

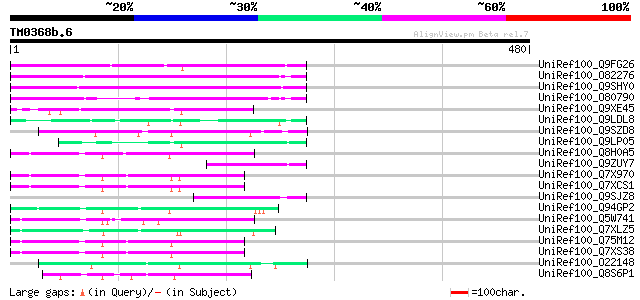
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0368b.6
(480 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like... 140 6e-32
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 123 1e-26
UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana] 104 6e-21
UniRef100_O80790 Putative non-LTR retroelement reverse transcrip... 101 4e-20
UniRef100_Q9XE45 Putative non-LTR retrolelement reverse transcri... 90 2e-16
UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis tha... 89 3e-16
UniRef100_Q9SZD8 Hypothetical protein F19B15.120 [Arabidopsis th... 81 6e-14
UniRef100_Q9LP05 F9C16.13 [Arabidopsis thaliana] 81 6e-14
UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa] 79 4e-13
UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis tha... 77 1e-12
UniRef100_Q7X970 BZIP-like protein [Oryza sativa] 75 4e-12
UniRef100_Q7XCS1 Putative reverse transcriptase [Oryza sativa] 75 5e-12
UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcrip... 74 9e-12
UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa] 73 2e-11
UniRef100_Q5W741 Hypothetical protein OJ1005_D04.5 [Oryza sativa] 72 3e-11
UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa] 70 2e-10
UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa] 68 7e-10
UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa] 68 7e-10
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 67 1e-09
UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa] 66 3e-09
>UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 676
Score = 140 bits (354), Expect = 6e-32
Identities = 88/279 (31%), Positives = 141/279 (49%), Gaps = 9/279 (3%)
Query: 1 LWVHVLRHKYGCGN-NVMSIVSHRASESSIWNGVR-NTWESFSCGIRWQIGDGKVVRF*Q 58
LW VLR KY G+ + + ++ + + S++W V E + GI W +GDGK++RF Q
Sbjct: 184 LWARVLRSKYKIGDIHDSAWMTPKGTWSALWRSVNVGLREVVNRGIGWVLGDGKIIRFWQ 243
Query: 59 DRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
DRWL S LL V+ Q RV D ++ G W+ +P + Q ++ +
Sbjct: 244 DRWLLSTPLLEWVSDQLPVEERGQRVADYWIEGVG-WDMERIAVFLPEFMRQRLLAVVIG 302
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGPDPM---WCLLWRWKGAPCVKMFL 175
W T +G + +SAY L +++++ M + +WR ++FL
Sbjct: 303 GCYGVEDKMSWVGTENGRFTVSSAY--LIQSVDEISKQCMSRFFDRVWRVMVPERARIFL 360
Query: 176 WKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESH 235
W V N ++ N +RV H+ADSD+CPLC SES+IH LRDCP + + + +P +
Sbjct: 361 WLVGNQVVLTNAERVRRHMADSDVCPLCKGASESLIHVLRDCPAMMGIWMRVVPVMEQRR 420
Query: 236 FFSMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
FF L W+ N+++ +R +W FA+ VW W+
Sbjct: 421 FFETSLLEWMYGNLKERSDS-ERRSWPTLFALTVWWGWK 458
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 123 bits (308), Expect = 1e-26
