

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
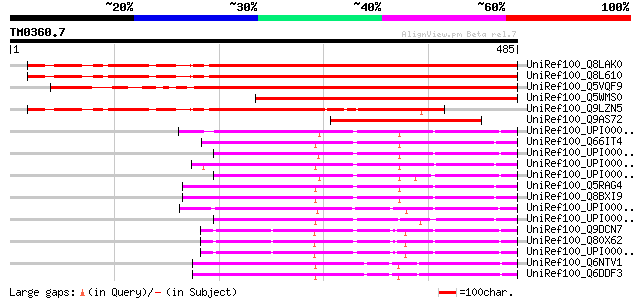
Query= TM0360.7
(485 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LAK0 Hypothetical protein [Arabidopsis thaliana] 536 e-151
UniRef100_Q8L610 Hypothetical protein At5g01960 [Arabidopsis tha... 536 e-151
UniRef100_Q5VQF9 Hypothetical protein OJ1276_B06.18 [Oryza sativa] 519 e-146
UniRef100_Q5WMS0 Hypothetical protein OJ1333_C12.12 [Oryza sativa] 421 e-116
UniRef100_Q9LZN5 Hypothetical protein T7H20_10 [Arabidopsis thal... 327 6e-88
UniRef100_Q9AS72 P0028E10.20 protein [Oryza sativa] 242 2e-62
UniRef100_UPI000024A180 UPI000024A180 UniRef100 entry 163 9e-39
UniRef100_Q66IT4 LOC446942 protein [Xenopus laevis] 157 5e-37
UniRef100_UPI0000365016 UPI0000365016 UniRef100 entry 157 8e-37
UniRef100_UPI00003AB1CD UPI00003AB1CD UniRef100 entry 154 4e-36
UniRef100_UPI0000438DA1 UPI0000438DA1 UniRef100 entry 154 5e-36
UniRef100_Q5RAG4 Hypothetical protein DKFZp459B2128 [Pongo pygma... 150 6e-35
UniRef100_Q8BXI9 Mus musculus 10 days neonate medulla oblongata ... 150 8e-35
UniRef100_UPI00003AB70F UPI00003AB70F UniRef100 entry 149 2e-34
UniRef100_UPI00003AB1CE UPI00003AB1CE UniRef100 entry 148 3e-34
UniRef100_Q9DCN7 Mus musculus adult male kidney cDNA, RIKEN full... 147 9e-34
UniRef100_Q80X62 0610013E23Rik protein [Mus musculus] 145 3e-33
UniRef100_UPI00001D0574 UPI00001D0574 UniRef100 entry 145 3e-33
UniRef100_Q6NTV1 MGC81516 protein [Xenopus laevis] 142 3e-32
UniRef100_Q6DDF3 MGC81516 protein [Xenopus laevis] 142 3e-32
>UniRef100_Q8LAK0 Hypothetical protein [Arabidopsis thaliana]
Length = 426
Score = 536 bits (1382), Expect = e-151
Identities = 281/470 (59%), Positives = 336/470 (70%), Gaps = 48/470 (10%)
Query: 18 SPSSSSGGFNSSDDHNNGSVSWYGMRLPSVNPLRSPLSLLLDYSGILTQTHDSGSVIVNN 77
+P +S F S N+ L + N +++P+S LL+YSG+
Sbjct: 3 TPGASHDSFRGSPRRNS--------ILSASNIIQAPISTLLEYSGLFRSRPS-------- 46
Query: 78 GVPASEIRSQVQMQPFDDAVGGSAGEVAIRIIGAGEHD-QNQIGEVGFDGCDNLMGERRG 136
P+ E + V DD+ G S GEVAIRIIG E D + + G L+G
Sbjct: 47 --PSHEAETLVS----DDSSGLSNGEVAIRIIGNTEQDAETDTNALREPGHSELLGS--- 97
Query: 137 AGMSPLDDDPEGRGGAAVNETVPLVSSSASLAGSVQGDGDAAGNGAESDSRGSSSYQRYD 196
S DP G ++ P GD AAG+ A DS YQRYD
Sbjct: 98 ---SATQVDPMGGASEGASQAAP-------------GD-PAAGDAASRDS----PYQRYD 136
Query: 197 IQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKVS 256
IQQ A+WIEQILPFSLLLL+VFIRQHLQGFFV IWI+AVMFKSN+I+KKQTALKG+R +S
Sbjct: 137 IQQAARWIEQILPFSLLLLVVFIRQHLQGFFVAIWIAAVMFKSNDILKKQTALKGERHIS 196
Query: 257 VLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPRPTP-FWHSVFIILVNDVLVRQAAMV 315
L+GIS AF V +YWW+R DDLLYPLIMLPP+ P FWH++FII+VND LVRQA+M+
Sbjct: 197 ALIGISVAFTAHVVGVYWWFRKDDLLYPLIMLPPKSIPPFWHAIFIIVVNDTLVRQASMI 256
Query: 316 FKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRDYGSLFSSLT 375
FKCFLL+YYKN +G N+R+Q Q+LTLVEY +LLYR+LLPT VWYRFFLN+DYGSLFSSL
Sbjct: 257 FKCFLLMYYKNSRGRNYRKQGQLLTLVEYFMLLYRSLLPTPVWYRFFLNKDYGSLFSSLM 316
Query: 376 TGLYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPV 435
TGLYLTFKLTS++EKV+ F +A KALSRK+VHYG YAT EQVNAAGDLCAICQEKMH+P+
Sbjct: 317 TGLYLTFKLTSVVEKVQSFFTALKALSRKEVHYGSYATTEQVNAAGDLCAICQEKMHTPI 376
Query: 436 LLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTSLFFQLF 485
LLRCKH+FCE+CVSEWFERERTCPLCRALVKPADL++FGDGSTSLFFQ+F
Sbjct: 377 LLRCKHMFCEDCVSEWFERERTCPLCRALVKPADLKSFGDGSTSLFFQIF 426
>UniRef100_Q8L610 Hypothetical protein At5g01960 [Arabidopsis thaliana]
Length = 426
Score = 536 bits (1381), Expect = e-151
Identities = 281/470 (59%), Positives = 336/470 (70%), Gaps = 48/470 (10%)
Query: 18 SPSSSSGGFNSSDDHNNGSVSWYGMRLPSVNPLRSPLSLLLDYSGILTQTHDSGSVIVNN 77
