

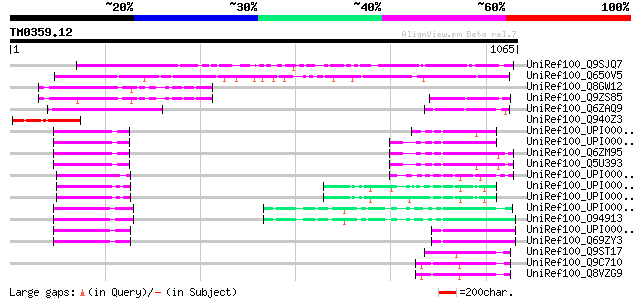
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0359.12
(1065 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SJQ7 Hypothetical protein At2g36480 [Arabidopsis tha... 431 e-119
UniRef100_Q650V5 Putative KIAA protein [Oryza sativa] 414 e-114
UniRef100_Q8GW12 Hypothetical protein At4g04885 [Arabidopsis tha... 198 8e-49
UniRef100_Q9ZS85 T4B21.1 protein [Arabidopsis thaliana] 185 7e-45
UniRef100_Q6ZAQ9 Putative S-locus protein 4 [Oryza sativa] 184 1e-44
UniRef100_Q940Z3 At2g36480/F1O11.11 [Arabidopsis thaliana] 113 3e-23
UniRef100_UPI000043663C UPI000043663C UniRef100 entry 99 9e-19
UniRef100_UPI000043663B UPI000043663B UniRef100 entry 99 9e-19
UniRef100_Q6ZM95 Novel protein similar to human pre-mRNA cleavag... 99 9e-19
UniRef100_Q5U393 Zgc:92226 [Brachydanio rerio] 99 9e-19
UniRef100_UPI000027C40A UPI000027C40A UniRef100 entry 98 1e-18
UniRef100_UPI0000361313 UPI0000361313 UniRef100 entry 98 2e-18
UniRef100_UPI0000361312 UPI0000361312 UniRef100 entry 98 2e-18
UniRef100_UPI000036EFCD UPI000036EFCD UniRef100 entry 98 2e-18
UniRef100_O94913 Pre-mRNA cleavage complex II protein Pcf11 [Hom... 98 2e-18
UniRef100_UPI00001CE95F UPI00001CE95F UniRef100 entry 97 2e-18
UniRef100_Q69ZY3 MKIAA0824 protein [Mus musculus] 97 2e-18
UniRef100_Q9ST17 S-locus protein 4 [Brassica campestris] 97 3e-18
UniRef100_Q9C710 Hypothetical protein F28G11.6 [Arabidopsis thal... 97 3e-18
UniRef100_Q8VZG9 At1g66500/F28G11_6 [Arabidopsis thaliana] 97 3e-18
>UniRef100_Q9SJQ7 Hypothetical protein At2g36480 [Arabidopsis thaliana]
Length = 828
Score = 431 bits (1107), Expect = e-119
Identities = 339/934 (36%), Positives = 467/934 (49%), Gaps = 134/934 (14%)
Query: 140 IEVPSEQKLPSLYLLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTW 199
++VPS+QKLP+LYLLDSIVKNIGR Y+KYF ARLPEVF KAYR VDP +H +MRHLFGTW
Sbjct: 1 MQVPSDQKLPTLYLLDSIVKNIGRDYIKYFGARLPEVFVKAYRQVDPPMHSNMRHLFGTW 60
Query: 200 RGVFPSQTLQAIEKELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLERQRLQQS 259
+GVF QTLQ IEKELGF +GS++ +T R++ QSQRPP SIHVNPKYLERQRLQQS
Sbjct: 61 KGVFHPQTLQLIEKELGFNAKSDGSAAVVSTARAEPQSQRPPHSIHVNPKYLERQRLQQS 120
Query: 260 SRTKG-VDDMSGAIANSNDDPEMPDRALGVKRPRA--DPSNN-QRAHRDAFNDSVREKSI 315
RTKG V D+ N D + +R + + P+ N +R RD ++ + EK I
Sbjct: 121 GRTKGMVTDVPETAPNLTRDSDRLERVSSIASGGSWVGPAKNIRRPQRDLLSEPLYEKDI 180
Query: 316 SASYGGNEYGSDLSRNLGMGTGRTGGRVAELGSDKSGYNKAAGAGVAGTISGQRNGLSIK 375
+ G +Y SDL N G R+ + G +K Y A IS QR+GL K
Sbjct: 181 ESIAGEYDYASDLPHNSRSVIKNVGSRITDDGCEKQWY--GATNRDPDLISDQRDGLHSK 238
Query: 376 NNFLNTEASMILDARNQ-PRQNINSLQRGVMSNSWKHSEEEEFMWDEMNAGLTGHDAASV 434
+ N + + + + P +NI GV +SWK+SEEEEFMWD M++ L+ D A++
Sbjct: 239 SRTSNYATARVENLESSGPSRNI-----GVPYDSWKNSEEEEFMWD-MHSRLSETDVATI 292
Query: 435 PNNLSKDSWTSDDENLEVEDNHHQSRNPFGANVDTEMSIESQATERKQLPAFRPHPSLSW 494
K+ + DE+ +E +H + P + +D P F P S +
Sbjct: 293 N---PKNELHAPDESERLESENHLLKRPRFSALD---------------PRFDPANSTNS 334
Query: 495 KLQEQHSIDELNQKPGHSDGFVSTLGALPTNTNSAATRMGNRSLLPKATIGLAGNLGQLH 554
EQ + GH F S TN S ATR G + P+ + +G L
Sbjct: 335 YSSEQKDPSSI----GHW-AFSS------TNATSTATRKG---IQPQPRVASSGIL---- 376
Query: 555 SVGDESPSRESSLRQQSPPPVSVHLPHLMQNLAEQDHPQTH-------KVSKFSGGLHSQ 607
PS S +QSP +H QN+ +QD + H + S+F Q
Sbjct: 377 ------PSSGSGSDRQSP----LHDSTSKQNVTKQDVRRAHSLPQRDPRASRFPA---KQ 423
Query: 608 NI-KDISPALPPNIQVGKFHRSQLRDLQSGSFSSMTFQPSHKQQLGSSHTEVTAKTEKPH 666
