

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
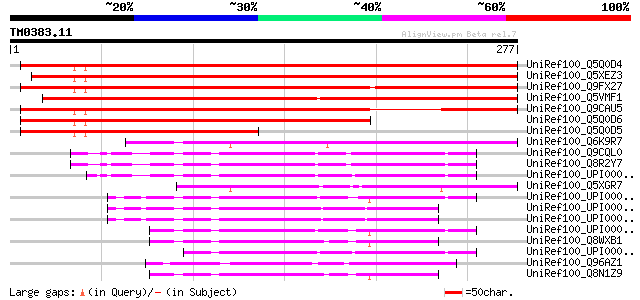
Query= TM0383.11
(277 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5Q0D4 Hypothetical protein [Arabidopsis thaliana] 351 1e-95
UniRef100_Q5XEZ3 At1g73320 [Arabidopsis thaliana] 349 5e-95
UniRef100_Q9FX27 Tumor-related protein, putative [Arabidopsis th... 344 1e-93
UniRef100_Q5VMF1 Tumor-related protein-like [Oryza sativa] 305 6e-82
UniRef100_Q9CAU5 Hypothetical protein T18K17.1 [Arabidopsis thal... 286 4e-76
UniRef100_Q5Q0D6 Hypothetical protein [Arabidopsis thaliana] 237 2e-61
UniRef100_Q5Q0D5 Hypothetical protein [Arabidopsis thaliana] 156 6e-37
UniRef100_Q6K9R7 DNA ligase-like [Oryza sativa] 134 3e-30
UniRef100_Q9CQL0 Mus musculus 10 days embryo whole body cDNA, RI... 106 6e-22
UniRef100_Q8R2Y7 RIKEN cDNA 2310038H17 [Mus musculus] 103 4e-21
UniRef100_UPI0000180E91 UPI0000180E91 UniRef100 entry 102 8e-21
UniRef100_Q5XGR7 Hypothetical protein [Xenopus laevis] 102 1e-20
UniRef100_UPI000024A8E7 UPI000024A8E7 UniRef100 entry 97 5e-19
UniRef100_UPI00002DA761 UPI00002DA761 UniRef100 entry 97 5e-19
UniRef100_UPI0000015242 UPI0000015242 UniRef100 entry 96 1e-18
UniRef100_UPI00003697CC UPI00003697CC UniRef100 entry 94 3e-18
UniRef100_Q8WXB1 Hepatocellular carcinoma-associated antigen HCA... 93 9e-18
UniRef100_UPI00003AE5D2 UPI00003AE5D2 UniRef100 entry 92 1e-17
UniRef100_Q96AZ1 Hepatocellularcarcinoma-associated antigen HCA5... 91 2e-17
UniRef100_Q8N1Z9 Hypothetical protein FLJ36493 [Homo sapiens] 91 3e-17
>UniRef100_Q5Q0D4 Hypothetical protein [Arabidopsis thaliana]
Length = 316
Score = 351 bits (901), Expect = 1e-95
Identities = 181/281 (64%), Positives = 215/281 (76%), Gaps = 10/281 (3%)
Query: 7 EEEEEEEEEDAPPPMVKLGSYGGEVRLV--VPGEES--------AAEETMLLWGIQQPTL 56
E E+ +E++A + ++ YGG+V +V P ES AA E M++W IQ PT
Sbjct: 21 EAEKMGKEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTS 80
Query: 57 SKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLG 116
PN V+QSSL+LRLD+CGHSLSILQSP SL PGVTG+VMWDSGVVLGKFLEHSVD
Sbjct: 81 FAPNTLVAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSK 140
Query: 117 TLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSAT 176
L L+ KKIVELGSGCGLVGCIAALLGG +LTDL DRLRLL+KNI+TN+ + RGSA
Sbjct: 141 VLSLEGKKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAI 200
Query: 177 ATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRND 236
EL WG+DPD +LI+P PD+V+GSDV+YSE AV LV+TL QL TTIFL+GELRND
Sbjct: 201 VQELVWGDDPDPDLIEPFPDYVLGSDVIYSEEAVHHLVKTLLQLCSDQTTIFLSGELRND 260
Query: 237 AILEYFLEAAMNDFTIGRVDQTLWHPEYRSNRVVLYVLVKK 277
A+LEYFLE A+ DF IGRV+QT WHP+YRS+RVVLYVL KK
Sbjct: 261 AVLEYFLETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 301
>UniRef100_Q5XEZ3 At1g73320 [Arabidopsis thaliana]
Length = 292
Score = 349 bits (895), Expect = 5e-95
Identities = 179/275 (65%), Positives = 212/275 (77%), Gaps = 10/275 (3%)
Query: 13 EEEDAPPPMVKLGSYGGEVRLV--VPGEES--------AAEETMLLWGIQQPTLSKPNAF 62