Identities = 81/277 (29%), Positives = 124/277 (44%), Gaps = 9/277 (3%)
Query: 1 LWVHVLRHKYGCGN-NVMSIVSHRASESSIWNGVR-NTWESFSCGIRWQIGDGKVVRF*Q 58
LW V+R KY G S + + SS W V E G+ W GDG +RF
Sbjct: 743 LWARVVRKKYKVGGVQDTSWLKPQPRWSSTWRSVAVGLREVVVKGVGWVPGDGCTIRFWL 802
Query: 59 DRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
DRWL L+ E+ IP G +V ++ W +P V + ++ + +
Sbjct: 803 DRWLLQEPLV-ELGTDMIPEGERIKVAADYWLPGSGWNLEILGLYLPETVKRRLLSVVVQ 861
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTENIEDT-GPDPMWCLLWRWKGAPCVKMFLWK 177
W T G + SAY LL ++ D + +W+ V++F+W
Sbjct: 862 VFLGNGDEISWKGTQDGAFTVRSAYSLLQGDVGDRPNMGSFFNRIWKLITPERVRVFIWL 921
Query: 178 VLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESHFF 237
V N +M NV+RV HL+++ +C +C E+++H LRDCP ++P+ LP FF
Sbjct: 922 VSQNVIMTNVERVRRHLSENAICSVCNGAEETILHVLRDCPAMEPIWRRLLPLRRHHEFF 981
Query: 238 SMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
S L WL NM D ++G W F + +W W+
Sbjct: 982 SQSLLEWLFTNM-DPVKG----IWPTLFGMGIWWAWK 1013
>UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana]
Length = 1055
Score = 104 bits (259), Expect = 6e-21
Identities = 82/277 (29%), Positives = 123/277 (43%), Gaps = 5/277 (1%)
Query: 1 LWVHVLRHKYGCGNNVMSI-VSHRASESSIWNGVR-NTWESFSCGIRWQIGDGKVVRF*Q 58
LW VL+ KY G S + + S SS W + + S G+ W GDG+ +RF
Sbjct: 237 LWTLVLQKKYHVGEIRDSRWLIPKGSWSSTWRSIAIGLRDVVSHGVGWIPGDGQQIRFWT 296
Query: 59 DRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
DRW+ SG L E+ P V + W+ A N E+ +
Sbjct: 297 DRWV-SGKPLLELDNGERPTDCDTVVAKDLWIPGRGWDFAKIDPYTTNNTRLELRAVVLD 355
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLT-ENIEDTGPDPMWCLLWRWKGAPCVKMFLWK 177
++ A W + G S SAYE+LT + + + LW+ + VK FLW
Sbjct: 356 LVTGARDRLSWKFSQDGQFSVRSAYEMLTVDEVPRPNMASFFNCLWKVRVPERVKTFLWL 415
Query: 178 VLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESHFF 237
V N +M +R HL+ S++C +C ES++H LRDCP + + +P + FF
Sbjct: 416 VGNQAVMTEEERHRRHLSASNVCQVCKGGVESMLHVLRDCPAQLGIWVRVVPQRRQQGFF 475
Query: 238 SMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
S L WL N+ D G + W+ FA+ +W W+
Sbjct: 476 SKSLFEWLYDNLGD-RSGCEDIPWSTIFAVIIWWGWK 511
>UniRef100_O80790 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 970
Score = 101 bits (252), Expect = 4e-20
Identities = 83/277 (29%), Positives = 117/277 (41%), Gaps = 52/277 (18%)
Query: 1 LWVHVLRHKYGCGNNVMSI-VSHRASESSIWNGVR-NTWESFSCGIRWQIGDGKVVRF*Q 58
LW VL KY G S + ++S SS W V E + G+RW GDGK + F
Sbjct: 525 LWARVLSKKYKVGEMQNSNWLKPKSSWSSTWRSVTVGLREVVAKGVRWVPGDGKTISFWS 584
Query: 59 DRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
DRWL L A VA + I +L L
Sbjct: 585 DRWLLQEQLSA-VAKESISADAL----------------------------------LSD 609