N+ +D S LP + +F + +R+L F S +A P
Sbjct: 424 NVPRDDSVRLPSS--SSQFKNTNMRELPVEIFDSK-----------------SAAENAPG 464
Query: 667 LSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSS 726
L+ T QP + A K G LSN + S D N P L + +
Sbjct: 465 LTLA--SEATGQPNMSDLLEAVMKSGILSNNSTCGAI-KEESHDEVN-PGAL----TLPA 516
Query: 727 GSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQ 786
S P L P+S +A+ L R K S+ P + A+S +VQ
Sbjct: 517 ASKPKTL--PIS-LATDNLLARLKVEQSSAPLV--------------SCAASLTGITSVQ 559
Query: 787 ASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGAS 846
S + A +P++ LL+SLV+KGLISA PS D S + + S S+ A
Sbjct: 560 TSKEKSKASDPLSCLLSSLVSKGLISASKTELPSAPSITQEHSPDHSTNSSMSVSVVPAD 619
Query: 847 VSGSA-VPVPSTKDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFD 905
S V PST +V G P SE++ +E ++ IG +F+ D IRELHPSVIS LFD
Sbjct: 620 AQPSVLVKGPSTAPKV-KGLAAP---SETSKSEPKDLIGLKFRADKIRELHPSVISSLFD 675
Query: 906 DFPHHCSICSLKLKFQEQFDRHLEWHATREREHSGLIKASR-WYLKSSDWVVGKVESLSE 964
D PH C+ CS++LK +E+ DRH+E H ++ E SG R W+ K +W+ K L E
Sbjct: 676 DLPHLCTSCSVRLKQKEELDRHMELHDKKKLELSGTNSKCRVWFPKVDNWIAAKAGEL-E 734
Query: 965 NEFADSVDRYGKETDRNQEDAM-VLADENQCLCVLCGELFEEFYCQENGEWMFKGAVYVP 1023
E+ + + E + ED V ADE QC C+LCGE+FE+++ QE +WMFKGA Y+
Sbjct: 735 PEYEEVL----SEPESAIEDCQAVAADETQCACILCGEVFEDYFSQEMAQWMFKGASYLT 790
Query: 1024 NSDINDDMGIRDASTGGGPIIHARCLSENPVSSV 1057
N N S GPI+H CL+ + + S+
Sbjct: 791 NPPAN--------SEASGPIVHTGCLTTSSLQSL 816
>UniRef100_Q650V5 Putative KIAA protein [Oryza sativa]
Length = 1062
Score = 414 bits (1065), Expect = e-114
Identities = 352/1058 (33%), Positives = 493/1058 (46%), Gaps = 156/1058 (14%)
Query: 94 ELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLYL 153
ELVAQY++AL ELTFNSKPIITNLTIIAGEN A + IA+ +CANI+EVPSEQKLPSLYL
Sbjct: 50 ELVAQYRTALGELTFNSKPIITNLTIIAGENLHAAKPIASLICANILEVPSEQKLPSLYL 109
Query: 154 LDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEK 213
LDSIVKNIG+ YVK+F+ARLPEVFCKAY+ VD ++H+SMRHLFGTW+GVF +LQ IEK
Sbjct: 110 LDSIVKNIGKDYVKHFSARLPEVFCKAYKQVDSSIHNSMRHLFGTWKGVFSPTSLQVIEK 169
Query: 214 ELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLE-RQRLQQSSRTKGV---DDMS 269
ELGF + NGSS +A + DSQS RP SIHVNPKYLE RQ+LQQ ++ +G+ +
Sbjct: 170 ELGFQSSTNGSSG-AAPSKPDSQSNRPSHSIHVNPKYLEARQQLQQPNKGQGILGAGAKT 228
Query: 270 GAIANSNDDPEMP-----DRALGVKRPRADPSNN-QRAHRDAFNDSVREKSISASYGGNE 323
I++S DD E DR G + +P N QRA RD F++ + EK
Sbjct: 229 TTISDSGDDIERTSRTAVDRGAGRRLDALNPRTNVQRAQRDPFSNPIHEKQ---DRDMRV 285
Query: 324 YG-SDLSRNLGMGTGRTGGRVAELGSDKS-GYNKAAGAGVAGTISGQRNGLSIKNNFLNT 381
G S++S+ +GTG R G D S G AG G + +R+ N +
Sbjct: 286 LGFSNISQQAVVGTGLV--RAKPKGQDGSGGPYYTAGVGSSEEQFDRRSNF-YANKDVRP 342
Query: 382 EASMILDAR--NQPRQNINSLQRGVMSNSWKHSEEEEFMWDEMNAGLTGHDAASVPNNLS 439
S+ LD P N + + R + SWK+SEEEE+MWD++ + + S +
Sbjct: 343 SGSVRLDGALLPTPVSNSDRIGRPSSNKSWKNSEEEEYMWDDVRSQGADYGGTS---STR 399
Query: 440 KDSWTSDDEN---------------LEVEDNHHQSRNPFGANVDTEMSIE---------- 474
K W DD N L+ + + S FG + I
Sbjct: 400 KREWIPDDGNVGSFQRVKWAEAGGPLDPDQHKLDSFQRFGNATGQDRRITPYMDHEEYLH 459
Query: 475 -----SQATERKQLPAFRPHPS---LSWKLQEQHSIDELNQKPGHS-------DGFVSTL 519
+R+ LP +P S S L + + ++ P S D
Sbjct: 460 GKHEVEPRIDREMLPEGQPFSSSRGSSLWLSHEKPLPDIVSDPRISAFSNQPADRPTIYA 519
Query: 520 GALPTNTNSA-----ATRMGNRSLLPKATIGLAGNLGQ-------LHSVGDESPSRESSL 567
G L T+ S+ + RS L A +GQ S +SPS S