+E++A + ++ YGG+V +V P ES AA E M++W IQ PT PN
Sbjct: 3 KEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTSFAPNTL 62
Query: 63 VSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQE 122
V+QSSL+LRLD+CGHSLSILQSP SL PGVTG+VMWDSGVVLGKFLEHSVD L L+
Sbjct: 63 VAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSKVLSLEG 122
Query: 123 KKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSATATELTW 182
KKIVELGSGCGLVGCIAALLGG +LTDL DRLRLL+KNI+TN+ + RGSA EL W
Sbjct: 123 KKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAIVQELVW 182
Query: 183 GEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRNDAILEYF 242
G+DPD +LI+P PD+V+GSDV+YSE AV LV+TL QL TTIFL+GELRNDA+LEYF
Sbjct: 183 GDDPDPDLIEPFPDYVLGSDVIYSEEAVHHLVKTLLQLCSDQTTIFLSGELRNDAVLEYF 242
Query: 243 LEAAMNDFTIGRVDQTLWHPEYRSNRVVLYVLVKK 277
LE A+ DF IGRV+QT WHP+YRS+RVVLYVL KK
Sbjct: 243 LETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 277
>UniRef100_Q9FX27 Tumor-related protein, putative [Arabidopsis thaliana]
Length = 314
Score = 344 bits (883), Expect = 1e-93
Identities = 180/281 (64%), Positives = 213/281 (75%), Gaps = 12/281 (4%)
Query: 7 EEEEEEEEEDAPPPMVKLGSYGGEVRLV--VPGEES--------AAEETMLLWGIQQPTL 56
E E+ +E++A + ++ YGG+V +V P ES AA E M++W IQ PT
Sbjct: 21 EAEKMGKEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTS 80
Query: 57 SKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLG 116
PN V+QSSL+LRLD+CGHSLSILQSP SL PGVTG+VMWDSGVVLGKFLEHSVD
Sbjct: 81 FAPNTLVAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSK 140
Query: 117 TLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSAT 176
L L+ KKIVELGSGCGLVGCIAALLGG +LTDL DRLRLL+KNI+TN+ + RGSA
Sbjct: 141 VLSLEGKKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAI 200
Query: 177 ATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRND 236
EL WG+DPD +LI+P PD+ GSDV+YSE AV LV+TL QL TTIFL+GELRND
Sbjct: 201 VQELVWGDDPDPDLIEPFPDY--GSDVIYSEEAVHHLVKTLLQLCSDQTTIFLSGELRND 258
Query: 237 AILEYFLEAAMNDFTIGRVDQTLWHPEYRSNRVVLYVLVKK 277
A+LEYFLE A+ DF IGRV+QT WHP+YRS+RVVLYVL KK
Sbjct: 259 AVLEYFLETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 299
>UniRef100_Q5VMF1 Tumor-related protein-like [Oryza sativa]
Length = 271
Score = 305 bits (782), Expect = 6e-82
Identities = 151/259 (58%), Positives = 191/259 (73%), Gaps = 1/259 (0%)
Query: 19 PPMVKLGSYGGEVRLVVPGEESAAEETMLLWGIQQPTLSKPNAFVSQSSLQLRLDSCGHS 78
PP + +G+Y G VR V G+ AA ETMLLW + QP + NAFV + L LD+CG
Sbjct: 5 PPRLGMGAYSGPVRPVGDGDGGAAGETMLLWALGQPAAQRHNAFVRHGAHSLTLDACGRR 64
Query: 79 LSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCI 138
LS+LQSPSS+ PGVTGAV+WDSGVVL KFLEH+VD G L L+ + +ELG+GCGL GC+
Sbjct: 65 LSLLQSPSSMSTPGVTGAVVWDSGVVLAKFLEHAVDSGLLTLRAARALELGAGCGLAGCV 124
Query: 139 AALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTPDFV 198
AALLG V+LTDL DRL+LLRKNI+ N+ RGSA +L W +DP +L++P D+V
Sbjct: 125 AALLGAHVLLTDLPDRLKLLRKNIDLNVGD-DARGSARVAQLVWADDPHPDLLNPPLDYV 183
Query: 199 VGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRVDQT 258
