

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371b.6
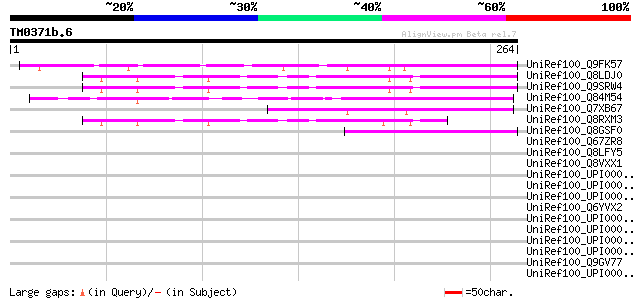
(264 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FK57 Gb|AAF00631.1 [Arabidopsis thaliana] 120 5e-26
UniRef100_Q8LDJ0 Hypothetical protein [Arabidopsis thaliana] 111 2e-23
UniRef100_Q9SRW4 F20H23.8 protein [Arabidopsis thaliana] 109 8e-23
UniRef100_Q84M54 Hypothetical protein OSJNBa0059E14.21 [Oryza sa... 106 5e-22
UniRef100_Q7XB67 Hypothetical protein P0431A02.22 [Oryza sativa] 89 9e-17
UniRef100_Q8RXM3 Hypothetical protein At3g03870 [Arabidopsis tha... 69 9e-11
UniRef100_Q8GSF0 Hypothetical protein At4g10080/T5L19_210 [Arabi... 47 7e-04
UniRef100_Q67ZR8 Hypothetical protein At4g13530 [Arabidopsis tha... 40 0.080
UniRef100_Q8LFY5 Hypothetical protein [Arabidopsis thaliana] 40 0.080
UniRef100_Q8VXX1 Hypothetical protein At4g13530 [Arabidopsis tha... 39 0.18
UniRef100_UPI00003B00B6 UPI00003B00B6 UniRef100 entry 38 0.30
UniRef100_UPI00003AB881 UPI00003AB881 UniRef100 entry 38 0.30
UniRef100_UPI00003AB880 UPI00003AB880 UniRef100 entry 38 0.30
UniRef100_Q6YVX2 Hypothetical protein OSJNBb0038F20.18 [Oryza sa... 37 0.68
UniRef100_UPI000046AE24 UPI000046AE24 UniRef100 entry 36 0.88
UniRef100_UPI00003AD93B UPI00003AD93B UniRef100 entry 35 1.5
UniRef100_UPI000021B24B UPI000021B24B UniRef100 entry 35 2.6
UniRef100_UPI00002F9BED UPI00002F9BED UniRef100 entry 34 3.4
UniRef100_Q9GV77 Embryonic blastocoelar extracellular matrix pro... 34 3.4
UniRef100_UPI0000365F7D UPI0000365F7D UniRef100 entry 34 4.4
>UniRef100_Q9FK57 Gb|AAF00631.1 [Arabidopsis thaliana]
Length = 280
Score = 120 bits (300), Expect = 5e-26
Identities = 96/291 (32%), Positives = 145/291 (48%), Gaps = 47/291 (16%)
Query: 6 DWVLLSDDG----SLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHD- 60
DW +L LD G E + K + FD +YF C P T+ LH
Sbjct: 5 DWEILPKINYKGLELDLGHEEDHEVTKMMRNTAKSFDSDYFIC--PIQDSVGKTEFLHQR 62
Query: 61 ---------------DPIQFVENTEDDHVGDNSDTTSEKIKAVVVEADEQETLSVSQFFL 105
+P+ V+ + DH SE ++ E+ ++ F
Sbjct: 63 SSVVPTQLLQIPITWEPLSPVD--DKDHNKYLDPDFSEPDPELLTESFPSPRIT---FKK 117
Query: 106 QLENKFADMKMDSPKSSPRVLFPPPLDAGALKFEDK-----GEAMEVMVSPRRKVENEMA 160
E +FADMK+DSP + F PL + D GE+ + ++ + ++++
Sbjct: 118 SKETEFADMKIDSPAAR----FTSPLPQNDERHSDSEGGLGGESYDEIMGSEVEESSDLS 173
Query: 161 TMDCDREEVDPTDG--FNFWKWSLTGVGAICSFGVAAA--TICVLVFG--SQQKNKFHQN 214