Sbjct: 520 GTLSTSITSSVPVGLSAAYAGRSSLESAATRSTETIGQQKNRYWSTSSPPVQSPS-ASFA 578
Query: 568 RQQSPPPV-----SVHLPHLMQNLAEQDHPQTHKVSKFSGGLHSQNIKDISPALPPNIQV 622
RQ SP PV S L QN E + +TH AL N+ +
Sbjct: 579 RQSSPSPVELDYSSKPFSQLGQNSLEDYNQRTH-------------------ALAQNLAL 619
Query: 623 GKFHRSQLRDLQSGSFSSMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQP--- 679
SQ R G+ PSH Q H + KPHL + P+ P
Sbjct: 620 -----SQGRPNLLGA-------PSHASQQIEKHPSLL--QSKPHLRTLDQPQANFSPENS 665
Query: 680 -----TTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSIL------GVRPSQSSGS 728
++ + + G G + + T S D + S+L G +P+ +
Sbjct: 666 SSVFKSSIQLPISVGVGHRQPEEVSLSSDSTLMSSDHLSASSLLAGLIKSGFKPNDPNDL 725
Query: 729 SPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRA--AQPPRTSALP-PASSNLKSAAV 785
+ PP+ + PP + +S+ +P +Q P + P P L S V
Sbjct: 726 ASLRAQPPLPS-GPPPHVSTSFSAASSSLHLPASDTLKSQAPNSLRPPLPPGLPLSSPFV 784
Query: 786 QASSSANNALNPIANLLNSLVAKGLISA-ETETTAKVPSEMLTRLEDQSDSITTSSSLPG 844
++ + P+++LL+SLVAKGLIS+ ++T +P + + D ++ SLP
Sbjct: 785 CPTTQTSEKAAPLSSLLSSLVAKGLISSPSADSTVAIPQQPCKSELNTPDDTASAPSLPF 844
Query: 845 ASVSGSAVPVPSTKDEVDDGAKT---------PISL-SESTSTEIRNFIGFEFKPDVIRE 894
PS K E + + P+ + E ++ + IGF+FKP+++R+
Sbjct: 845 VQ--------PSVKKETSNQNSSAPSKVLVHQPVEIKKEPAEIKMVDLIGFDFKPEMLRK 896
Query: 895 LHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEWHATRERE-HSGLIKASRWYLKSSD 953
H VIS LFDD H C+ C L+ +E+ H H +++ E I +WY ++
Sbjct: 897 YHAHVISTLFDDQSHQCNTCGLRFSLEEELSVHTACHGSKQSETRKTGIAPEKWYPSKNN 956
Query: 954 WVVGKVE-SLSENEFADSVDRYGKETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENG 1012
WV E S E A SV E + + MV ADE+Q +C LCGE F++ Y E
Sbjct: 957 WVDRSHEVQNSALESASSVADLSSEEE--VCEFMVPADESQIICALCGESFDDIYSIEKD 1014
Query: 1013 EWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCLS 1050
WM+K AVY +S G S PI+HARC+S
Sbjct: 1015 NWMYKDAVYFDSSKTEGSSGDSAESKERVPIVHARCMS 1052
>UniRef100_Q8GW12 Hypothetical protein At4g04885 [Arabidopsis thaliana]
Length = 614
Score = 198 bits (503), Expect = 8e-49
Identities = 136/376 (36%), Positives = 185/376 (49%), Gaps = 60/376 (15%)
Query: 61 RLRAAANSRDLESSDFGRGDGGGGYHPRPPPFQELVAQYKSALAELTFNSKPIITNLTII 120
R +A N R+ E GGG PP E+V Y+ L ELTFNSKPIIT+LTII
Sbjct: 39 RFKALLNQREDEF--------GGGEEVLPPSMDEIVQLYEVVLGELTFNSKPIITDLTII 90
Query: 121 AGENQAAEQAIAATVCANIIEVPSEQKLPSLYLLDSIVKNIGRGYVKYFAARLPEVFCKA 180
AGE + + IA +C I+E P EQKLPSLYLLDSIVKNIGR Y +YF++RLPEVFC A
Sbjct: 91 AGEQREHGEGIANAICTRILEAPVEQKLPSLYLLDSIVKNIGRDYGRYFSSRLPEVFCLA 150
Query: 181 YRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEKELGFTPAVNGSSSTSATLRSDSQSQRP 240
YR P++H SMRHLFGTW VFP L+ I+ +L + A N SS + S+ +P
Sbjct: 151 YRQAHPSLHPSMRHLFGTWSSVFPPPVLRKIDMQLQLSSAANQSSVGA------SEPSQP 204
Query: 241 PPSIHVNPKYLER----------QRLQQSSRTKGVDDMSGAIANSNDDPEMPDRALGVKR 290
IHVNPKYL R + + S+R G + + G + D E P
Sbjct: 205 TRGIHVNPKYLRRLEPSAAENNLRGINSSARVYGQNSL-GGYNDFEDQLESPSSL----- 258
Query: 291 PRADPSNNQRAHRDAFNDSVREKSISASYGGNEYGSDLSRNLGMGTGRTGGRVAELGSDK 350
+ P R D N S+ + N GM GR +
Sbjct: 259 -SSTPDGFTRRSNDGANP-----------------SNQAFNYGM------GRATSRDDEH 294
Query: 351 SGYNKAAGAGVAGTISGQRNGLSIKNNFLNTEASMILDARNQPRQNINSLQRGVMSNSWK 410
+ + G +R I ++T + + N+P +++N + M W+
Sbjct: 295 MEWRRKENLGQGN--DHERPRALIDAYGVDTSKHVTI---NKPIRDMNGM-HSKMVTPWQ 348
Query: 411 HSEEEEFMWDEMNAGL 426
++EEEEF W++M+ L
Sbjct: 349 NTEEEEFDWEDMSPTL 364
>UniRef100_Q9ZS85 T4B21.1 protein [Arabidopsis thaliana]