+GSDV+YSE AV DL+ TL LS P+TTI LA ELRNDA+LE FLEAAM DF +G ++Q
Sbjct: 184 LGSDVIYSEEAVDDLLLTLKHLSAPHTTIILAAELRNDAVLECFLEAAMADFQVGCIEQQ 243
Query: 259 LWHPEYRSNRVVLYVLVKK 277
WHP++RS RV L++L+KK
Sbjct: 244 QWHPDFRSTRVALFILLKK 262
>UniRef100_Q9CAU5 Hypothetical protein T18K17.1 [Arabidopsis thaliana]
Length = 273
Score = 286 bits (732), Expect = 4e-76
Identities = 155/281 (55%), Positives = 185/281 (65%), Gaps = 48/281 (17%)
Query: 7 EEEEEEEEEDAPPPMVKLGSYGGEVRLV--VPGEES--------AAEETMLLWGIQQPTL 56
E E+ +E++A + ++ YGG+V +V P ES AA E M++W IQ PT
Sbjct: 16 EAEKMGKEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTS 75
Query: 57 SKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLG 116
PN V+QSSL+LRLD+CGHSLSILQSP SL PGVTG+VMWDSGVVLGKFLEHSVD
Sbjct: 76 FAPNTLVAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSK 135
Query: 117 TLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSAT 176
L L+ KKIVELGSGCGLVGCIAALLGG +LTDL DRLRLL+KNI+TN+ + RGSA
Sbjct: 136 VLSLEGKKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAI 195
Query: 177 ATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRND 236
EL WG+DPD +LI+P PD+ D
Sbjct: 196 VQELVWGDDPDPDLIEPFPDY--------------------------------------D 217
Query: 237 AILEYFLEAAMNDFTIGRVDQTLWHPEYRSNRVVLYVLVKK 277
A+LEYFLE A+ DF IGRV+QT WHP+YRS+RVVLYVL KK
Sbjct: 218 AVLEYFLETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 258
>UniRef100_Q5Q0D6 Hypothetical protein [Arabidopsis thaliana]
Length = 247
Score = 237 bits (605), Expect = 2e-61
Identities = 123/201 (61%), Positives = 148/201 (73%), Gaps = 10/201 (4%)
Query: 7 EEEEEEEEEDAPPPMVKLGSYGGEVRLV--VPGEES--------AAEETMLLWGIQQPTL 56
E E+ +E++A + ++ YGG+V +V P ES AA E M++W IQ PT
Sbjct: 21 EAEKMGKEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTS 80
Query: 57 SKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLG 116
PN V+QSSL+LRLD+CGHSLSILQSP SL PGVTG+VMWDSGVVLGKFLEHSVD
Sbjct: 81 FAPNTLVAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSK 140
Query: 117 TLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSAT 176
L L+ KKIVELGSGCGLVGCIAALLGG +LTDL DRLRLL+KNI+TN+ + RGSA
Sbjct: 141 VLSLEGKKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAI 200
Query: 177 ATELTWGEDPDRELIDPTPDF 197
EL WG+DPD +LI+P PD+
Sbjct: 201 VQELVWGDDPDPDLIEPFPDY 221
>UniRef100_Q5Q0D5 Hypothetical protein [Arabidopsis thaliana]
Length = 170
Score = 156 bits (394), Expect = 6e-37
Identities = 84/140 (60%), Positives = 100/140 (71%), Gaps = 10/140 (7%)
Query: 7 EEEEEEEEEDAPPPMVKLGSYGGEVRLV--VPGEES--------AAEETMLLWGIQQPTL 56
E E+ +E++A + ++ YGG+V +V P ES AA E M++W IQ PT
Sbjct: 21 EAEKMGKEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTS 80
Query: 57 SKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLG 116
PN V+QSSL+LRLD+CGHSLSILQSP SL PGVTG+VMWDSGVVLGKFLEHSVD
Sbjct: 81 FAPNTLVAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSK 140
Query: 117 TLVLQEKKIVELGSGCGLVG 136
L L+ KKIVELGSGCGLVG
Sbjct: 141 VLSLEGKKIVELGSGCGLVG 160