+P +S F S N+ L + N +++P+S LL+YSG+
Sbjct: 3 TPGASHDSFRGSPRRNS--------ILSASNIIQAPISTLLEYSGLFRARPS-------- 46
Query: 78 GVPASEIRSQVQMQPFDDAVGGSAGEVAIRIIGAGEHD-QNQIGEVGFDGCDNLMGERRG 136
P+ E + V DD+ G S GEVAIRIIG E D + + G L+G
Sbjct: 47 --PSHEAETLVS----DDSSGLSNGEVAIRIIGNTEQDAETDTNALREPGHSELLGS--- 97
Query: 137 AGMSPLDDDPEGRGGAAVNETVPLVSSSASLAGSVQGDGDAAGNGAESDSRGSSSYQRYD 196
S DP G ++ P GD AAG+ A DS YQRYD
Sbjct: 98 ---SATQVDPMGGASEGASQAAP-------------GD-PAAGDAASRDS----PYQRYD 136
Query: 197 IQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKVS 256
IQQ A+WIEQILPFSLLLL+VFIRQHLQGFFV IWI+AVMFKSN+I+KKQTALKG+R +S
Sbjct: 137 IQQAARWIEQILPFSLLLLVVFIRQHLQGFFVAIWIAAVMFKSNDILKKQTALKGERHIS 196
Query: 257 VLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPRPTP-FWHSVFIILVNDVLVRQAAMV 315
L+GIS AF V +YWW+R DDLLYPLIMLPP+ P FWH++FII+VND LVRQA+M+
Sbjct: 197 ALIGISVAFTAHVVGVYWWFRKDDLLYPLIMLPPKSIPPFWHAIFIIVVNDTLVRQASMI 256
Query: 316 FKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRDYGSLFSSLT 375
FKCFLL+YYKN +G N+R+Q Q+LTLVEY +LLYR+LLPT VWYRFFLN+DYGSLFSSL
Sbjct: 257 FKCFLLMYYKNSRGRNYRKQGQLLTLVEYFMLLYRSLLPTPVWYRFFLNKDYGSLFSSLM 316
Query: 376 TGLYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPV 435
TGLYLTFKLTS++EKV+ F +A KALSRK+VHYG YAT EQVNAAGDLCAICQEKMH+P+
Sbjct: 317 TGLYLTFKLTSVVEKVQSFFTALKALSRKEVHYGSYATTEQVNAAGDLCAICQEKMHTPI 376
Query: 436 LLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTSLFFQLF 485
LLRCKH+FCE+CVSEWFERERTCPLCRALVKPADL++FGDGSTSLFFQ+F
Sbjct: 377 LLRCKHMFCEDCVSEWFERERTCPLCRALVKPADLKSFGDGSTSLFFQIF 426
>UniRef100_Q5VQF9 Hypothetical protein OJ1276_B06.18 [Oryza sativa]
Length = 427
Score = 519 bits (1337), Expect = e-146
Identities = 268/447 (59%), Positives = 322/447 (71%), Gaps = 49/447 (10%)
Query: 40 YGMRLPSVNPLRSPLSLLLDYSGILTQTHDSGSVIVNNGVPASEIRSQVQMQPFDDAVGG 99
YG+ + + +++PLS LL+YSGIL G V G GG
Sbjct: 29 YGVHFSASSFIQAPLSALLEYSGILRADPGGGPHQVGGGG------------------GG 70
Query: 100 SAGEVAIRIIGAGEHDQNQIGEVGFDGCDNLMGERRGAGMSPLDDDPEGRGGAAVNETVP 159
GEV+IRI+G+GE ER G+ E GAA P
Sbjct: 71 GGGEVSIRIVGSGE-------------AAGAASERGEEGVV------EDEAGAA-----P 106
Query: 160 LVSSSASLAGSVQGDGDAAGNGAESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFI 219
+ S S A + A G E+ SSSYQRYDIQQVA+W+EQILPFSLLLL+VFI
Sbjct: 107 QANPSTSAAAA------ATAGGGEAGRESSSSYQRYDIQQVARWVEQILPFSLLLLVVFI 160
Query: 220 RQHLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRND 279
RQHLQGFFVTIWI+AVMFKSN+I++KQTALKG+RK+SVLVGI+ F++ V +YW Y+N
Sbjct: 161 RQHLQGFFVTIWIAAVMFKSNDILRKQTALKGERKMSVLVGITILFVVHVFGVYWCYKNG 220
Query: 280 DLLYPLIMLPPRPTP-FWHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQM 338
DL+ PL+ L P+ P FWH++FIILVND +VRQ AM+ KC LL+YYKN KG ++RRQ QM
Sbjct: 221 DLVRPLVALAPKEIPPFWHAIFIILVNDTMVRQTAMIIKCMLLMYYKNSKGRSYRRQGQM 280
Query: 339 LTLVEYMLLLYRALLPTLVWYRFFLNRDYGSLFSSLTTGLYLTFKLTSIIEKVRGFISAF 398
LT+VEY LLLYRALLP VWYRFFLN++YGSLFSSLTTGLYLTFKLTS++EKV+ F++A
Sbjct: 281 LTVVEYFLLLYRALLPAPVWYRFFLNKEYGSLFSSLTTGLYLTFKLTSVVEKVQSFLTAL 340
Query: 399 KALSRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTC 458
+ALS KD HYG YAT EQV+A GD+CAICQEKMH+P+LLRCKHIFCE+CVSEWFERERTC
Sbjct: 341 RALSHKDFHYGSYATSEQVSATGDMCAICQEKMHTPILLRCKHIFCEDCVSEWFERERTC 400
Query: 459 PLCRALVKPADLQTFGDGSTSLFFQLF 485
PLCRALVKPADL++FGDGSTSLFFQLF
Sbjct: 401 PLCRALVKPADLRSFGDGSTSLFFQLF 427
>UniRef100_Q5WMS0 Hypothetical protein OJ1333_C12.12 [Oryza sativa]
Length = 251
Score = 421 bits (1081), Expect = e-116
Identities = 195/251 (77%), Positives = 227/251 (89%), Gaps = 1/251 (0%)
Query: 236 MFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPRPTP- 294