+ ++EVD +G N WK L G+GAICSFGVAAA TICV G S + ++N
Sbjct: 174 S----KKEVDWDEGERTNLWKKGLNGIGAICSFGVAAAAATICVFFLGHNSSIQGGRNKN 229
Query: 215 HKIQFQIYTDD-KRIKQVVQQATKLNEAIAATRGIPLSRAHITYGGYYEAL 264
++FQIY+DD KR+ +VV+ ATKLNEAI+ +G+P++RA I++GGYY+AL
Sbjct: 230 QILRFQIYSDDNKRMNEVVKHATKLNEAISVMKGLPVARAQISFGGYYDAL 280
>UniRef100_Q8LDJ0 Hypothetical protein [Arabidopsis thaliana]
Length = 235
Score = 111 bits (278), Expect = 2e-23
Identities = 88/252 (34%), Positives = 129/252 (50%), Gaps = 43/252 (17%)
Query: 39 MNYFYCTS-----PKSRDSSTTQLLHDDPIQF------VENTEDDHVGDNSDTTSEKIKA 87
M+YF C S PK+ ++++ +Q V + ++ + +N D +
Sbjct: 1 MDYFLCPSTKDPLPKTESPPRSKVVPKKILQVPIAWEAVLDQDNTKIPNNPDLEPDPHP- 59
Query: 88 VVVEADEQETLSVSQ-------FFLQLENKFADMKMDSPKSSPRVLFP-PPLDAGALKFE 139
+ D + TLS F + + +FADMK+D P R+ P P +DA A K
Sbjct: 60 ---DPDSKPTLSTDSVSSPRVSFKIMKDTEFADMKIDPPA---RITSPLPQIDAAASKPS 113
Query: 140 DKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKWSLTGVGAICSFGVAAA--T 197
D +E R K E ++ + E+ + N WK L G+GAICSFGVAAA T
Sbjct: 114 D----LEGGDDLRDKKE---VVLEEEDEDGSGGERLNLWKAGLNGIGAICSFGVAAAAAT 166
Query: 198 ICVLVFGSQQ----KNKFHQNHKIQFQIYTDD-KRIKQVVQQATKLNEAIAATRGIPLSR 252
+CV G KNK N ++FQIY+DD KR+K+VV ATKLNEAI +G+P+ R
Sbjct: 167 VCVFFLGHNNIKICKNK---NQMLRFQIYSDDNKRMKEVVNHATKLNEAIFGMKGVPVVR 223
Query: 253 AHITYGGYYEAL 264
A I++GG+Y+ L
Sbjct: 224 AQISFGGHYDGL 235
>UniRef100_Q9SRW4 F20H23.8 protein [Arabidopsis thaliana]
Length = 266
Score = 109 bits (272), Expect = 8e-23
Identities = 87/252 (34%), Positives = 128/252 (50%), Gaps = 43/252 (17%)
Query: 39 MNYFYCTS-----PKSRDSSTTQLLHDDPIQF------VENTEDDHVGDNSDTTSEKIKA 87
M+YF C S PK+ ++++ +Q V + ++ + +N D +
Sbjct: 32 MDYFLCPSTKDPLPKTESPPRSKVVPKKILQVPIAWEAVLDQDNTKIPNNPDLEPDPHP- 90
Query: 88 VVVEADEQETLSVSQ-------FFLQLENKFADMKMDSPKSSPRVLFP-PPLDAGALKFE 139
+ D + TLS F + + +FADMK+D P R+ P P +D A K
Sbjct: 91 ---DPDSKPTLSTDSVSSPRVSFKIMKDTEFADMKIDPPA---RITSPLPQIDDAASKPS 144
Query: 140 DKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKWSLTGVGAICSFGVAAA--T 197
D +E R K E ++ + E+ + N WK L G+GAICSFGVAAA T
Sbjct: 145 D----LEGGDDLRDKKE---VVLEEEDEDGSGGERLNLWKAGLNGIGAICSFGVAAAAAT 197
Query: 198 ICVLVFGSQQ----KNKFHQNHKIQFQIYTDD-KRIKQVVQQATKLNEAIAATRGIPLSR 252
+CV G KNK N ++FQIY+DD KR+K+VV ATKLNEAI +G+P+ R
Sbjct: 198 VCVFFLGHNNIKICKNK---NQMLRFQIYSDDNKRMKEVVNHATKLNEAIFGMKGVPVVR 254