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGPDPMWCL-LWRWKGAPCVKMFLWK 177
LS W T +G + SAYELL E+ + +W+ V++F+W
Sbjct: 610 ELS-------WKGTQNGDFTVRSAYELLKPEAEERPLIGSFLKQIWKLVAPERVRVFIWL 662
Query: 178 VLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESHFF 237
V + +M NV+RV HL+D C +C ES++H LRDCP + P+ LP ++ FF
Sbjct: 663 VSHMVIMTNVERVRRHLSDIATCSVCNGADESILHVLRDCPAMTPIWQRLLPQRRQNEFF 722
Query: 238 SMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
S WL N+ D +G W F++ +W W+
Sbjct: 723 SQ--FEWLFTNL-DPAKG----DWPTLFSMGIWWAWK 752
>UniRef100_Q9XE45 Putative non-LTR retrolelement reverse transcriptase [Arabidopsis
thaliana]
Length = 732
Score = 89.7 bits (221), Expect = 2e-16
Identities = 64/241 (26%), Positives = 102/241 (41%), Gaps = 30/241 (12%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRN----TWESFSCGIR--------WQI 48
LW ++R C V + W VR+ TW S + G+R W I
Sbjct: 405 LWARIMR----CNYRVQDV------RDGAWTKVRSVCSSTWRSVALGMREVVIPGLSWVI 454
Query: 49 GDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIV 108
GDG+ + F D+WL + I LAE+ VQ P G + + W+ A + +
Sbjct: 455 GDGREILFWMDKWLTN-IPLAELLVQEPPAGWKGMRARDLRRNGIGWDMATIAPYISSYN 513
Query: 109 FQEIMESLPRHLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGPDP----MWCLLWR 164
++ + ++ A WG + G ++ Y LT D P + +W
Sbjct: 514 RLQLQSFVLDDITGARDRISWGESQDGRFKVSTTYSFLTR---DEAPRQDMKRFFDRVWS 570
Query: 165 WKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVR 224
V++FLW V +M N +R HL+ + +C +C ES+IH LRDCP ++ +
Sbjct: 571 VTAPERVRLFLWLVAQQAIMTNQERHRRHLSTTTICQVCKGAKESIIHVLRDCPAMEGIW 630
Query: 225 L 225
L
Sbjct: 631 L 631
>UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis thaliana]
Length = 947
Score = 89.0 bits (219), Expect = 3e-16
Identities = 74/281 (26%), Positives = 113/281 (39%), Gaps = 53/281 (18%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
LW +LR KY G E S G RW +G+G+ + F D
Sbjct: 557 LWARILRSKYRVGLR----------------------EVVSRGSRWVVGNGRDILFWSDN 594
Query: 61 WLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPRHL 120
WL+ L+ ++ IP V + + W+ ++ P I + +E +
Sbjct: 595 WLSHEALINRAVIE-IPNSEKELRVKDLWANGLGWKLD---KIEPYISYHTRLELAAVVV 650
Query: 121 SSAPGLP---VWGRTTSGINSTASAYELLTENIEDTGPD--PMWCLLWRWKGAPCVKMFL 175
S G WG + G+ + SAY LLTE+ D P+ + LWR VK FL
Sbjct: 651 DSVTGARDRLSWGYSADGVFTVKSAYRLLTED-HDPRPNMAAFFDRLWRVVALERVKTFL 709
Query: 176 WKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESH 235
W H+ D+ +C +C E+++H L+DCP + + +
Sbjct: 710 W----------------HIGDTSVCQVCKGGDETILHVLKDCPSIAGIWRRLVQVQRSYD 753
Query: 236 FFSMELQAWLLRN--MRDGLQGWDRDAWTKRFAINVWCLWR 274
FF+ L WL N M++ G+ AW FAI VW W+
Sbjct: 754 FFNGSLFGWLYVNLGMKNAETGY---AWATLFAIVVWWSWK 791
>UniRef100_Q9SZD8 Hypothetical protein F19B15.120 [Arabidopsis thaliana]