MFKSN+I++KQTALKG+RK++VLVGI+ F++ V +YWWYRNDDLL PL MLPP+ P
Sbjct: 1 MFKSNDILRKQTALKGERKIAVLVGITVIFMIHVFGVYWWYRNDDLLRPLFMLPPKEIPP 60
Query: 295 FWHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLP 354
FWH++FII+VND +VRQAAM KC LL+YYKN +G N+R+Q QMLTLVEY+LLLYRALLP
Sbjct: 61 FWHAIFIIMVNDTMVRQAAMAIKCMLLMYYKNSRGRNYRKQGQMLTLVEYLLLLYRALLP 120
Query: 355 TLVWYRFFLNRDYGSLFSSLTTGLYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVYATM 414
T VWYRFFLN++YGSLFSSLTTGLYLTFKLTS++EKV+ F++A KALSRKDVHYG YAT
Sbjct: 121 TPVWYRFFLNKEYGSLFSSLTTGLYLTFKLTSVVEKVQSFLAAVKALSRKDVHYGSYATA 180
Query: 415 EQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFG 474
EQV AAGD+CAICQEKMH PVLLRCKHIFCE+CVSEWFERERTCPLCRALVKPAD+++FG
Sbjct: 181 EQVIAAGDMCAICQEKMHVPVLLRCKHIFCEDCVSEWFERERTCPLCRALVKPADIRSFG 240
Query: 475 DGSTSLFFQLF 485
DGSTSLFFQLF
Sbjct: 241 DGSTSLFFQLF 251
>UniRef100_Q9LZN5 Hypothetical protein T7H20_10 [Arabidopsis thaliana]
Length = 413
Score = 327 bits (837), Expect = 6e-88
Identities = 205/410 (50%), Positives = 247/410 (60%), Gaps = 61/410 (14%)
Query: 18 SPSSSSGGFNSSDDHNNGSVSWYGMRLPSVNPLRSPLSLLLDYSGILTQTHDSGSVIVNN 77
+P +S F S N+ L + N +++P+S LL+YSG+
Sbjct: 3 TPGASHDSFRGSPRRNS--------ILSASNIIQAPISTLLEYSGLFRARPS-------- 46
Query: 78 GVPASEIRSQVQMQPFDDAVGGSAGEVAIRIIGAGEHD-QNQIGEVGFDGCDNLMGERRG 136
P+ E + V DD+ G S GEVAIRIIG E D + + G L+G
Sbjct: 47 --PSHEAETLVS----DDSSGLSNGEVAIRIIGNTEQDAETDTNALREPGHSELLGS--- 97
Query: 137 AGMSPLDDDPEGRGGAAVNETVPLVSSSASLAGSVQGDGDAAGNGAESDSRGSSSYQRYD 196
S DP G ++ P GD AAG+ A DS YQRYD
Sbjct: 98 ---SATQVDPMGGASEGASQAAP-------------GD-PAAGDAASRDS----PYQRYD 136
Query: 197 IQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKVS 256
IQQ A+WIEQILPFSLLLL+VFIRQHLQGFFV IWI+AVMFKSN+I+KKQTALKG+R +S
Sbjct: 137 IQQAARWIEQILPFSLLLLVVFIRQHLQGFFVAIWIAAVMFKSNDILKKQTALKGERHIS 196
Query: 257 VLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPRP-TPFWHSVFIILVNDVLVRQAAMV 315
L+GIS AF V +YWW+R DDLLYPLIMLPP+ PFWH++FII V V A +
Sbjct: 197 ALIGISVAFTAHVVGVYWWFRKDDLLYPLIMLPPKSIPPFWHAIFII-VYPVGSFGAYSL 255
Query: 316 FKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRDYGSLFSSLT 375
FL Y G N Q Q+LTLVEY +LLYR+LLPT VWYRFFLN+DYGSLFSSL
Sbjct: 256 VPIFL--RYTCPAGLN-DFQGQLLTLVEYFMLLYRSLLPTPVWYRFFLNKDYGSLFSSLM 312
Query: 376 TGLYLTFKLTSIIEKVR---------GFISAFKALSRKDVHYGVYATMEQ 416
TGLYLTFKLTS++EK+R F +A KALSRK+VHYG YAT EQ
Sbjct: 313 TGLYLTFKLTSVVEKLREIGNEIQVQSFFTALKALSRKEVHYGSYATTEQ 362
>UniRef100_Q9AS72 P0028E10.20 protein [Oryza sativa]
Length = 659
Score = 242 bits (618), Expect = 2e-62
Identities = 109/144 (75%), Positives = 128/144 (88%)
Query: 308 LVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRDY 367
+VRQ AM+ KC LL+YYKN KG ++RRQ QMLT+VEY LLLYRALLP VWYRFFLN++Y
Sbjct: 1 MVRQTAMIIKCMLLMYYKNSKGRSYRRQGQMLTVVEYFLLLYRALLPAPVWYRFFLNKEY 60
Query: 368 GSLFSSLTTGLYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVYATMEQVNAAGDLCAIC 427
GSLFSSLTTGLYLTFKLTS++EKV+ F++A +ALS KD HYG YAT EQV+A GD+CAIC
Sbjct: 61 GSLFSSLTTGLYLTFKLTSVVEKVQSFLTALRALSHKDFHYGSYATSEQVSATGDMCAIC 120
Query: 428 QEKMHSPVLLRCKHIFCEECVSEW 451
QEKMH+P+LLRCKHIFCE+CVSEW
Sbjct: 121 QEKMHTPILLRCKHIFCEDCVSEW 144
>UniRef100_UPI000024A180 UPI000024A180 UniRef100 entry
Length = 359
Score = 163 bits (413), Expect = 9e-39
Identities = 99/328 (30%), Positives = 168/328 (51%), Gaps = 17/328 (5%)
Query: 162 SSSASLAGSVQGDGDAAGNGAESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQ 221
++SA GS + G+ A A + S +++ V W+++ PF L+LL Q
Sbjct: 45 ATSAERHGSREDGGEDASTPAPALS---------ELKAVVTWLQRGFPFILILLAKVCFQ 95
Query: 222 HLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDL 281
