMVYVGDSLNRGQWVSMVCMVSSVITNPKAMY 159
Query: 192 EVNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLV-LQSRPPTGVSGKIRTTLKVDKMDWN 250
N + FK ++N T++YY AP LV S PT R +++ ++ +
Sbjct: 160 MHNNG-----SNLITFKALEYNATIDYYWAPLLVESNSDDPTNHRFPDR-IVRIQSIEKH 213
Query: 251 SLKWSNADILILNTGHWWN--YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQ 308
+ W+N+DI++ N+ WW + K++ G F++ + ++ + Y+ +++T+ W++
Sbjct: 214 ARHWTNSDIIVFNSYLWWRMPHIKSLWGS--FEKLDGIYKEVEMVRVYEMALQTLSQWLE 271
Query: 309 SSVNPSKTQVFFRTLAPVHFRGGDWRNGG----NCHSE-TLPDLGSSLVPNDNWSRFKAA 363
VNP+ T++FF +++P H R +W GG NC+ E +L D + +
Sbjct: 272 VHVNPNITKLFFMSMSPTHERAEEW--GGILNQNCYGEASLIDKEGYTGRGSDPKMMRVL 329
Query: 364 NAVLSSHTNASEIMKIKVLNITHMTAQRKDGHSSIY--YLGRVAG---PAPLHRQDCSHW 418
VL N + ++++NIT ++ RK+GH SIY G V P DC HW
Sbjct: 330 ENVLDGLKNRG--LNMQMINITQLSEYRKEGHPSIYRKQWGTVKENEISNPSSNADCIHW 387
Query: 419 CLPGVPDTWNELLYAMLLKH 438
CLPGVPD WNELLYA +L H
Sbjct: 388 CLPGVPDVWNELLYAYILDH 407
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 813,948,116
Number of Sequences: 2790947
Number of extensions: 35865050
Number of successful extensions: 66227
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 133
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 65634
Number of HSP's gapped (non-prelim): 155
length of query: 449
length of database: 848,049,833
effective HSP length: 131
effective length of query: 318
effective length of database: 482,435,776
effective search space: 153414576768
effective search space used: 153414576768
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0387.8