Length = 575
Score = 81.3 bits (199), Expect = 6e-14
Identities = 68/270 (25%), Positives = 115/270 (42%), Gaps = 31/270 (11%)
Query: 27 SSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPP------GS 80
S +W + + E G R +G+G+ + + +WL S A + +Q +PP S
Sbjct: 108 SFVWKSIHASQEILRQGARAVVGNGEDIIIWRHKWLDSKPASAALRMQRVPPQEYASVSS 167
Query: 81 LNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLP---RHLSSAPGLPVWGRTTSGIN 137
+ +V D S W L P + + I E P R L S W T+SG
Sbjct: 168 ILKVSDLIDESGREWRKDVIEMLFPEVERKLIGELRPGGRRILDSY----TWDYTSSGDY 223
Query: 138 STASAYELLTE---------NIEDTGPDPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVK 188
+ S Y +LT+ + + +P++ +W+ + +P ++ FLWK L+N L V
Sbjct: 224 TVKSGYWVLTQIINKRSSPQEVSEPSLNPIYQKIWKSQTSPKIQHFLWKCLSNSLPVAGA 283
Query: 189 RVHNHLADSDLCPLCGTLSESVIHALRDCPMVK---PVRLANLPTGGESHFFSMELQAWL 245
+ HL+ C C + E+V H L C + + +P GGE S+ + +
Sbjct: 284 LAYRHLSKESACIRCPSCKETVNHLLFKCTFARLTWAISSIPIPLGGE-WADSIYVNLYW 342
Query: 246 LRNMRDGLQGWDRDAWTKRFAINVWCLWRM 275
+ N+ +G W K + W LWR+
Sbjct: 343 VFNLGNG-----NPQWEKASQLVPWLLWRL 367
>UniRef100_Q9LP05 F9C16.13 [Arabidopsis thaliana]
Length = 1172
Score = 81.3 bits (199), Expect = 6e-14
Identities = 59/233 (25%), Positives = 93/233 (39%), Gaps = 52/233 (22%)
Query: 46 WQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMP 105
W +GDG+ +RF +D+WL + + SL + D G+
Sbjct: 3 WVVGDGRKIRFWKDKWLFNKTV------------SLLAIEDLHAGARDCM---------- 40
Query: 106 NIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGPDP----MWCL 161
WG T G S SA+ L+T D P P ++
Sbjct: 41 ----------------------AWGETVDGEFSVRSAHALVTR---DLSPQPNVSALFSR 75
Query: 162 LWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVK 221
+W KMFL V N LM +V+R H +++C +C SE+++H L+DCP +
Sbjct: 76 IWGVVATERAKMFLCLVGNQALMTDVERFWRHRCITEVCQMCKGGSETILHMLQDCPAMA 135
Query: 222 PVRLANLPTGGESHFFSMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
+ +P FF L WL N+ D + ++ W+ F + VW W+
Sbjct: 136 GLWNQIIPRRRRRLFFEQSLIEWLYSNLGDDQETYE-SVWSTVFTMAVWWAWK 187
>UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa]
Length = 1557
Score = 78.6 bits (192), Expect = 4e-13
Identities = 66/237 (27%), Positives = 101/237 (41%), Gaps = 19/237 (8%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L VL+ KY +++ S + S W + + E GI W+IG+G+ VR QD
Sbjct: 1249 LCARVLKAKYYPNGSIVD-TSFGGNASPGWQAIEHGLELVKKGIIWRIGNGRSVRVWQDP 1307
Query: 61 WLASGILLAEVAVQPIPPGSLNRV--VDEFLGSNGTWETAGFVEL-MPNIVFQEIMESLP 117
WL +++ +PI P + R+ V + + NG W+ ++ +P V EI+ L
Sbjct: 1308 WLPR-----DLSRRPITPKNNCRIKWVADLMLDNGMWDANKINQIFLP--VDVEIILKLR 1360
Query: 118 RHLSSAPGLPVWGRTTSGINSTASAYELL--------TENIEDTGPDPMWCLLWRWKGAP 169
W G S +AY L + + D W LLW+