H G V I + + +N +K Q AL+ +R V + I + +Y+ + ++L
Sbjct: 96 HKLGIAVCIGMISTFAFANSTLKHQVALREERSVFTTLWIMVFLAGNIVYVYYTFSTEEL 155
Query: 282 LYPLIMLPPRPTPF--WHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQML 339
LI P F + ++ + + D +++ + KCF+L K F+ + +
Sbjct: 156 HNSLIFAKPNINSFDFFDLIWAVGITDFVLKYFTIELKCFILFLPKMILA--FKSRGKFY 213
Query: 340 TLVEYMLLLYRALLPTLVWYRFFLNRDYGSLF--SSLTTGLYLTFKLTSIIEKVRGFISA 397
L+E + L+RAL+P +WY++ + D S + ++ +Y K + ++ G A
Sbjct: 214 LLIEELSQLFRALVPIQLWYKYIMGEDPSSSYFLGAMVIIIYSLCKSFDLCGRISGIRKA 273
Query: 398 FKALSRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERT 457
F L +YG+ A+ +Q AGD+CAICQ + PV L C+H+FCEEC+ WF+RERT
Sbjct: 274 FIILCSSQ-NYGMRASAQQCGEAGDVCAICQAEFREPVALLCQHVFCEECLCLWFDRERT 332
Query: 458 CPLCRALVKPADLQTFGDGSTSLFFQLF 485
CPLCR+ V L+ + DG+TS FQ++
Sbjct: 333 CPLCRSTVVET-LRNWKDGTTSAHFQIY 359
>UniRef100_Q66IT4 LOC446942 protein [Xenopus laevis]
Length = 390
Score = 157 bits (398), Expect = 5e-37
Identities = 92/306 (30%), Positives = 160/306 (52%), Gaps = 7/306 (2%)
Query: 184 SDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIV 243
SD G+ + +++ + W+++ LPF L+L+ QH G V I +++ +N V
Sbjct: 88 SDEHGTPAPALSELRAIVSWLQKGLPFILILMAKACFQHKLGIAVCIGMASTFAYANSSV 147
Query: 244 KKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPR--PTPFWHSVFI 301
+ Q +L+ R V ++ I + + + ++ L Y LI L P F+ ++I
Sbjct: 148 RHQVSLQEKRSVFAVMWIVVFLMANTLYLLYSIQSQQLQYSLIFLKPNLENLDFFDLMWI 207
Query: 302 ILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRF 361
+ ++D +++ + KC +L + + + + L+E + L R+L+P +WY++
Sbjct: 208 VGISDFVLKYVTVSVKCLVLALPRIILA--VKSKGKFYLLIEELSQLVRSLVPIQLWYKY 265
Query: 362 FLNRDYGSLF--SSLTTGLYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVYATMEQVNA 419
+ D + + S+ LY K I ++ G A K L YGV A+ +Q
Sbjct: 266 IIGDDPSNTYFLGSILIILYSLCKSFDICGRIGGLRKAVKLLCHPQQKYGVRASSQQCAE 325
Query: 420 AGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTS 479
AGD+CAICQ P++L C+H+FCEEC+ WF+RERTCPLCR+ V ++ + DGSTS
Sbjct: 326 AGDVCAICQGDFKDPLILICQHVFCEECLCLWFDRERTCPLCRS-VAVETVRLWKDGSTS 384
Query: 480 LFFQLF 485
FQ++
Sbjct: 385 AHFQVY 390
>UniRef100_UPI0000365016 UPI0000365016 UniRef100 entry
Length = 366
Score = 157 bits (396), Expect = 8e-37
Identities = 89/294 (30%), Positives = 155/294 (52%), Gaps = 8/294 (2%)
Query: 196 DIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKV 255
+++ V W+++ PF L+LL QH G V + +++ +N + Q +L+ +R V
Sbjct: 77 ELKAVVTWLQRGFPFILILLAKVCFQHKLGIAVAVGLASTFTYANSTFRHQVSLREERSV 136
Query: 256 SVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPRPTP--FWHSVFIILVNDVLVRQAA 313
V + I + +Y+ + ++L LI P F+ ++++ + D +++
Sbjct: 137 FVALWIIIFLAGNIVYVYYTFSQEELHNSLIFARPNLNSYDFFDLIWVVGITDFVLKFIT 196
Query: 314 MVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRDYGSLF-- 371
+ KCF+L K F+ + + L+E + L+RAL+P +WY++ + D + +
Sbjct: 197 IGLKCFVLFLPKILLA--FKSRGKFYLLLEELSQLFRALVPIQLWYKYIVGEDPSNSYFL 254
Query: 372 SSLTTGLYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVYATMEQVNAAGDLCAICQEKM 431
++ +Y K I +V A L YGV A +Q + AGD+CAICQ
Sbjct: 255 GAMLIIIYSICKSFDICGRVSAIRKALVMLCSSQ-SYGVRAGSQQCSEAGDICAICQADF 313
Query: 432 HSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTSLFFQLF 485
PVLL C+H+FCEEC+ WF+RERTCPLCR+ + + L+ + DG+TS FQ++
Sbjct: 314 RDPVLLLCQHVFCEECLCLWFDRERTCPLCRSTITES-LRCWKDGTTSAHFQIY 366
>UniRef100_UPI00003AB1CD UPI00003AB1CD UniRef100 entry
Length = 354
Score = 154 bits (390), Expect = 4e-36
Identities = 95/317 (29%), Positives = 162/317 (50%), Gaps = 10/317 (3%)
Query: 175 GDAAGNGAES--DSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWI 232
GD G+ E + G+ + +++ V W+++ LPF L+LL QH G V I +