>UniRef100_Q6K9R7 DNA ligase-like [Oryza sativa]
Length = 489
Score = 134 bits (336), Expect = 3e-30
Identities = 79/224 (35%), Positives = 123/224 (54%), Gaps = 14/224 (6%)
Query: 64 SQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLV---L 120
S S+ + L+ GH L I Q P+S G +WD+ +V KFLE + G L
Sbjct: 23 SPSTSAISLELLGHRLHISQDPNSKHL----GTTVWDASMVFVKFLEKNSRKGRFSPSKL 78
Query: 121 QEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLR-------G 173
+ K+++ELG+GCGL G ALLG +V+ TD ++ L LL +N+E N +S G
Sbjct: 79 KGKRVIELGAGCGLAGLGMALLGCDVVTTDQVEVLPLLLRNVERNKSWISQSNSDSGSIG 138
Query: 174 SATATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGEL 233
S T EL WG +DP D+++G+DVVYSE+ + L+ET+ LSGP T I L E+
Sbjct: 139 SVTVAELDWGNKDHIRAVDPPFDYIIGTDVVYSEHLLQPLMETIIALSGPKTKIMLGYEI 198
Query: 234 RNDAILEYFLEAAMNDFTIGRVDQTLWHPEYRSNRVVLYVLVKK 277
R+ + E ++ ++F + V ++ +Y+ + LY++ K
Sbjct: 199 RSTTVHEQMMQMWKSNFNVKTVSKSKMDVKYQHPSIHLYIMDPK 242
>UniRef100_Q9CQL0 Mus musculus 10 days embryo whole body cDNA, RIKEN full-length
enriched library, clone:2610017G06
product:HEPATOCELLULAR CARCINOMA- ASSOCIATED ANTIGEN
HCA557B homolog [Mus musculus]
Length = 218
Score = 106 bits (265), Expect = 6e-22
Identities = 81/223 (36%), Positives = 117/223 (52%), Gaps = 30/223 (13%)
Query: 34 VVPGEESAAEETMLLWGIQQPTLSKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGV 93
+VP EESAA G+Q+ KP A S ++ H++ I Q LG
Sbjct: 3 LVPYEESAAI------GLQK--FHKPLATFSFAN---------HTIQIRQDWRQLGV--- 42
Query: 94 TGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLD 153
AV+WD+ VVL +LE +G + L+ VELG+G GLVG +AALLG +V +TD
Sbjct: 43 -AAVVWDAAVVLSMYLE----MGAVELRGCSAVELGAGTGLVGIVAALLGAQVTITDRKV 97
Query: 154 RLRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTP-DFVVGSDVVYSENAVVD 212
L L+ N+E N+ ++ A ELTWG+ + E P D ++G+DV+Y E+ D
Sbjct: 98 ALEFLKSNVEANLP-PHIQPKAVVKELTWGQ--NLESFSPGEFDLILGADVIYLEDTFTD 154
Query: 213 LVETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRV 255
L++TLG L N+ I LA +R + FL FT+ +V
Sbjct: 155 LLQTLGHLCSNNSVILLACRIRYERD-SNFLTMLERQFTVSKV 196
>UniRef100_Q8R2Y7 RIKEN cDNA 2310038H17 [Mus musculus]
Length = 218
Score = 103 bits (258), Expect = 4e-21
Identities = 80/223 (35%), Positives = 116/223 (51%), Gaps = 30/223 (13%)
Query: 34 VVPGEESAAEETMLLWGIQQPTLSKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGV 93
+VP EESAA G+Q+ KP A S ++ H++ I Q LG
Sbjct: 3 LVPYEESAAI------GLQK--FHKPLATFSFAN---------HTIQIRQDWRQLGV--- 42
Query: 94 TGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLD 153
AV+WD+ VVL +LE +G + L+ VELG+G GLVG +AAL G +V +TD
Sbjct: 43 -AAVVWDAAVVLSMYLE----MGAVELRGCSAVELGAGTGLVGIVAALPGAQVTITDRKV 97
Query: 154 RLRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTP-DFVVGSDVVYSENAVVD 212
L L+ N+E N+ ++ A ELTWG+ + E P D ++G+DV+Y E+ D
Sbjct: 98 ALEFLKSNVEANLP-PHIQPKAVVKELTWGQ--NLESFSPGEFDLILGADVIYLEDTFTD 154
Query: 213 LVETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRV 255
L++TLG L N+ I LA +R + FL FT+ +V
Sbjct: 155 LLQTLGHLCSNNSVILLACRIRYERD-SNFLTMLERQFTVSKV 196