Sbjct: 1361 TSSRDEEDFIAWHPDKLGNFSVRTAYRLAENWAKEEASSSSSDVNIRKAWELLWKCNVPS 1420
Query: 170 CVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLA 226
VK+F W+ +N L + +L SD C +CG E +HAL CP K + LA
Sbjct: 1421 KVKIFTWRATSNCLPTWDNKKKRNLEISDTCVICGMEKEDTMHALCRCPQAKHLWLA 1477
>UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 314
Score = 76.6 bits (187), Expect = 1e-12
Identities = 32/92 (34%), Positives = 53/92 (56%), Gaps = 1/92 (1%)
Query: 183 LMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESHFFSMELQ 242
LM N +R HL+DSD+C +C +++IH LRDCP ++ + + +P G FF+ L
Sbjct: 5 LMTNAERRRRHLSDSDICQICKGAEKTIIHILRDCPAMEGIWIRLVPAGKRREFFTQSLL 64
Query: 243 AWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
WL N+ D + + W+ FA+++W W+
Sbjct: 65 EWLFANLGDRRKTCE-STWSTLFALSIWWAWK 95
>UniRef100_Q7X970 BZIP-like protein [Oryza sativa]
Length = 2367
Score = 75.1 bits (183), Expect = 4e-12
Identities = 57/227 (25%), Positives = 102/227 (44%), Gaps = 17/227 (7%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L VL+ KY +++ S ++ S W + + GI W++G+G +R +D
Sbjct: 1614 LCARVLKAKYFPHGSLID-TSFGSNSSPAWRSIEYGLDLLKKGIIWRVGNGNSIRIWRDS 1672
Query: 61 WLASGILLAEVAVQPIPPGSLNRV--VDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
WL + + +PI + R+ V + + +G+W+ + N+ E++ ++
Sbjct: 1673 WLPR-----DHSRRPITGKANCRLKWVSDLITEDGSWDVPKIHQYFHNLD-AEVILNICI 1726
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTE--NIEDTGP------DPMWCLLWRWKGAPC 170
S W +G+ S SAY L + NIE++ + W ++W+ K
Sbjct: 1727 SSRSEEDFIAWHPDKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKAWEMIWKCKVPQK 1786
Query: 171 VKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDC 217
VK+F W+V +N L V + L SD+C +C +E HAL C
Sbjct: 1787 VKIFAWRVASNCLATMVNKKKRKLEQSDMCQICDRENEDDAHALCRC 1833
>UniRef100_Q7XCS1 Putative reverse transcriptase [Oryza sativa]
Length = 791
Score = 74.7 bits (182), Expect = 5e-12
Identities = 57/227 (25%), Positives = 102/227 (44%), Gaps = 17/227 (7%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L VL+ KY +++ S ++ S W + + GI W++G+G +R +D
Sbjct: 290 LCARVLKAKYFPHGSLID-TSFGSNSSPAWRSIEYGLDLLKKGIIWRVGNGNSIRIWRDP 348
Query: 61 WLASGILLAEVAVQPIPPGSLNRV--VDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
WL + + +PI + R+ V + + +G+W+ + N+ E++ ++
Sbjct: 349 WLPR-----DHSRRPITGKANCRLKWVSDLITEDGSWDVPKIHQYFHNLD-AEVILNICI 402
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTE--NIEDTGP------DPMWCLLWRWKGAPC 170
S W +G+ S SAY L + NIE++ + W ++W+ K
Sbjct: 403 SSRSEEDFIAWHPDKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKAWEMIWKCKVPQK 462
Query: 171 VKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDC 217
VK+F W+V +N L V + L SD+C +C +E HAL C
Sbjct: 463 VKIFAWRVASNCLATMVNKKKRKLEQSDMCQICDRENEDDAHALCRC 509
>UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 321
Score = 73.9 bits (180), Expect = 9e-12
Identities = 33/104 (31%), Positives = 57/104 (54%), Gaps = 6/104 (5%)
Query: 171 VKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPT 230
V++FLW V+ ++ NV+R HL+D+ +C +C E+++H LRDCP + + +P
Sbjct: 6 VRVFLWLVVQQVIITNVERYRRHLSDTRVCQICQGGEETILHVLRDCPAMAGIWSRLVPR 65
Query: 231 GGESHFFSMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
FF+ L W+ +N+R +R +W F + VW W+
Sbjct: 66 DQIRQFFTASLLEWIYKNLR------ERGSWPTVFVMAVWWGWK 103
>UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa]
Length = 1833
Score = 73.2 bits (178), Expect = 2e-11
Identities = 66/289 (22%), Positives = 116/289 (39%), Gaps = 49/289 (16%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L V + KY N++ S SS+W G+++ + G W++G+G ++ + +
Sbjct: 792 LCARVFKAKYYPNGNLVDTAFPTVS-SSVWKGIQHGLDLLKQGTIWRVGNGHNIKIWRHK 850
Query: 61 WLASGILLAEVAVQPIPPGSLNRV--VDEFLGS-NGTWETAGFVELMPNIVFQEIME-SL 116
W+ G + + NR+ V+E + N W ++ E+++ L
Sbjct: 851 WVPHG-----EHINVLEKKGRNRLIYVNELINEENRCWNETLVRHVLKEEDANEVLKIRL 905
Query: 117 PRHLSSAPGLPVWGRTTSGINSTASAYELL-----------TENIEDTGPDPMWCLLWRW 165
P H P W SG+ + SAY+L + + +G +W +W
Sbjct: 906 PNH--QMDDFPAWHHEKSGLFTVKSAYKLAWNLSGKGVVQSSSSTATSGERKIWSRVWNA 963
Query: 166 KGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRL 225
K VK+F+WK+ + L + + + CP+CGT E+ HA +C + +R
Sbjct: 964 KVQAKVKIFIWKLAQDKLPTWENKRRRKIEMNGTCPVCGTKGENSYHATVECTKARALRE 1023
Query: 226 A-----NLP-------TGGE--------------SHFFSMELQAWLLRN 248
A +LP TG + +H M +AW LRN
Sbjct: 1024 ALRAVWHLPGEDKFLWTGPDWLLILLDGVNEEQRTHIMYMLWRAWYLRN 1072
>UniRef100_Q5W741 Hypothetical protein OJ1005_D04.5 [Oryza sativa]
Length = 697
Score = 72.4 bits (176), Expect = 3e-11
Identities = 60/239 (25%), Positives = 107/239 (44%), Gaps = 24/239 (10%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L +L+ KY N ++ + ++ S W G+ + E G+ W+IGD + + ++R
Sbjct: 314 LCARLLKAKY-YPNGSLTDTAFPSNTSPTWKGIEHGLELLKKGLIWRIGDRRTTKIWRNR 372
Query: 61 WLASGILLAEVAVQPIPPGSLNRV--VDEFL--GSNGTWETAGFVELMPNIVFQEIMESL 116
W+A G V+ I + NR+ V E L G+N W A L+ ++ +E ++
Sbjct: 373 WVAHG-----EKVEVIQKKNWNRLTYVHELLIPGTNA-WNEA----LIRHVATEEDANAI 422
Query: 117 PR-HLSS--APGLPVWGRTTSGI------NSTASAYELLTENIEDTGPDPMWCLLWRWKG 167
+ H+ + P W +G+ N T SA + G +W +W+
Sbjct: 423 LKIHVPNQETSHFPAWHYEKTGLFSGVAWNLTKSAQVQAASSTATNGERKLWENVWKADV 482
Query: 168 APCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLA 226
P VK+F WK+ ++ L + + +CP+CG E+ HA +C M + +R A
Sbjct: 483 QPKVKIFAWKLAHDRLPTWENKRKRRIQPVGICPICGVKEENGFHATVECTMARSLREA 541
>UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa]
Length = 1205
Score = 69.7 bits (169), Expect = 2e-10