Length = 827
Score = 185 bits (469), Expect = 7e-45
Identities = 136/399 (34%), Positives = 185/399 (46%), Gaps = 83/399 (20%)
Query: 61 RLRAAANSRDLESSDFGRGDGGGGYHPRPPPFQELVAQYKSALAELTFNSKPIITNLTII 120
R +A N R+ E GGG PP E+V Y+ L ELTFNSKPIIT+LTII
Sbjct: 39 RFKALLNQREDEF--------GGGEEVLPPSMDEIVQLYEVVLGELTFNSKPIITDLTII 90
Query: 121 AGENQAAEQAIAATVCANIIE-----------------------VPSEQKLPSLYLLDSI 157
AGE + + IA +C I+E P EQKLPSLYLLDSI
Sbjct: 91 AGEQREHGEGIANAICTRILERLVFGLWFSEMAASELPVGVYMQAPVEQKLPSLYLLDSI 150
Query: 158 VKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEKELGF 217
VKNIGR Y +YF++RLPEVFC AYR P++H SMRHLFGTW VFP L+ I+ +L
Sbjct: 151 VKNIGRDYGRYFSSRLPEVFCLAYRQAHPSLHPSMRHLFGTWSSVFPPPVLRKIDMQLQL 210
Query: 218 TPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLER----------QRLQQSSRTKGVDD 267
+ A N SS + S+ +P IHVNPKYL R + + S+R G +
Sbjct: 211 SSAANQSSVGA------SEPSQPTRGIHVNPKYLRRLEPSAAENNLRGINSSARVYGQNS 264
Query: 268 MSGAIANSNDDPEMPDRALGVKRPRADPSNNQRAHRDAFNDSVREKSISASYGGNEYGSD 327
+ G + D E P + P R D N S+
Sbjct: 265 L-GGYNDFEDQLESPSSL------SSTPDGFTRRSNDGANP-----------------SN 300
Query: 328 LSRNLGMGTGRTGGRVAELGSDKSGYNKAAGAGVAGTISGQRNGLSIKNNFLNTEASMIL 387
+ N GM GR + + + G +R I ++T + +
Sbjct: 301 QAFNYGM------GRATSRDDEHMEWRRKENLGQGN--DHERPRALIDAYGVDTSKHVTI 352
Query: 388 DARNQPRQNINSLQRGVMSNSWKHSEEEEFMWDEMNAGL 426
N+P +++N + M W+++EEEEF W++M+ L
Sbjct: 353 ---NKPIRDMNGM-HSKMVTPWQNTEEEEFDWEDMSPTL 387
Score = 118 bits (295), Expect = 1e-24
Identities = 63/174 (36%), Positives = 95/174 (54%), Gaps = 10/174 (5%)
Query: 883 IGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEWHATRER--EHSG 940
+G EF D+++ + S IS L+ D P C+ C L+ K QE+ +H++WH T+ R ++
Sbjct: 648 LGLEFDADMLKIRNESAISALYGDLPRQCTTCGLRFKCQEEHSKHMDWHVTKNRMSKNHK 707
Query: 941 LIKASRWYLKSSDWVVGKVESLSENEFADSVDRYGKETDRNQEDAMVLADENQCLCVLCG 1000
+ +W++ +S W+ G E+L + ++ ED V ADE+Q C LCG
Sbjct: 708 QNPSRKWFVSASMWLSG-AEALGAEAVPGFLPTEPTTEKKDDEDMAVPADEDQTSCALCG 766
Query: 1001 ELFEEFYCQENGEWMFKGAVYV--PNSDINDDMGIRDASTGGGPIIHARCLSEN 1052
E FE+FY E EWM+KGAVY+ P D D S GPI+HA+C E+
Sbjct: 767 EPFEDFYSDETEEWMYKGAVYMNAPEESTTD----MDKSQ-LGPIVHAKCRPES 815
>UniRef100_Q6ZAQ9 Putative S-locus protein 4 [Oryza sativa]
Length = 971
Score = 184 bits (467), Expect = 1e-44
Identities = 115/246 (46%), Positives = 145/246 (58%), Gaps = 9/246 (3%)
Query: 80 DGGGGYHPRPPPFQELVAQYKSALAELTFNSKPIITNLTIIAGENQA-AEQAIAATVCAN 138
+GGGG +V Y L+ELTFN KPIIT LTIIAG++ A A + IA +CA
Sbjct: 32 EGGGG----EVAAAAVVRVYVEVLSELTFNCKPIITELTIIAGQHAALAARGIADAICAR 87
Query: 139 IIEVPSEQKLPSLYLLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGT 198
I EV ++QKLPSLYLLDSIVKNIGR YV +FAARL +VFC AYR V H +MRHLFGT
Sbjct: 88 IAEVSADQKLPSLYLLDSIVKNIGREYVGHFAARLQKVFCDAYRKVHRNQHAAMRHLFGT 147
Query: 199 WRGVFPSQTLQAIEKELGFTPAVNGSSSTSATLR-SDSQSQRPPPSIHVNPKYLERQRLQ 257
W VFPS L+ IE EL F+P N S+T+ +R S+S S R +IHVNPKYLE Q+ Q
Sbjct: 148 WSQVFPSSVLRGIEDELQFSPLENKRSATATDIRQSESISPRLSHAIHVNPKYLEAQQ-Q 206
Query: 258 QSSRTKGVDDMSGAIANSNDDPEMPDRALGVKRPRADPSNNQRAHRDA--FNDSVREKSI 315
T ++ ND E L K R P+ N + + + D + ++
Sbjct: 207 FKQSTSVHQPITRGNRQMNDVEEDQINGLTSKSSRGWPATNSKLQKSTMLYADDLDQQEA 266
Query: 316 SASYGG 321
S+ G
Sbjct: 267 FCSHTG 272
Score = 106 bits (265), Expect = 3e-21
Identities = 62/188 (32%), Positives = 95/188 (49%), Gaps = 20/188 (10%)
Query: 871 LSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEW 930