Sbjct: 42 GDRHGHAEEGSEEQAGTPAPALSELKAVVGWLQKGLPFILILLAKVCFQHKLGIAVCIGM 101
Query: 233 SAVMFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPR- 291
++ +N +++Q ALK R V V+ I + + + + L LI L P
Sbjct: 102 ASTFAYANSTLREQVALKEKRSVLVVFWILAFLTGNTLYLLYTFSSQQLYNSLIFLKPNI 161
Query: 292 -PTPFWHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYR 350
F+ ++I+ + D +++ + KC ++ K + + + ++E + L+R
Sbjct: 162 ERLDFFDLMWIVGITDFVLKYLTIALKCLIVALPKIILA--VKSKGKFYLVIEELSQLFR 219
Query: 351 ALLPTLVWYRFFLNRDYGSLF--SSLTTGLYLTFKLTSIIEKVRGFISAFKALSRKDVHY 408
+L+P +WY++ + D S + + LY K I +V A K L Y
Sbjct: 220 SLVPIQLWYKYIMGDDPSSSYFLGGILIILYSLCKSFDICGRVGSVRKALKVLCTPQT-Y 278
Query: 409 GVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPA 468
G AT +Q + AGD+CAICQ + P++L C+H+FCEEC+ WF+RE+TCPLCR+ V
Sbjct: 279 GARATSQQCSEAGDICAICQAEFREPLILMCQHVFCEECLCLWFDREKTCPLCRS-VTVE 337
Query: 469 DLQTFGDGSTSLFFQLF 485
L+ + DG+TS FQ++
Sbjct: 338 TLRCWKDGTTSAHFQVY 354
>UniRef100_UPI0000438DA1 UPI0000438DA1 UniRef100 entry
Length = 303
Score = 154 bits (389), Expect = 5e-36
Identities = 90/296 (30%), Positives = 156/296 (52%), Gaps = 12/296 (4%)
Query: 196 DIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKV 255
+++ V W+++ PF L+LL QH G V I + + +N +K Q AL+ +R V
Sbjct: 14 ELKAVVTWLQRGFPFILILLAKVCFQHKLGIAVCIGMISTFAFANSTLKHQVALREERSV 73
Query: 256 SVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPRPTPF--WHSVFIILVNDVLVRQAA 313
+ I + +Y+ + ++L LI P F + ++ + + D +++
Sbjct: 74 FTTLWIMVFLAGNIVYVYYTFSTEELHNSLIFAKPNINSFDFFDLIWAVGITDFVLKYFT 133
Query: 314 MVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRDYGSLF-- 371
+ KCF+L K F+ + + L+E + L+RAL+P +WY++ + D S +
Sbjct: 134 IELKCFILFLPKMILA--FKSRGKFYLLIEELSQLFRALVPIQLWYKYIMGEDPSSSYFL 191
Query: 372 SSLTTGLYLTFKLTS--IIEKVRGFISAFKALSRKDVHYGVYATMEQVNAAGDLCAICQE 429
++ +Y K+ S +I F + + + +YG+ A+ +Q AGD+CAICQ
Sbjct: 192 GAMVIIIYSLCKVHSCFLIYNFYCFCCVYVCVLQ---NYGMRASAQQCGEAGDVCAICQA 248
Query: 430 KMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTSLFFQLF 485
+ PV L C+H+FCEEC+ WF+RERTCPLCR+ V L+ + DG+TS FQ++
Sbjct: 249 EFREPVALLCQHVFCEECLCLWFDRERTCPLCRSTVVET-LRNWKDGTTSAHFQIY 303
>UniRef100_Q5RAG4 Hypothetical protein DKFZp459B2128 [Pongo pygmaeus]
Length = 444
Score = 150 bits (380), Expect = 6e-35
Identities = 95/324 (29%), Positives = 163/324 (49%), Gaps = 8/324 (2%)
Query: 166 SLAGSVQGDGDAAGNGAESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQG 225
SL V GD + G+ + +++ V W+++ LPF L+LL QH G
Sbjct: 125 SLLQHVGGDHRGHSEEGGDEQPGTPAPALSELKAVICWLQKGLPFILILLAKLCFQHKLG 184
Query: 226 FFVTIWISAVMFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLLYPL 285
V I +++ +N +++Q +LK R V V++ I + + + + L L
Sbjct: 185 IAVCIGMASTFAYANSTLREQVSLKEKRSVLVILWILAFLAGNTLYVLYTFSSQQLYNSL 244
Query: 286 IMLPPR--PTPFWHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVE 343
I L P F+ ++I+ + D +++ + KC ++ K + + + ++E
Sbjct: 245 IFLKPNLETLDFFDLLWIVGIADFVLKYITIALKCLIVALPKIILA--VKSKGKFYLVIE 302
Query: 344 YMLLLYRALLPTLVWYRFFLNRDYGSLF--SSLTTGLYLTFKLTSIIEKVRGFISAFKAL 401
+ L+R+L+P +WY++ + D + + + LY K I +V G A K L
Sbjct: 303 ELSQLFRSLVPIQLWYKYIMGDDSSNSYFLGGVLIVLYSLCKSFDICGRVGGVRKALKLL 362
Query: 402 SRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLC 461
+YGV AT +Q AGD+CAICQ + P++L C+H+FCEEC+ W +RERTCPL
Sbjct: 363 CTSQ-NYGVRATGQQCTEAGDICAICQAEFREPLILLCQHVFCEECLCLWLDRERTCPLS 421
Query: 462 RALVKPADLQTFGDGSTSLFFQLF 485
R+ V L+ + DG+TS FQ++
Sbjct: 422 RS-VAVDTLRCWKDGATSAHFQVY 444
>UniRef100_Q8BXI9 Mus musculus 10 days neonate medulla oblongata cDNA, RIKEN
full-length enriched library, clone:B830028P19
product:CDNA FLJ14627 FIS, CLONE NT2RP2000289 homolog