Query: 253 AHITYGGYYEAL 264
A I++GG+Y+ L
Sbjct: 255 AQISFGGHYDGL 266
>UniRef100_Q84M54 Hypothetical protein OSJNBa0059E14.21 [Oryza sativa]
Length = 255
Score = 106 bits (265), Expect = 5e-22
Identities = 85/257 (33%), Positives = 137/257 (53%), Gaps = 34/257 (13%)
Query: 11 SDDGSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHDDPIQF---VE 67
SD G++ G D+ +F S +M++F +P S S +L D+ +
Sbjct: 26 SDHGTVVGKDDQFLF-----GASLIMINMDHF---TPASHPSPYDCILDDETKNIFLPIH 77
Query: 68 NTEDDHVGDNSDTTSEKIKAVVVEADEQETL-SVSQFFLQLENKFADMKMDSPKSSPRVL 126
+E+ +VGD + I V +E+ +E + V++ F A+ + + KS
Sbjct: 78 LSEEVYVGDPVIKFKD-IDVVKIESYREEFVPKVTEIF------DAEEEAEMIKS----- 125
Query: 127 FPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKWSLTGVG 186
P+ A + +D E M MV+P + VE E D+E +GF+ K + GVG
Sbjct: 126 ---PVSAEEVDVDDDDEVM-AMVAPDQCVEEEEGAQK-DKEH----NGFSVGKLRVNGVG 176
Query: 187 AICSFGVAAATICVLVFGSQQKNKFH-QNHKIQFQIYTDDKRIKQVVQQATKLNEAIAAT 245
A+CSFGVAAAT+C+ + G +Q+ QN K FQ+Y D++RI+QVVQQA++LN+A++
Sbjct: 177 ALCSFGVAAATLCIFLLGGRQQQLHKTQNQKTPFQMYADNERIQQVVQQASRLNQAVSTV 236
Query: 246 RGIPLSRAHITYGGYYE 262
G +RA I++GGYY+
Sbjct: 237 MGGASTRASISFGGYYD 253
>UniRef100_Q7XB67 Hypothetical protein P0431A02.22 [Oryza sativa]
Length = 229
Score = 89.4 bits (220), Expect = 9e-17
Identities = 59/154 (38%), Positives = 82/154 (52%), Gaps = 26/154 (16%)
Query: 135 ALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDG-------------------- 174
A+ E K E M + + E E A D + VD D
Sbjct: 74 AVPAETKTSQEETMAAKVTEEEEEEAFQGSDAKVVDGDDDGGGEEEEEEEEEGKKAGAEC 133
Query: 175 --FNFWKWSLTGVGAICSFGVAAA-TICVLVFGS--QQKNKFHQNHKIQFQIYTDDKRIK 229
F K + G+GA+CSFGVAAA T+CV + G Q ++ Q HKIQ Q+Y DDKR++
Sbjct: 134 VVFRVGKLRVNGIGALCSFGVAAAATVCVFLVGGRLQHHHRQQQQHKIQLQLYGDDKRMQ 193
Query: 230 QVVQQATKLNEAIAATRGIPLS-RAHITYGGYYE 262
QVVQQ ++LN+A+++ G S RA+I++GGYYE
Sbjct: 194 QVVQQTSRLNQAMSSVMGGGGSTRANISFGGYYE 227
>UniRef100_Q8RXM3 Hypothetical protein At3g03870 [Arabidopsis thaliana]
Length = 234
Score = 69.3 bits (168), Expect = 9e-11
Identities = 65/215 (30%), Positives = 99/215 (45%), Gaps = 42/215 (19%)
Query: 39 MNYFYCTS-----PKSRDSSTTQLLHDDPIQF------VENTEDDHVGDNSDTTSEKIKA 87
M+YF C S PK+ ++++ +Q V + ++ + +N D +
Sbjct: 32 MDYFLCPSTKDPLPKTESPPRSKVVPKKILQVPIAWEAVLDQDNTKIPNNPDLEPDPHP- 90