+S ++ ++ +G +F D ++ + SVI+ L+ D P C C L+ K QE+ H++W
Sbjct: 791 ISLQPPSQPQDSVGVDFNVD-LKVRNESVINALYQDLPRQCKTCGLRFKCQEEHRAHMDW 849
Query: 931 HATREREHSGLIKASRWYLKS-SDWVVGKVESLSENEFADSVDRYGKETDRNQEDAMVL- 988
H T+ R + SR Y + +W+ + N+ S + D +E + +
Sbjct: 850 HVTKNRNSKNRKQTSRKYFVTVGEWL--RAAETVGNDGVPSFEPAEPVADAKEEKELAVP 907
Query: 989 ADENQCLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGG------GP 1042
ADE+Q C LC E FE+FY E EWM+KGAVY M D + GG GP
Sbjct: 908 ADEDQTTCALCQEPFEDFYSDETEEWMYKGAVY---------MNAPDGNIGGLERSQLGP 958
Query: 1043 IIHARCLS 1050
I+HA+CLS
Sbjct: 959 IVHAKCLS 966
>UniRef100_Q940Z3 At2g36480/F1O11.11 [Arabidopsis thaliana]
Length = 158
Score = 113 bits (283), Expect = 3e-23
Identities = 72/146 (49%), Positives = 93/146 (63%), Gaps = 9/146 (6%)
Query: 6 RRPFDRSREPG-LKRPRMMEEPERVAAAQNPSSRQFLPQRQQQVAASGISTLSSAARLRA 64
RRPFDRSR+PG +K+PR+ EE R N ++RQFL QR A + ++ +++R R
Sbjct: 5 RRPFDRSRDPGPMKKPRLSEESIRPV---NSNARQFLSQRTLGTATA-VTVPPASSRFRV 60
Query: 65 AANSRDLESSDFGRGDGGGGYHPRPP-PFQELVAQYKSALAELTFNSKPIITNLTIIAGE 123
+ R+ ESS Y P+P P ELV QYKSALAELTFNSKPIITNLTIIAGE
Sbjct: 61 SG--RETESSIVS-DPSREAYQPQPVHPHYELVNQYKSALAELTFNSKPIITNLTIIAGE 117
Query: 124 NQAAEQAIAATVCANIIEVPSEQKLP 149
N A +A+ +C NI+EV ++ P
Sbjct: 118 NVHAAKAVVTAICNNILEVNTQFSCP 143
>UniRef100_UPI000043663C UPI000043663C UniRef100 entry
Length = 1331
Score = 98.6 bits (244), Expect = 9e-19
Identities = 59/160 (36%), Positives = 84/160 (51%), Gaps = 9/160 (5%)
Query: 93 QELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLY 152
++ +Y+S+L +LTFNSKP I LTI+A EN + I A + A I + P +KLP LY
Sbjct: 4 EDACREYQSSLEDLTFNSKPHINMLTILAEENIQFTKDIVAIIEAQIAKAPPVEKLPVLY 63
Query: 153 LLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIE 212
L+DSIVKN+G Y++ FA L F + VD S+ L TW +FP + L A++
Sbjct: 64 LVDSIVKNVGGAYLEVFAKNLVNSFICVFEKVDEGTRKSLFKLRSTWDEIFPLKKLYALD 123
Query: 213 KELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLE 252
+ +S SIHVNPK+L+
Sbjct: 124 VRV---------NSVDPAWPIKPLPPNVNASIHVNPKFLK 154
Score = 57.8 bits (138), Expect = 2e-06
Identities = 42/193 (21%), Positives = 85/193 (43%), Gaps = 26/193 (13%)
Query: 845 ASVSGSAVPVPSTKDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLF 904
++ + + VP ++E ++ + + + GF D +++ H S+I+ L+
Sbjct: 1133 STAASQTISVPEEEEEEEEQ-------EQEEDDSVPDLTGFIL--DDMKQRHESIITKLY 1183
Query: 905 DDFPHHCSICSLKLKFQEQ--FDRHLEWHATREREHSGLIKA---SRWYLKSSDWV---- 955
C C ++ + + HL+WH + R + K RWY +DW+
Sbjct: 1184 TGI--QCYSCGMRFTASQTDVYADHLDWHYRQNRSEKDISKKVTHRRWYYSLTDWIEFEE 1241
Query: 956 VGKVESLSENEFADSV-DRYGKET-----DRNQEDAMVLADENQCLCVLCGELFEEFYCQ 1009
+ +E ++++F + V + ++T +R + AD LC +C E FE ++ +
Sbjct: 1242 IADLEERAKSQFFEKVHEEVVQKTQEAAKEREFQSVKAAADVVHELCEICQEQFEMYWEE 1301
Query: 1010 ENGEWMFKGAVYV 1022
E EW K A+ V
Sbjct: 1302 EEEEWHLKNAIRV 1314
>UniRef100_UPI000043663B UPI000043663B UniRef100 entry
Length = 1323
Score = 98.6 bits (244), Expect = 9e-19
Identities = 59/160 (36%), Positives = 84/160 (51%), Gaps = 9/160 (5%)
Query: 93 QELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLY 152
++ +Y+S+L +LTFNSKP I LTI+A EN + I A + A I + P +KLP LY
Sbjct: 8 EDACREYQSSLEDLTFNSKPHINMLTILAEENIQFTKDIVAIIEAQIAKAPPVEKLPVLY 67
Query: 153 LLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIE 212
L+DSIVKN+G Y++ FA L F + VD S+ L TW +FP + L A++
Sbjct: 68 LVDSIVKNVGGAYLEVFAKNLVNSFICVFEKVDEGTRKSLFKLRSTWDEIFPLKKLYALD 127