[Mus musculus]
Length = 355
Score = 150 bits (379), Expect = 8e-35
Identities = 94/324 (29%), Positives = 162/324 (49%), Gaps = 8/324 (2%)
Query: 166 SLAGSVQGDGDAAGNGAESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQG 225
SL V GD + G+ + +++ V W+++ LPF L+LL QH G
Sbjct: 36 SLLQHVGGDHRGHSEEGVDEQPGTPAPALSELKAVISWLQKGLPFILILLAKLCFQHKLG 95
Query: 226 FFVTIWISAVMFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLLYPL 285
V I +++ +N +++Q +LK R V V++ I + + + + L L
Sbjct: 96 IAVCIGMASTFAYANSTLREQVSLKEKRSVLVILWILAFLAGNTMYVLYTFSSQQLYSSL 155
Query: 286 IMLPPR--PTPFWHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVE 343
I L P F+ ++I+ + D +++ + KC ++ K + + + ++E
Sbjct: 156 IFLKPNLETLDFFDLLWIVGIADFVLKYITIALKCLIVALPKIILA--VKSKGKFYLVIE 213
Query: 344 YMLLLYRALLPTLVWYRFFLNRDYGSLF--SSLTTGLYLTFKLTSIIEKVRGFISAFKAL 401
+ L+R+L+P +WY++ + D + + + LY K I +V G A K L
Sbjct: 214 ELSQLFRSLVPIQLWYKYIMGDDSSNSYFLGGVLIVLYSLCKSFDICGRVGGLRKALKLL 273
Query: 402 SRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLC 461
+YGV AT +Q AG +CAICQ + P++L C+H+FCEEC+ W +RERTCPLC
Sbjct: 274 CTSQ-NYGVRATGQQCTEAGAVCAICQAEFRDPMILLCQHVFCEECLCLWLDRERTCPLC 332
Query: 462 RALVKPADLQTFGDGSTSLFFQLF 485
R+ V L+ + DG+TS Q++
Sbjct: 333 RS-VAVDTLRCWKDGATSAHLQVY 355
>UniRef100_UPI00003AB70F UPI00003AB70F UniRef100 entry
Length = 383
Score = 149 bits (375), Expect = 2e-34
Identities = 97/332 (29%), Positives = 160/332 (47%), Gaps = 18/332 (5%)
Query: 163 SSASLAGSVQGDGDAAGNGAESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQH 222
SS S + G +A + A+ + SS + ++ V +W+ + LP+ +L I + QH
Sbjct: 61 SSQSRSHGHSEAGGSADSAADPEEHSSSISE---LRCVLQWLHRSLPYLFILGIKLVVQH 117
Query: 223 LQGFFVTIWISAVMFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLL 282
+ G + I + +N+ + Q L+ + +Y+ + + L
Sbjct: 118 MIGISLGIGLLTTYMYANKSIVNQVFLQEQCSKLQCAWLLLYLTGSSLLLYYTFHSQSLY 177
Query: 283 YPLIMLPPRPT--PFWHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLT 340
Y LI L P FW ++I+ V D +++ M FKCF+L+ +F+ +
Sbjct: 178 YSLIFLNPTVDFKNFWDVLWIVGVTDFVLKFLFMGFKCFILLVPSFMM--SFKSKGYWYM 235
Query: 341 LVEYMLLLYRALLPTLVWYRFFLNRDYGSLFSSLTTGL-------YLTFKLTSIIEKVRG 393
L+E + YR +P VW+R+ + YG S L L YL KL S + R
Sbjct: 236 LLEELCQYYRMFVPIPVWFRYLIG--YGEPDSVLGWTLGTLLGLSYLILKLLSFFGQWRN 293
Query: 394 FISAFKALSRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFE 453
F + + HYGV A+ Q + + D+C+IC + P+LL C+H FCEEC+S WF
Sbjct: 294 FRQVLRIFCTRP-HYGVTASKRQCSESDDICSICHAEFQKPILLICQHTFCEECISLWFN 352
Query: 454 RERTCPLCRALVKPADLQTFGDGSTSLFFQLF 485
RE+TCPLCR ++ + + DG+TS+ Q+F
Sbjct: 353 REKTCPLCRTVISD-HVNKWKDGATSMRLQIF 383
>UniRef100_UPI00003AB1CE UPI00003AB1CE UniRef100 entry
Length = 288
Score = 148 bits (374), Expect = 3e-34
Identities = 91/294 (30%), Positives = 155/294 (51%), Gaps = 13/294 (4%)
Query: 196 DIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKV 255
+++ V W+++ LPF L+LL QH G V I +++ +N +++Q ALK R V
Sbjct: 4 ELKAVVGWLQKGLPFILILLAKVCFQHKLGIAVCIGMASTFAYANSTLREQVALKEKRSV 63
Query: 256 SVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPR--PTPFWHSVFIILVNDVLVRQAA 313
V+ I + + + + L LI L P F+ ++I+ + D +++
Sbjct: 64 LVVFWILAFLTGNTLYLLYTFSSQQLYNSLIFLKPNIERLDFFDLMWIVGITDFVLKYLT 123
Query: 314 MVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRDYGSLFSS 373
+ KC ++ K + + + ++E + L+R+L+P +WY++ + D S +
Sbjct: 124 IALKCLIVALPKIILA--VKSKGKFYLVIEELSQLFRSLVPIQLWYKYIMGDDPSSSYF- 180
Query: 374 LTTGLYLTFKLTSIIEKV--RGFISAFKALSRKDVHYGVYATMEQVNAAGDLCAICQEKM 431
L L + + L II K GF S + YG AT +Q + AGD+CAICQ +
Sbjct: 181 LGGILIILYSLCKIIFKPFHSGFFSPILPNT-----YGARATSQQCSEAGDICAICQAEF 235
Query: 432 HSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTSLFFQLF 485