Query: 88 VVVEADEQETLSVSQ-------FFLQLENKFADMKMDSPKSSPRVLFP-PPLDAGALKFE 139
+ D + TLS F + + +FADMK+D P R+ P P +D A K
Sbjct: 91 ---DPDSKPTLSTDSVSSPRVSFKIMKDTEFADMKIDPPA---RITSPLPQIDDAASKPS 144
Query: 140 DKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKWSLTGVGAICSFGV--AAAT 197
D +E R K E ++ + E+ + N WK L G+GAICSFGV AAAT
Sbjct: 145 D----LEGGDDLRDKKE---VVLEEEDEDGSGGERLNLWKAGLNGIGAICSFGVAAAAAT 197
Query: 198 ICVLVFGSQQ----KNKFHQNHKIQFQIYTDDKRI 228
+CV G KNK N ++FQIY+DD ++
Sbjct: 198 VCVFFLGHNNIKICKNK---NQMLRFQIYSDDNKV 229
>UniRef100_Q8GSF0 Hypothetical protein At4g10080/T5L19_210 [Arabidopsis thaliana]
Length = 325
Score = 46.6 bits (109), Expect = 7e-04
Identities = 24/90 (26%), Positives = 46/90 (50%)
Query: 175 FNFWKWSLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQ 234
F K+S+ +G + S +AAA + +++ G + N + + ++ DDK+ +V+ Q
Sbjct: 233 FVLLKYSVFRIGPVWSVSMAAAVMGLVLLGRRLYNMKKKAQRFHLKVTIDDKKASRVMSQ 292
Query: 235 ATKLNEAIAATRGIPLSRAHITYGGYYEAL 264
A +LNE R +P+ R + G + L
Sbjct: 293 AARLNEVFTEVRRVPVIRPALPSPGAWPVL 322
>UniRef100_Q67ZR8 Hypothetical protein At4g13530 [Arabidopsis thaliana]
Length = 270
Score = 39.7 bits (91), Expect = 0.080
Identities = 32/133 (24%), Positives = 60/133 (45%), Gaps = 22/133 (16%)
Query: 139 EDKGEAMEVMVSPRRKVENEMATMDCDREEV--DP----------TDGFNFWK------- 179
E + M+++ S RK E+ + ++ + V DP GF +WK
Sbjct: 123 ESIAQDMDLISSDERKEESLLHPVEGEGNSVSIDPGVKSGGGGGEEKGFVWWKIPIEVLK 182
Query: 180 WSLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQATKLN 239
+ + + I S +AAA + ++ G + N + +Q ++ DDK+ V A + N
Sbjct: 183 YCVLKINPIWSLSMAAAFVGFVMLGRRLYNMKKKTRSLQLKVLLDDKK---VANHAARWN 239
Query: 240 EAIAATRGIPLSR 252
EAI+ + +P+ R
Sbjct: 240 EAISVVKRVPIIR 252
>UniRef100_Q8LFY5 Hypothetical protein [Arabidopsis thaliana]
Length = 270
Score = 39.7 bits (91), Expect = 0.080
Identities = 32/133 (24%), Positives = 60/133 (45%), Gaps = 22/133 (16%)
Query: 139 EDKGEAMEVMVSPRRKVENEMATMDCDREEV--DP----------TDGFNFWK------- 179
E + M+++ S RK E+ + ++ + V DP GF +WK
Sbjct: 123 ESIAQDMDLISSDERKEESLLHPVEGEGNSVSIDPGVKSGGGGGEEKGFVWWKIPIEVLK 182
Query: 180 WSLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQATKLN 239
+ + + I S +AAA + ++ G + N + +Q ++ DDK+ V A + N
Sbjct: 183 YCVLKINPIWSLSMAAAFVGFVMLGRRLYNMKKKTRSLQLKVLLDDKK---VANHAARWN 239
Query: 240 EAIAATRGIPLSR 252
EAI+ + +P+ R
Sbjct: 240 EAISVVKRVPIIR 252