Query: 213 KELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLE 252
+ +S SIHVNPK+L+
Sbjct: 128 VRV---------NSVDPAWPIKPLPPNVNASIHVNPKFLK 158
Score = 52.4 bits (124), Expect = 7e-05
Identities = 50/239 (20%), Positives = 101/239 (41%), Gaps = 50/239 (20%)
Query: 798 IANLLNSLVAKGLIS-AETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPS 856
+ ++++ L++ G+I A T++TA PS T + VP
Sbjct: 1104 VNDVMSKLISTGIIKPAPTDSTADSPSASQT------------------------ISVPE 1139
Query: 857 TKDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSL 916
++E ++ + + + GF D +++ H S+I+ L+ C C +
Sbjct: 1140 EEEEEEEQ-------EQEEDDSVPDLTGFIL--DDMKQRHESIITKLYTGI--QCYSCGM 1188
Query: 917 KLKFQEQ--FDRHLEWHATREREHSGLIKA---SRWYLKSSDWV----VGKVESLSENEF 967
+ + + HL+WH + R + K RWY +DW+ + +E ++++F
Sbjct: 1189 RFTASQTDVYADHLDWHYRQNRSEKDISKKVTHRRWYYSLTDWIEFEEIADLEERAKSQF 1248
Query: 968 ADSVD----RYGKETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKGAVYV 1022
+ V + +E + +E V A + + +C E FE ++ +E EW K A+ V
Sbjct: 1249 FEKVHEEVVQKTQEAAKEREFQSVKAAAD-VVHEICQEQFEMYWEEEEEEWHLKNAIRV 1306
>UniRef100_Q6ZM95 Novel protein similar to human pre-mRNA cleavage complex II protein
Pcf11 [Brachydanio rerio]
Length = 1420
Score = 98.6 bits (244), Expect = 9e-19
Identities = 59/160 (36%), Positives = 84/160 (51%), Gaps = 9/160 (5%)
Query: 93 QELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLY 152
++ +Y+S+L +LTFNSKP I LTI+A EN + I A + A I + P +KLP LY
Sbjct: 8 EDACREYQSSLEDLTFNSKPHINMLTILAEENIQFTKDIVAIIEAQIAKAPPVEKLPVLY 67
Query: 153 LLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIE 212
L+DSIVKN+G Y++ FA L F + VD S+ L TW +FP + L A++
Sbjct: 68 LVDSIVKNVGGAYLEVFAKNLVNSFICVFEKVDEGTRKSLFKLRSTWDEIFPLKKLYALD 127
Query: 213 KELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLE 252
+ +S SIHVNPK+L+
Sbjct: 128 VRV---------NSVDPAWPIKPLPPNVNASIHVNPKFLK 158
Score = 53.9 bits (128), Expect = 3e-05
Identities = 58/283 (20%), Positives = 115/283 (40%), Gaps = 63/283 (22%)
Query: 798 IANLLNSLVAKGLIS-AETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPS 856
+ +L++ L++ G+I A T++TA PS T + VP
Sbjct: 1133 VNDLMSKLISTGIIKPAPTDSTADSPSASQT------------------------ISVPE 1168
Query: 857 TKDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSL 916
++E ++ + + + GF D +++ H S+I+ L+ C C +
Sbjct: 1169 EEEEEEEQ-------EQEEDDSVPDLTGFIL--DDMKQRHESIITKLYTGI--QCYSCGM 1217
Query: 917 KLKFQEQ--FDRHLEWHATREREHSGLIKA---SRWYLKSSDWV----VGKVESLSENEF 967
+ + + HL+WH + R + K RWY +DW+ + +E ++++F
Sbjct: 1218 RFTASQTDVYADHLDWHYRQNRSEKDISKKVTHRRWYYSLTDWIEFEEIADLEERAKSQF 1277
Query: 968 ADSVD----RYGKETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKGAVYVP 1023
+ V + +E + +E V A + + +C E FE ++ +E EW K A+ V
Sbjct: 1278 FEKVHEEVVQKTQEAAKEREFQSVKAAAD-VVHEICQEQFEMYWEEEEEEWHLKNAIRVD 1336
Query: 1024 N--------SDINDDMGIRDASTGGGPIIHARCLSENPVSSVL 1058
D + D + + L+ENP+++ L
Sbjct: 1337 EKTYHPSCYEDYKNTSSFMDTTPSPN-----KMLTENPLTAFL 1374
>UniRef100_Q5U393 Zgc:92226 [Brachydanio rerio]
Length = 1457
Score = 98.6 bits (244), Expect = 9e-19
Identities = 59/160 (36%), Positives = 84/160 (51%), Gaps = 9/160 (5%)
Query: 93 QELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLY 152
++ +Y+S+L +LTFNSKP I LTI+A EN + I A + A I + P +KLP LY
Sbjct: 8 EDACREYQSSLEDLTFNSKPHINMLTILAEENIQFTKDIVAIIEAQIAKAPPVEKLPVLY 67
Query: 153 LLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIE 212