P++L C+H+FCEEC+ WF+RE+TCPLCR+ V L+ + DG+TS FQ++
Sbjct: 236 REPLILMCQHVFCEECLCLWFDREKTCPLCRS-VTVETLRCWKDGTTSAHFQVY 288
>UniRef100_Q9DCN7 Mus musculus adult male kidney cDNA, RIKEN full-length enriched
library, clone:0610013E23 product:similar to PTD016
PROTEIN [Mus musculus]
Length = 395
Score = 147 bits (370), Expect = 9e-34
Identities = 93/314 (29%), Positives = 155/314 (48%), Gaps = 21/314 (6%)
Query: 183 ESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEI 242
ES GS S+ + + + KW+++ LP+ L+L I + QH+ G + I + +N+
Sbjct: 92 ESGEHGSGSFSEF--RYLFKWLQKSLPYILILGIKLVMQHITGISLGIGLLTTFMYANKS 149
Query: 243 VKKQTALKGDRKVSVLVGISFAFIL-QVTCIYWWYRNDDLLYPLIMLPP--RPTPFWHSV 299
+ Q L+ +R + F+ +Y+ + + L Y LI L P FW +
Sbjct: 150 IVNQVFLR-ERSSKLRCAWLLVFLAGSSVLLYYTFHSQSLHYSLIFLNPTLEQLSFWEVL 208
Query: 300 FIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWY 359
+I+ + D +++ M KC +L+ F+ + L+E + YR +P VW+
Sbjct: 209 WIVGITDFILKFFFMGLKCLILLVPSFIMP--FKSKGYWYMLLEELCQYYRIFVPIPVWF 266
Query: 360 RFFLNRDYGSLFSSLTTG--------LYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVY 411
R+ ++ YG F ++TT LYL KL +R F + + YGV
Sbjct: 267 RYLIS--YGE-FGNVTTWSLGILLALLYLILKLLDFFGHLRTFRQVLRVFFTRP-SYGVP 322
Query: 412 ATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQ 471
A+ Q + +C ICQ + PVLL C+HIFCEEC++ WF RE+TCPLCR ++ +
Sbjct: 323 ASKRQCSDMDGICTICQAEFQKPVLLFCQHIFCEECITLWFNREKTCPLCRTVISEC-IN 381
Query: 472 TFGDGSTSLFFQLF 485
+ DG+TS Q++
Sbjct: 382 KWKDGATSSHLQMY 395
>UniRef100_Q80X62 0610013E23Rik protein [Mus musculus]
Length = 395
Score = 145 bits (366), Expect = 3e-33
Identities = 92/314 (29%), Positives = 155/314 (49%), Gaps = 21/314 (6%)
Query: 183 ESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEI 242
+S GS S+ + + + KW+++ LP+ L+L I + QH+ G + I + +N+
Sbjct: 92 KSGEHGSGSFSEF--RYLFKWLQKSLPYILILGIKLVMQHITGISLGIGLLTTFMYANKS 149
Query: 243 VKKQTALKGDRKVSVLVGISFAFIL-QVTCIYWWYRNDDLLYPLIMLPP--RPTPFWHSV 299
+ Q L+ +R + F+ +Y+ + + L Y LI L P FW +
Sbjct: 150 IVNQVFLR-ERSSKLRCAWLLVFLAGSSVLLYYTFHSQSLHYSLIFLNPTLEQLSFWEVL 208
Query: 300 FIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWY 359
+I+ + D +++ M KC +L+ F+ + L+E + YR +P VW+
Sbjct: 209 WIVGITDFILKFFFMGLKCLILLVPSFIMP--FKSKGYWYMLLEELCQYYRIFVPIPVWF 266
Query: 360 RFFLNRDYGSLFSSLTTG--------LYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVY 411
R+ ++ YG F ++TT LYL KL +R F + + YGV
Sbjct: 267 RYLIS--YGE-FGNVTTWSLGILLALLYLILKLLDFFGHLRTFRQVLRVFFTRP-SYGVP 322
Query: 412 ATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQ 471
A+ Q + +C ICQ + PVLL C+HIFCEEC++ WF RE+TCPLCR ++ +
Sbjct: 323 ASKRQCSDMDGICTICQAEFQKPVLLFCQHIFCEECITLWFNREKTCPLCRTVISEC-IN 381
Query: 472 TFGDGSTSLFFQLF 485
+ DG+TS Q++
Sbjct: 382 KWKDGATSSHLQMY 395
>UniRef100_UPI00001D0574 UPI00001D0574 UniRef100 entry
Length = 577
Score = 145 bits (365), Expect = 3e-33
Identities = 93/314 (29%), Positives = 155/314 (48%), Gaps = 21/314 (6%)
Query: 183 ESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEI 242
ES GS S+ + + + KW+++ LP+ L+L I + QH+ G + I + +N+
Sbjct: 274 ESGEHGSGSFSEF--RYLFKWLQKSLPYILILGIKLVMQHITGISLGIGLLTTFMYANKS 331
Query: 243 VKKQTALKGDRKVSVLVGISFAFIL-QVTCIYWWYRNDDLLYPLIMLPP--RPTPFWHSV 299
+ Q L+ +R + F+ +Y+ + + L Y LI L P FW +
Sbjct: 332 IVNQVFLR-ERSSKLRCAWLLVFLAGSSVLLYYTFHSQSLHYSLIFLNPTLEQLSFWEVL 390
Query: 300 FIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWY 359
+I+ + D +++ M KC +L+ F+ + L+E + YR +P VW+
Sbjct: 391 WIVGITDFILKFFFMGLKCLVLLVPSFIVP--FKSKGYWYMLLEELCQYYRIFVPIPVWF 448
Query: 360 RFFLNRDYGSLFSSLTTG--------LYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVY 411