Identities = 65/263 (24%), Positives = 106/263 (39%), Gaps = 25/263 (9%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L VL+ KY N ++ + + S W G+ + E G+ W+IGDG + ++
Sbjct: 868 LCARVLKAKY-YPNGTITDTAFPSVSSPTWKGIVHGLELLKKGLIWRIGDGSKTKIWRNH 926
Query: 61 WLASGILLAEVAVQPIPPGSLNRVV---DEFLGSNGTWETAGFVELMPNIVFQEIME-SL 116
W+A G L + + + NRV+ + + TW ++ EI++ +
Sbjct: 927 WVAHGENL-----KILEKKTWNRVIYVRELIVTDTKTWNEPLIRHIIREEDADEILKIRI 981
Query: 117 PRHLSSAPGLPVWGRTTSGINSTASAYELLTENIEDT---------GPD--PMWCLLWRW 165
P+ P W +GI S S Y L T G D +W +W+
Sbjct: 982 PQR--EEEDFPAWHYEKTGIFSVRSVYRLAWNLARKTSEQASSSSGGADGRKIWDNVWKA 1039
Query: 166 KGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVR- 224
P V++F WK+ + L + + CP+CG E+ HA +C + K +R
Sbjct: 1040 NVQPKVRVFAWKLAQDRLATWENKKKRKIEMFGTCPICGQKEETGFHATVECTLAKALRA 1099
Query: 225 -LANLPTGGESHFFSMELQAWLL 246
L T + FSM WLL
Sbjct: 1100 SLREHWTLPDESLFSMTGPDWLL 1122
>UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa]
Length = 1936
Score = 67.8 bits (164), Expect = 7e-10
Identities = 53/227 (23%), Positives = 99/227 (43%), Gaps = 17/227 (7%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L VL+ KY + ++ + A+ S W+G+ + GI W+IG+G VR +D
Sbjct: 1520 LCAQVLKAKY-FPHGSLTDTTFSANASPTWHGIEYGLDLLKKGIIWRIGNGNSVRIWRDP 1578
Query: 61 WLASGILLAEVAVQPIPPGSLNRV--VDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
W+ +++ +P+ + R+ V + + +GTW++A + I +I++ +
Sbjct: 1579 WIPR-----DLSRRPVSSKANCRLKWVSDLIAEDGTWDSAKINQYFLKID-ADIIQKICI 1632
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTE--------NIEDTGPDPMWCLLWRWKGAPC 170
W +G S SAY+L + + + + W L+W+
Sbjct: 1633 SARLEEDFIAWHPDKTGRFSVRSAYKLALQLADMNNCSSSSSSRLNKSWELIWKCNVPQK 1692
Query: 171 VKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDC 217
V++F W+V +N L + +L D+C +C E HAL C
Sbjct: 1693 VRIFAWRVASNSLATMENKKKRNLERFDVCGICDREKEDAGHALCRC 1739
>UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa]
Length = 1711
Score = 67.8 bits (164), Expect = 7e-10
Identities = 53/227 (23%), Positives = 99/227 (43%), Gaps = 17/227 (7%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L VL+ KY + ++ + A+ S W+G+ + GI W+IG+G VR +D
Sbjct: 1364 LCAQVLKAKY-FPHGSLTDTTFSANASPTWHGIEYGLDLLKKGIIWRIGNGNSVRIWRDP 1422
Query: 61 WLASGILLAEVAVQPIPPGSLNRV--VDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
W+ +++ +P+ + R+ V + + +GTW++A + I +I++ +
Sbjct: 1423 WIPK-----DLSRRPVSSKANCRLKWVSDLIAEDGTWDSAKINQYFLKID-ADIIQKICI 1476
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTE--------NIEDTGPDPMWCLLWRWKGAPC 170
W +G S SAY+L + + + + W L+W+
Sbjct: 1477 SARLEEDFIAWHPDKTGRFSVRSAYKLALQLADMNNCSSSSSSRLNKSWELIWKCNVPQK 1536
Query: 171 VKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDC 217
V++F W+V +N L + +L D+C +C E HAL C