L+DSIVKN+G Y++ FA L F + VD S+ L TW +FP + L A++
Sbjct: 68 LVDSIVKNVGGAYLEVFAKNLVNSFICVFEKVDEGTRKSLFKLRSTWDEIFPLKKLYALD 127
Query: 213 KELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLE 252
+ +S SIHVNPK+L+
Sbjct: 128 VRV---------NSVDPAWPIKPLPPNVNASIHVNPKFLK 158
Score = 60.8 bits (146), Expect = 2e-07
Identities = 60/285 (21%), Positives = 116/285 (40%), Gaps = 64/285 (22%)
Query: 798 IANLLNSLVAKGLIS-AETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPS 856
+ +L++ L++ G+I A T++TA PS T + VP
Sbjct: 1167 VNDLMSKLISTGIIKPAPTDSTADSPSASQT------------------------ISVPE 1202
Query: 857 TKDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSL 916
++E ++ + + + GF D +++ H S+I+ L+ C C +
Sbjct: 1203 EEEEEEEQ-------EQEEDDSVPDLTGFIL--DDMKQRHESIITKLYTGI--QCYSCGM 1251
Query: 917 KLKFQEQ--FDRHLEWHATREREHSGLIKA---SRWYLKSSDWV----VGKVESLSENEF 967
+ + + HL+WH + R + K RWY +DW+ + +E ++++F
Sbjct: 1252 RFTASQTDVYADHLDWHYRQNRSEKDISKKVTHRRWYYSLTDWIEFEEIADLEERAKSQF 1311
Query: 968 ADSV-DRYGKET-----DRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKGAVY 1021
+ V + ++T +R + AD LC +C E FE ++ +E EW K A+
Sbjct: 1312 FEKVHEEVVQKTQEAAKEREFQSVKAAADVVHELCEICQEQFEMYWEEEEEEWHLKNAIR 1371
Query: 1022 VPN--------SDINDDMGIRDASTGGGPIIHARCLSENPVSSVL 1058
V D + D + + L+ENP+++ L
Sbjct: 1372 VDEKTYHPSCYEDYKNTSSFMDTTPSPN-----KMLTENPLTAFL 1411
>UniRef100_UPI000027C40A UPI000027C40A UniRef100 entry
Length = 1517
Score = 98.2 bits (243), Expect = 1e-18
Identities = 61/156 (39%), Positives = 84/156 (53%), Gaps = 9/156 (5%)
Query: 98 QYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLYLLDSI 157
+Y+S+L +LTFNSKP I LTI+A EN + I A + A I + P+ +KLP LYL+DSI
Sbjct: 13 EYQSSLEDLTFNSKPHINMLTILAEENINFAKDIVAIIEAQISKAPTTEKLPVLYLVDSI 72
Query: 158 VKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEKELGF 217
VKNIG Y+ FA L F + VD S+ L TW VFP + L A++ +
Sbjct: 73 VKNIGGEYLAVFAKNLITSFICVFEKVDENTRKSLFKLRSTWDEVFPLKKLYALDVRVNS 132
Query: 218 TPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLER 253
T+ + SIHVNPK+L++
Sbjct: 133 VDPAWPIKPLPPTVNA---------SIHVNPKFLKQ 159
Score = 62.8 bits (151), Expect = 5e-08
Identities = 59/280 (21%), Positives = 121/280 (43%), Gaps = 70/280 (25%)
Query: 798 IANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPST 857
+ +LL+ L++ G+I A Q ++ TS + P S P
Sbjct: 1244 VNDLLSKLISNGIIKAA-----------------QPEATQTSGTAPAGS--------PLA 1278
Query: 858 KDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLK 917
+D+ +D + L ++ ++R+F + +++ S+++ L+ C +CS++
Sbjct: 1279 EDDEEDDPE----LLDNDVPDLRSFT-----IETMKQRFESIVTKLYSGT--QCGLCSMR 1327
Query: 918 LKFQEQ--FDRHLEWHATREREHSGLIKAS-----RWYLKSSDWV----VGKVESLSENE 966
++ + HL+WH + H+G + + RWY DW+ + +E ++++
Sbjct: 1328 FSSRQPDLYGDHLDWHY--RQNHNGKVPSKKVTHRRWYYGLRDWIEFEEIADLEERAKSQ 1385
Query: 967 FADSVDRYGKETDRNQEDAM--------VLADENQCLCVLCGELFEEFYCQENGEWMFKG 1018
F + + E +NQ A D+ + LC +C E FE ++ +E +W K
Sbjct: 1386 FFEKENEV--EVQKNQAAAKEKEIQCVKATKDQVEELCEICQEQFETYWVEEEEDWFLKN 1443
Query: 1019 AVYVPNSDINDDMGIRDASTGGGPIIHARCLSENPVSSVL 1058
A+ V DD DA+ + L+++P+SS +
Sbjct: 1444 AIRV------DDKSYIDATPSPN-----KLLTDHPLSSFI 1472
>UniRef100_UPI0000361313 UPI0000361313 UniRef100 entry
Length = 1279
Score = 97.8 bits (242), Expect = 2e-18
Identities = 63/159 (39%), Positives = 88/159 (54%), Gaps = 15/159 (9%)
Query: 98 QYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLYLLDSI 157