>UniRef100_Q8VXX1 Hypothetical protein At4g13530 [Arabidopsis thaliana]
Length = 269
Score = 38.5 bits (88), Expect = 0.18
Identities = 32/133 (24%), Positives = 59/133 (44%), Gaps = 23/133 (17%)
Query: 139 EDKGEAMEVMVSPRRKVENEMATMDCDREEV--DP----------TDGFNFWK------- 179
E + M+++ S RK E+ + ++ + V DP GF +WK
Sbjct: 123 ESIAQDMDLISSDERKEESLLHPVEGEGNSVSIDPGVKSGGGGGEEKGFVWWKIPIEVLK 182
Query: 180 WSLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQATKLN 239
+ + + I S +AAA + ++ G + N + +Q ++ DDK V A + N
Sbjct: 183 YCVLKINPIWSLSMAAAFVGFVMLGRRLYNMKKKTRSLQLKVLLDDK----VANHAARWN 238
Query: 240 EAIAATRGIPLSR 252
EAI+ + +P+ R
Sbjct: 239 EAISVVKRVPIIR 251
>UniRef100_UPI00003B00B6 UPI00003B00B6 UniRef100 entry
Length = 4746
Score = 37.7 bits (86), Expect = 0.30
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query: 118 SPKSSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNF 177
SP SSP ++FPP +A K E K + ++V V + K++ +++D R G +
Sbjct: 4310 SPSSSPPIIFPPAFEAA--KVEAKPDELKVTVKLKPKLKAIHSSLDDCRSPSKKWKGMKW 4367
Query: 178 WKWSL 182
KWS+
Sbjct: 4368 KKWSI 4372
>UniRef100_UPI00003AB881 UPI00003AB881 UniRef100 entry
Length = 1342
Score = 37.7 bits (86), Expect = 0.30
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query: 118 SPKSSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNF 177
SP SSP ++FPP +A K E K + ++V V + K++ +++D R G +
Sbjct: 921 SPSSSPPIIFPPAFEAA--KVEAKPDELKVTVKLKPKLKAIHSSLDDCRSPSKKWKGMKW 978
Query: 178 WKWSL 182
KWS+
Sbjct: 979 KKWSI 983
>UniRef100_UPI00003AB880 UPI00003AB880 UniRef100 entry
Length = 3488
Score = 37.7 bits (86), Expect = 0.30
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query: 118 SPKSSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNF 177
SP SSP ++FPP +A K E K + ++V V + K++ +++D R G +
Sbjct: 3067 SPSSSPPIIFPPAFEAA--KVEAKPDELKVTVKLKPKLKAIHSSLDDCRSPSKKWKGMKW 3124
Query: 178 WKWSL 182
KWS+
Sbjct: 3125 KKWSI 3129
>UniRef100_Q6YVX2 Hypothetical protein OSJNBb0038F20.18 [Oryza sativa]
Length = 353
Score = 36.6 bits (83), Expect = 0.68
Identities = 23/69 (33%), Positives = 33/69 (47%), Gaps = 1/69 (1%)
Query: 185 VGAICSFGVAAATICVLVFGSQQKNKFHQNHKI-QFQIYTDDKRIKQVVQQATKLNEAIA 243
V I SF +AAA + + V G + + + +I DDKR Q + +LNEA
Sbjct: 272 VKPIWSFSIAAALLGLFVLGRRMYRMRRKARGLPHIKIAFDDKRASQFADRTARLNEAFF 331
Query: 244 ATRGIPLSR 252
R IP+ R
Sbjct: 332 VARHIPMLR 340