R+ ++ YG F ++TT LYL KL +R F + + YGV
Sbjct: 449 RYLIS--YGE-FGNVTTWSLGILLALLYLILKLLDFFGHLRTFRQVLRIFFTRP-SYGVP 504
Query: 412 ATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQ 471
A+ Q + +C ICQ + PVLL C+HIFCEEC++ WF RE+TCPLCR ++ +
Sbjct: 505 ASKRQCSDMDGICPICQAEFQKPVLLFCQHIFCEECITLWFNREKTCPLCRTVISER-IN 563
Query: 472 TFGDGSTSLFFQLF 485
+ DG+TS Q++
Sbjct: 564 KWKDGATSSHLQMY 577
>UniRef100_Q6NTV1 MGC81516 protein [Xenopus laevis]
Length = 416
Score = 142 bits (357), Expect = 3e-32
Identities = 90/319 (28%), Positives = 151/319 (47%), Gaps = 15/319 (4%)
Query: 176 DAAGNGAESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAV 235
DA+G +D S ++ + KW+E+ P+ L+ + QH+ G V I +
Sbjct: 104 DASGPEDANDDSREQSNSISEVFHLYKWLEKSFPYILIFSAKLVVQHITGISVGIGLLTT 163
Query: 236 MFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPR--PT 293
+N+ + Q L+ + + + +Y+ + + L Y L+ + P P
Sbjct: 164 FLYANKCIVNQVFLRDKCSKLQCLWVLVFLLFSSFLLYYTFSSQALYYSLVFMNPSLGPL 223
Query: 294 PFWHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALL 353
F+ +++++ + D + + M KC +L+ H + M +E + Y L+
Sbjct: 224 HFFDALWVVGITDFIGKFFFMGLKCIILLVPSFVMSHKSKGYWYMA--LEELAQCYCTLV 281
Query: 354 PTLVWYRFFLNRDYGSL-------FSSLTTGLYLTFKLTSIIEKVRGFISAFKALSRKDV 406
T VW+R+ + DYG+L F L LYL K+ I + R S L
Sbjct: 282 STPVWFRYLI--DYGNLDSGAEWHFGILLALLYLILKILIIFGQ-RKTSSDCLRLFLSQP 338
Query: 407 HYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVK 466
+YG AT Q + A +CAICQ + P+ L C+H+FCEEC+S WF +E+TCPLCR L+
Sbjct: 339 NYGTTATKRQCSEADGMCAICQAEFTKPIALICQHVFCEECISSWFNKEKTCPLCRTLIS 398
Query: 467 PADLQTFGDGSTSLFFQLF 485
+ DG+TSL ++F
Sbjct: 399 NHS-HKWKDGATSLQLRIF 416
>UniRef100_Q6DDF3 MGC81516 protein [Xenopus laevis]
Length = 391
Score = 142 bits (357), Expect = 3e-32
Identities = 90/319 (28%), Positives = 151/319 (47%), Gaps = 15/319 (4%)
Query: 176 DAAGNGAESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAV 235
DA+G +D S ++ + KW+E+ P+ L+ + QH+ G V I +
Sbjct: 79 DASGPEDANDDSREQSNSISEVFHLYKWLEKSFPYILIFSAKLVVQHITGISVGIGLLTT 138
Query: 236 MFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPR--PT 293
+N+ + Q L+ + + + +Y+ + + L Y L+ + P P
Sbjct: 139 FLYANKCIVNQVFLRDKCSKLQCLWVLVFLLFSSFLLYYTFSSQALYYSLVFMNPSLGPL 198
Query: 294 PFWHSVFIILVNDVLVRQAAMVFKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALL 353
F+ +++++ + D + + M KC +L+ H + M +E + Y L+
Sbjct: 199 HFFDALWVVGITDFIGKFFFMGLKCIILLVPSFVMSHKSKGYWYMA--LEELAQCYCTLV 256
Query: 354 PTLVWYRFFLNRDYGSL-------FSSLTTGLYLTFKLTSIIEKVRGFISAFKALSRKDV 406
T VW+R+ + DYG+L F L LYL K+ I + R S L
Sbjct: 257 STPVWFRYLI--DYGNLDSGAEWHFGILLALLYLILKILIIFGQ-RKTSSDCLRLFLSQP 313
Query: 407 HYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVK 466
+YG AT Q + A +CAICQ + P+ L C+H+FCEEC+S WF +E+TCPLCR L+
Sbjct: 314 NYGTTATKRQCSEADGMCAICQAEFTKPIALICQHVFCEECISSWFNKEKTCPLCRTLIS 373
Query: 467 PADLQTFGDGSTSLFFQLF 485
+ DG+TSL ++F
Sbjct: 374 NHS-HKWKDGATSLQLRIF 391
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 825,120,982
Number of Sequences: 2790947
Number of extensions: 36063389
Number of successful extensions: 113439
Number of sequences better than 10.0: 3893
Number of HSP's better than 10.0 without gapping: 1832
Number of HSP's successfully gapped in prelim test: 2080
Number of HSP's that attempted gapping in prelim test: 109553
Number of HSP's gapped (non-prelim): 5016
length of query: 485
length of database: 848,049,833
effective HSP length: 131
effective length of query: 354
effective length of database: 482,435,776
effective search space: 170782264704
effective search space used: 170782264704
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0360.7