>UniRef100_UPI0000180E91 UPI0000180E91 UniRef100 entry
Length = 218
Score = 102 bits (255), Expect = 8e-21
Identities = 74/213 (34%), Positives = 112/213 (51%), Gaps = 23/213 (10%)
Query: 43 EETMLLWGIQQPTLSKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSG 102
EE+M + G+Q+ L KP A S ++ H++ I Q LG AV+WD+
Sbjct: 7 EESMSI-GLQK--LHKPLATFSFAN---------HTIQIRQDWGRLGV----AAVVWDAA 50
Query: 103 VVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNI 162
+VL +LE +G + L+ VELG+G GLVG +AALLG V +TD L L+ N+
Sbjct: 51 IVLSTYLE----MGAVELRGCSAVELGAGTGLVGIVAALLGAHVTITDRKVALEFLKSNV 106
Query: 163 ETNMKHVSLRGSATATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSG 222
E N+ ++ ELTWG++ D D ++G+D++Y E+ DL++TLG L
Sbjct: 107 EANLP-PHIQPKVVVKELTWGQNLD-SFSPGEFDLILGADIIYLEDTFTDLLQTLGHLCS 164
Query: 223 PNTTIFLAGELRNDAILEYFLEAAMNDFTIGRV 255
N+ I LA +R + FL FT+ +V
Sbjct: 165 NNSVILLACRIRYERD-NNFLTMLERQFTVSKV 196
>UniRef100_Q5XGR7 Hypothetical protein [Xenopus laevis]
Length = 229
Score = 102 bits (253), Expect = 1e-20
Identities = 69/192 (35%), Positives = 103/192 (52%), Gaps = 10/192 (5%)
Query: 92 GVTGAVMWDSGVVLGKFLEHSVDLGTLV--LQEKKIVELGSGCGLVGCIAALLGGEVILT 149
G G V+WD+ +VL KFLE + L K ++ELG+G G+VG +AA G V++T
Sbjct: 41 GDVGCVVWDAAIVLSKFLESREFMCPEGHRLSGKCVLELGAGTGIVGIMAATQGANVMVT 100
Query: 150 DLLDRLRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTPDFVVGSDVVYSENA 209
DL D L++ NIE+N + RGS A L WGE+ +EL+ P PD+++ +D +Y E +
Sbjct: 101 DLEDLQELMKTNIESNSHFI--RGSCQAKVLKWGEEV-KELV-PKPDYILLADCIYYEES 156
Query: 210 VVDLVETLGQLSGPNTTIFLAGELR----NDAILEYFLEAAMNDFTIGRVDQTLWHPEYR 265
+ L++TL L+G +T I E R N I + F E F V EYR
Sbjct: 157 LEPLLKTLRDLTGSDTCILCCYEQRTMGKNPQIEKRFFELLAEHFKYEEVPLEKHDKEYR 216
Query: 266 SNRVVLYVLVKK 277
S + + + +K
Sbjct: 217 SEDIHILQIYRK 228
>UniRef100_UPI000024A8E7 UPI000024A8E7 UniRef100 entry
Length = 218
Score = 97.1 bits (240), Expect = 5e-19
Identities = 72/206 (34%), Positives = 107/206 (50%), Gaps = 25/206 (12%)
Query: 54 PTLSKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSV 113
P LSK + QSS + L H + + Q LG AV+WD+ VVL FLE
Sbjct: 12 PALSK----LHQSSAEFTL--ANHRIRLSQDWKRLGV----AAVVWDAAVVLCMFLE--- 58
Query: 114 DLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRG 173
+G + L+ K+++ELG+G GLVG +AALLG V +TD L L N+ N+ +
Sbjct: 59 -MGKVDLKGKRVIELGAGTGLVGIVAALLGANVTITDREPALEFLTANVHENIPQ-GRQK 116
Query: 174 SATATELTWGEDPDRELIDPTP----DFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFL 229
+ +ELTWGE+ +D P D ++G+D+VY E L++TL LS +T + L
Sbjct: 117 AVQVSELTWGEN-----LDLYPQGGYDLILGADIVYLEETFPALLQTLEHLSSGDTVVLL 171
Query: 230 AGELRNDAILEYFLEAAMNDFTIGRV 255
+ +R + E FL F++ V
Sbjct: 172 SCRIRYERD-ERFLTELRQRFSVQEV 196
>UniRef100_UPI00002DA761 UPI00002DA761 UniRef100 entry
Length = 209
Score = 97.1 bits (240), Expect = 5e-19
Identities = 67/181 (37%), Positives = 98/181 (54%), Gaps = 16/181 (8%)
Query: 54 PTLSKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSV 113
P LSK + SS R L + Q LG AV+WD+ VV+ +LE