>UniRef100_UPI000046AE24 UPI000046AE24 UniRef100 entry
Length = 308
Score = 36.2 bits (82), Expect = 0.88
Identities = 46/229 (20%), Positives = 91/229 (39%), Gaps = 23/229 (10%)
Query: 11 SDDGSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQL-LHDDPIQFVENT 69
SDDG+ ++I L + E+KS+ D ++ RD + ++P +F +
Sbjct: 62 SDDGNKTKNKNDKITLPVNKKEAKSN-DEDFNMINGKTKRDYPNYKNGAKENPNEFNYSE 120
Query: 70 EDDHVGDNSDTTS-------------EKIKAVVVEADEQETLSVSQFFLQLENKFADMKM 116
++D+ DNS+ + KIK ++ QETL+ Q N F++
Sbjct: 121 KEDNHSDNSEDNTASAPDAVSNDWFFSKIKEIIEPNINQETLN--QLITNNYNSFSEANT 178
Query: 117 DSPKSSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRR-KVENEMATMDCDREEVDPTDGF 175
+V+ ++ A + + P+ + + +MD V P D +
Sbjct: 179 GIYDLIDKVIRTNNNNSNAYDNNSQNNISIINKQPQHGNIYDHYNSMD-----VSPNDNY 233
Query: 176 NFWKWSLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTD 224
NF +S+ + S V I ++ + NK+ ++ + +Y D
Sbjct: 234 NFINYSIYCINKELSANVETEAINMMKIYAHTFNKYKKDINYKDDLYKD 282
>UniRef100_UPI00003AD93B UPI00003AD93B UniRef100 entry
Length = 651
Score = 35.4 bits (80), Expect = 1.5
Identities = 31/126 (24%), Positives = 57/126 (44%), Gaps = 5/126 (3%)
Query: 66 VENTEDDHVGDNSDTTSEKIKAVVVE-ADEQETLSVSQFFLQLENKFADMKMDSPKSSPR 124
V + E D++ + D +EKI + A++++T++ + L D D S R
Sbjct: 233 VLDKEKDNLQETVDEKTEKIACLDDNLANKEKTITHLRLTLSELESSTDQLKDLLSSRDR 292
Query: 125 VL--FPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKWSL 182
+ LDA + + G E+ + R++++++ATM RE P N W +
Sbjct: 293 EISSLRRQLDASHTEIAETGRVKEIALKENRRLQDDLATM--TRENQVPRYIINCWHFLT 350
Query: 183 TGVGAI 188
GV A+
Sbjct: 351 DGVVAV 356
>UniRef100_UPI000021B24B UPI000021B24B UniRef100 entry
Length = 1289
Score = 34.7 bits (78), Expect = 2.6
Identities = 42/151 (27%), Positives = 62/151 (40%), Gaps = 24/151 (15%)
Query: 27 GKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHDDPIQFVENTEDDHVGDNSDTTSEKIK 86
G+ P S D M +SP R T + DD + V N + DT+ E +K
Sbjct: 430 GEEAPTSGDDLTMRLG--SSPPERRLEITNISDDDGFEEVINAAEMR----EDTSHEFVK 483
Query: 87 AVVVEADEQETLSVSQFFLQLENKFADMKMDSPKSSPRVLFPPPLDAGALKFEDKGEAME 146
A E + ++L S F + E+ F D + SP+ L+ K E++
Sbjct: 484 A---EEEIPQSLPASSFLDKDEDDFVDEPLSSPR----------LNVSDSKVEEQHFEEP 530
Query: 147 VMV----SPRRKVENEMA-TMDCDREEVDPT 172
MV SPR+ E+A T D E +PT
Sbjct: 531 DMVVQPLSPRKLATAELAKTEPTDSPESNPT 561
>UniRef100_UPI00002F9BED UPI00002F9BED UniRef100 entry