Sbjct: 1537 VRIFAWRVASNSLATMENKKKRNLERFDVCGICDREKEDAGHALCCC 1583
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 67.0 bits (162), Expect = 1e-09
Identities = 68/271 (25%), Positives = 100/271 (36%), Gaps = 33/271 (12%)
Query: 27 SSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQ------PIPPGS 80
S W + GIR IG+G+ + D W+ + A AV+ S
Sbjct: 905 SFAWKSIYEAQVLIKQGIRAVIGNGETINVWTDPWIGAKPAKAAQAVKRSHLVSQYAANS 964
Query: 81 LNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTA 140
++ V D L W L P+ + I+ P + W + SG S
Sbjct: 965 IHVVKDLLLPDGRDWNWNLVSLLFPDNTQENILALRPGGKETRDRF-TWEYSRSGHYSVK 1023
Query: 141 SAYELLTENIEDTGP---------DPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVH 191
S Y ++TE I DP++ +W+ P + FLW+ +NN L V +
Sbjct: 1024 SGYWVMTEIINQRNNPQEVLQPSLDPIFQQIWKLDVPPKIHHFLWRCVNNCLSVASNLAY 1083
Query: 192 NHLADSDLCPLCGTLSESVIHALRDCPMVK---PVRLANLPTGGESHFFSMELQAW---L 245
HLA C C + E+V H L CP + + P GGE W L
Sbjct: 1084 RHLAREKSCVRCPSHGETVNHLLFKCPFARLTWAISPLPAPPGGE----------WAESL 1133
Query: 246 LRNMRDGLQ-GWDRDAWTKRFAINVWCLWRM 275
RNM L + + A+ W LWR+
Sbjct: 1134 FRNMHHVLSVHKSQPEESDHHALIPWILWRL 1164
>UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa]
Length = 1509
Score = 65.9 bits (159), Expect = 3e-09
Identities = 55/219 (25%), Positives = 89/219 (40%), Gaps = 36/219 (16%)
Query: 31 NGVRNTWESFSCGIR-------WQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNR 83
+ V TW S G+R W++GDG + D W+ G + +P+ P N
Sbjct: 1103 SNVSYTWRSIQKGLRVLQNGMIWRVGDGSKINIWADPWIPRGW-----SRKPMTPRGANL 1157
Query: 84 V--VDEFLGS-NGTWETAGFVELMPNIVFQE---IMESLPRHLSSAPGLPVWGRTTSGIN 137
V V+E + GTW+ +L+ ++E ++S+P H+ L W G
Sbjct: 1158 VTKVEELIDPYTGTWDE----DLLSQTFWEEDVAAIKSIPVHVEMEDVL-AWHFDARGCF 1212
Query: 138 STASAYELLTENIE-------------DTGPDPMWCLLWRWKGAPCVKMFLWKVLNNGLM 184
+ SAY++ E ++G D W LW+ +K FLW++ +N L
Sbjct: 1213 TVKSAYKVQREMERRASRNGCPGVSNWESGDDDFWKKLWKLGVPGKIKHFLWRMCHNTLA 1272
Query: 185 VNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPV 223
+ H + C +CG +E H C VK V
Sbjct: 1273 LRANLHHRGMDVDTRCVMCGRYNEDAGHLFFKCKPVKKV 1311
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.334 0.143 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 790,746,571
Number of Sequences: 2790947
Number of extensions: 31093623
Number of successful extensions: 102188
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 102009
Number of HSP's gapped (non-prelim): 138
length of query: 480
length of database: 848,049,833
effective HSP length: 131
effective length of query: 349
effective length of database: 482,435,776
effective search space: 168370085824
effective search space used: 168370085824
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0368b.6