Sbjct: 12 PVLSK----LHSSSAHFRF--ADRDLRLAQDWRRLGV----AAVVWDAAVVMCMYLE--- 58
Query: 114 DLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRG 173
LG + ++EK+++ELG+G GLVG +AAL+G +TD L LL N+ N+ S G
Sbjct: 59 -LGKVDVKEKEVIELGAGTGLVGIVAALMGARATITDRKPALELLSANVRANLPADS-PG 116
Query: 174 SATATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGEL 233
SA +EL+WGE +R D V+G+D++Y E+ V L+ TL L +T + LA ++
Sbjct: 117 SAAVSELSWGEGLERYPAGGF-DLVLGADIIYLEDTFVPLLRTLEHLCSGSTLLLLACKI 175
Query: 234 R 234
R
Sbjct: 176 R 176
>UniRef100_UPI0000015242 UPI0000015242 UniRef100 entry
Length = 218
Score = 95.9 bits (237), Expect = 1e-18
Identities = 68/181 (37%), Positives = 98/181 (53%), Gaps = 16/181 (8%)
Query: 54 PTLSKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSV 113
P LSK + SS + R L + Q LG AV+WD+ VVL +LE
Sbjct: 12 PVLSK----LHSSSAEFRF--VDRDLRLAQDWKRLGV----AAVVWDAAVVLCMYLE--- 58
Query: 114 DLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRG 173
LG + L+EK+++ELG+G GLVG +AAL+G +TD L L N++ N+ S G
Sbjct: 59 -LGQVDLKEKEVIELGAGTGLVGIVAALMGARATITDREPALDFLSANVKANLPADS-PG 116
Query: 174 SATATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGEL 233
SA +EL+WGE DR D V+G+D++Y ++ L+ TL L +T I LA ++
Sbjct: 117 SAVVSELSWGEGLDRYPAGGF-DLVLGADIIYLKDTFGPLLRTLEHLCSESTLILLACKI 175
Query: 234 R 234
R
Sbjct: 176 R 176
>UniRef100_UPI00003697CC UPI00003697CC UniRef100 entry
Length = 218
Score = 94.4 bits (233), Expect = 3e-18
Identities = 62/182 (34%), Positives = 97/182 (53%), Gaps = 17/182 (9%)
Query: 77 HSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVG 136
H++ I Q LG AV+WD+ +VL +LE +G + L+ + VELG+G GLVG
Sbjct: 29 HTIQIRQDWRHLGV----AAVVWDAAIVLSTYLE----MGAVELRGRSAVELGAGTGLVG 80
Query: 137 CIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTP- 195
+AALLG V +TD L L+ N++ N+ ++ ELTWG++ L +P
Sbjct: 81 IVAALLGAHVTITDRKVALEFLKSNVQANLP-PHIQPKTVVKELTWGQN----LGSFSPG 135
Query: 196 --DFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIG 253
D ++G+D++Y E DL++TL L ++ I LA +R + FL FT+
Sbjct: 136 EFDLILGADIIYLEETFTDLLQTLEHLCSNHSVILLACRIRYERD-NNFLAMLERQFTVR 194
Query: 254 RV 255
+V
Sbjct: 195 KV 196
>UniRef100_Q8WXB1 Hepatocellular carcinoma-associated antigen HCA557b [Homo sapiens]
Length = 218
Score = 92.8 bits (229), Expect = 9e-18
Identities = 58/162 (35%), Positives = 90/162 (54%), Gaps = 18/162 (11%)
Query: 77 HSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVG 136
H++ I Q LG AV+WD+ +VL +LE +G + L+ + VELG+G GLVG
Sbjct: 29 HTIQIRQDWRHLGV----AAVVWDAAIVLSTYLE----MGAVELRGRSAVELGAGTGLVG 80
Query: 137 CIAALLGGEVILTDLLDRLRLLRKNIETNM-KHVSLRGSATATELTWGEDPDRELIDPTP 195
+AALLG V +TD L L+ N++ N+ H+ + ELTWG++ L +P
Sbjct: 81 IVAALLGAHVTITDRKVALEFLKSNVQANLPPHIQTK--TVVKELTWGQN----LGSFSP 134
Query: 196 ---DFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELR 234
D ++G+D++Y E DL++TL L ++ I LA +R
Sbjct: 135 GEFDLILGADIIYLEETFTDLLQTLEHLCSNHSVILLACRIR 176
>UniRef100_UPI00003AE5D2 UPI00003AE5D2 UniRef100 entry
Length = 215
Score = 92.4 bits (228), Expect = 1e-17