Length = 1196
Score = 34.3 bits (77), Expect = 3.4
Identities = 28/114 (24%), Positives = 49/114 (42%), Gaps = 6/114 (5%)
Query: 67 ENTEDDHVGDNSDTTSEKIKAVV---VEADEQETLSVSQFFLQ---LENKFADMKMDSPK 120
E +ED+ D + S KIK E E++ + L+ L+ AD+K ++ K
Sbjct: 209 EESEDEEEQDGEEAGSFKIKTAAQKKAEKKERDRKKKEEERLKQKKLKEASADVKKEAEK 268
Query: 121 SSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDG 174
+P PP A A++ E+K E + K + + ++EE + G
Sbjct: 269 KTPAAEATPPPPAAAMEEEEKAEPAAEEEADGDKKKKDKKKKKGEKEEKEKKKG 322
>UniRef100_Q9GV77 Embryonic blastocoelar extracellular matrix protein precursor
[Lytechinus variegatus]
Length = 3103
Score = 34.3 bits (77), Expect = 3.4
Identities = 30/155 (19%), Positives = 66/155 (42%), Gaps = 11/155 (7%)
Query: 38 DMNYFYCTSPKSRDSSTTQLLHDDPI------QFVENTEDDHVGDNSDTTSEKIKAVVVE 91
++++F T P+ + T+L D PI Q + ++ D+S+TT + ++ +
Sbjct: 1180 ELHFFIVTPPQHGTITYTRLEGDIPILNFTMDQIANGNDIKYIHDDSETTEDSFTVLLTD 1239
Query: 92 ADEQETLSVSQFFLQLENKFADMK----MDSPKSSPRVLFPPPLDAGALKFEDKGEAMEV 147
+ T ++ L+++++ + +D R++ L A L D V
Sbjct: 1240 GKYEITKEITITILEVDDETPRLTINDGIDIEIGESRIISNRILKATDLDSADSNLTYTV 1299
Query: 148 MVSPRRKVENEMATMDCDREEVDPTDGFNFWKWSL 182
+P + + ++ D E + T G NF +W +
Sbjct: 1300 RYAPEKGLLQRLSKFDGSVVE-NITLGMNFTQWEV 1333
>UniRef100_UPI0000365F7D UPI0000365F7D UniRef100 entry
Length = 579
Score = 33.9 bits (76), Expect = 4.4
Identities = 28/108 (25%), Positives = 47/108 (42%), Gaps = 15/108 (13%)
Query: 46 SPKSRDSSTTQLLHDDPIQFVENTEDDHVGD--NSDTTSEKIKAVVVEADEQETLSVSQF 103
S +SRDSS++ L H E+DHV N +S+ A + A + +
Sbjct: 104 SSRSRDSSSSPLPHP---------EEDHVYSFPNKQKSSDSSSAAMTSALGSNLSELDRL 154
Query: 104 FLQLENKFADMKMDSPKSSPRVLFPPPLDAGALKFEDKGEAMEVMVSP 151
L+L ++ SP PPL + ++ + G ++VMV+P
Sbjct: 155 LLELNA----VQQSSPSFPTTEEAAPPLPSCSITHYENGGGLDVMVTP 198
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 455,242,623
Number of Sequences: 2790947
Number of extensions: 18339632
Number of successful extensions: 51039
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 51008
Number of HSP's gapped (non-prelim): 37
length of query: 264
length of database: 848,049,833
effective HSP length: 125
effective length of query: 139
effective length of database: 499,181,458
effective search space: 69386222662
effective search space used: 69386222662
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0371b.6