Identities = 55/160 (34%), Positives = 85/160 (52%), Gaps = 7/160 (4%)
Query: 96 AVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRL 155
AV+WD+ VVL +LE +G + L+++ ++ELG+G GL+G +A LLG V +TD L
Sbjct: 42 AVVWDAAVVLSAYLE----MGGIDLRDRSVIELGAGTGLLGIVATLLGARVTITDREPAL 97
Query: 156 RLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVE 215
L N+ N+ L A ELTWG+D DF++G+D++Y E +L+
Sbjct: 98 EFLESNVWANLPS-ELHARAVVKELTWGKDLG-SFPPGAFDFILGADIIYLEETFAELLR 155
Query: 216 TLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRV 255
TL L T I L+ +R + FL+ F++ V
Sbjct: 156 TLEHLCSEQTVILLSCRIRYERD-NNFLKMLKGRFSVSEV 194
>UniRef100_Q96AZ1 Hepatocellularcarcinoma-associated antigen HCA557a, isoform a [Homo
sapiens]
Length = 226
Score = 91.3 bits (225), Expect = 2e-17
Identities = 61/171 (35%), Positives = 93/171 (53%), Gaps = 14/171 (8%)
Query: 75 CGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLE-HSVDLGTLVLQEKKIVELGSGCG 133
CGH L+I Q+ G+ A +WD+ + L + E +VD + KK++ELG+G G
Sbjct: 36 CGHVLTITQN---FGSRLGVAARVWDAALSLCNYFESQNVDF-----RGKKVIELGAGTG 87
Query: 134 LVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDP 193
+VG +AAL GG+V +TDL L ++ N++ N V G A L+WG D +
Sbjct: 88 IVGILAALQGGDVTITDLPLALEQIQGNVQAN---VPAGGQAQVRALSWG--IDHHVFPA 142
Query: 194 TPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRNDAILEYFLE 244
D V+G+D+VY E L+ TL L P+ TI+LA ++R + E F +
Sbjct: 143 NYDLVLGADIVYLEPTFPLLLGTLQHLCRPHGTIYLASKMRKEHGTESFFQ 193
>UniRef100_Q8N1Z9 Hypothetical protein FLJ36493 [Homo sapiens]
Length = 218
Score = 90.9 bits (224), Expect = 3e-17
Identities = 57/162 (35%), Positives = 89/162 (54%), Gaps = 18/162 (11%)
Query: 77 HSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVG 136
H++ Q LG AV+WD+ +VL +LE +G + L+ + VELG+G GLVG
Sbjct: 29 HTIQTRQDWRHLGV----AAVVWDAAIVLSTYLE----MGAVELRGRSAVELGAGTGLVG 80
Query: 137 CIAALLGGEVILTDLLDRLRLLRKNIETNM-KHVSLRGSATATELTWGEDPDRELIDPTP 195
+AALLG V +TD L L+ N++ N+ H+ + ELTWG++ L +P
Sbjct: 81 IVAALLGAHVTITDRKVALEFLKSNVQANLPPHIQTK--TVVKELTWGQN----LGSFSP 134
Query: 196 ---DFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELR 234
D ++G+D++Y E DL++TL L ++ I LA +R
Sbjct: 135 GEFDLILGADIIYLEETFTDLLQTLEHLCSNHSVILLACRIR 176
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.135 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 470,551,943
Number of Sequences: 2790947
Number of extensions: 19656892
Number of successful extensions: 140983
Number of sequences better than 10.0: 319
Number of HSP's better than 10.0 without gapping: 117
Number of HSP's successfully gapped in prelim test: 206
Number of HSP's that attempted gapping in prelim test: 139977
Number of HSP's gapped (non-prelim): 615
length of query: 277
length of database: 848,049,833
effective HSP length: 126
effective length of query: 151
effective length of database: 496,390,511
effective search space: 74954967161
effective search space used: 74954967161
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0383.11