+Y+S+L +LTFNSKP I LTI+A EN + I A + A I + P +KLP LYL+DSI
Sbjct: 8 EYQSSLEDLTFNSKPHINMLTILAEENINFAKDIVAIIEAQISKAPPTEKLPLLYLVDSI 67
Query: 158 VKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEKELGF 217
VKN+G Y+ FA L F + VD S+ L TW VFP + L A++ +
Sbjct: 68 VKNVGGEYLAVFAKNLITSFICVFEKVDENTRKSLFKLRSTWDEVFPLKKLYALDVRV-- 125
Query: 218 TPAVNGSSSTSATLRSDSQSQRPP---PSIHVNPKYLER 253
S+ T+ ++ PP SIHVNPK+L++
Sbjct: 126 -----NSADTAWPIK-----PLPPTVNASIHVNPKFLKQ 154
Score = 57.0 bits (136), Expect = 3e-06
Identities = 90/409 (22%), Positives = 159/409 (38%), Gaps = 96/409 (23%)
Query: 659 TAKTEKPHLSKVPL----PRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNL 714
+ + ++P + PL P E P RF+ G + +MP P + + +
Sbjct: 904 SGQNQRPMRFETPLNQMGPMRFEGPGPRRFDGPMQAGPRF-DMPHQGGAPVYDPVHSQQV 962
Query: 715 PSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSAL-------PKIPPRRAAQP 767
P G P Q + L PP+ I PP P L + P+
Sbjct: 963 P---GRFPPQHN------LQPPIRPIV-PPMYENPMGPQQNLNMGQHFPEPVDPQFPVGQ 1012
Query: 768 PRTSALPPASSNLKSAAVQASSSANNALN-PIA------------NLLNSLVAKGLISAE 814
SA PA + SA+ S+ ++ PIA LL+ L++ G+I A
Sbjct: 1013 MAYSAQAPAFNQQGSASFYNPSAPGLSMQQPIAPTENHFGQMDVNELLSKLISNGIIKAA 1072
Query: 815 TETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPSTKDEVDDGAKTPISLSES 874
Q ++ TS + G + +GS P+ DE D L ++
Sbjct: 1073 -----------------QPEATLTSGT--GTAPAGS--PLAEEDDEDDP------ELLDN 1105
Query: 875 TSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQ--FDRHLEWHA 932
++R+F + ++ + SV++ L+ C +CS++ ++ + HL+WH
Sbjct: 1106 DVPDLRSFT-----IEAMKLRYESVVTKLYSGT--QCGLCSMRFSSRQPDLYGDHLDWHY 1158
Query: 933 TREREHSGLIKAS-----RWYLKSSDWV----VGKVESLSENEFADSVDRYGKETDRNQE 983
+ H+G + + RWY DW+ + +E +++ F + + E +NQ
Sbjct: 1159 --RQNHNGKVPSKKVTHRRWYYGLRDWIEFEEIADLEERAKSLFFEKENEV--EVQKNQ- 1213
Query: 984 DAMVLADENQCL----------CVLCGELFEEFYCQENGEWMFKGAVYV 1022
A E QC+ C +C E FE ++ +E +W K A+ V
Sbjct: 1214 -AAAKEKEVQCVKATKDQVGESCEICQEQFETYWVEEEEDWFLKNAIRV 1261
>UniRef100_UPI0000361312 UPI0000361312 UniRef100 entry
Length = 1363
Score = 97.8 bits (242), Expect = 2e-18
Identities = 63/159 (39%), Positives = 88/159 (54%), Gaps = 15/159 (9%)
Query: 98 QYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLYLLDSI 157
+Y+S+L +LTFNSKP I LTI+A EN + I A + A I + P +KLP LYL+DSI
Sbjct: 13 EYQSSLEDLTFNSKPHINMLTILAEENINFAKDIVAIIEAQISKAPPTEKLPLLYLVDSI 72
Query: 158 VKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEKELGF 217
VKN+G Y+ FA L F + VD S+ L TW VFP + L A++ +
Sbjct: 73 VKNVGGEYLAVFAKNLITSFICVFEKVDENTRKSLFKLRSTWDEVFPLKKLYALDVRV-- 130
Query: 218 TPAVNGSSSTSATLRSDSQSQRPP---PSIHVNPKYLER 253
S+ T+ ++ PP SIHVNPK+L++
Sbjct: 131 -----NSADTAWPIK-----PLPPTVNASIHVNPKFLKQ 159
Score = 59.3 bits (142), Expect = 6e-07
Identities = 83/407 (20%), Positives = 160/407 (38%), Gaps = 67/407 (16%)
Query: 659 TAKTEKPHLSKVPL----PRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNL 714
+ + ++P + PL P E P RF+ G + +MP P + + +
Sbjct: 913 SGQNQRPMRFETPLNQMGPMRFEGPGPRRFDGPMQAGPRF-DMPHQGGAPVYDPVHSQQV 971
Query: 715 PSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSAL-------PKIPPRRAAQP 767
P G P Q + L PP+ I PP P L + P+
Sbjct: 972 P---GRFPPQHN------LQPPIRPIV-PPMYENPMGPQQNLNMGQHFPEPVDPQFPVGQ 1021
Query: 768 PRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSL----VAKGLISAETETTAKVPS 823
SA PA + SA+ S+ ++ NLL SL ++ + +T P+
Sbjct: 1022 MAYSAQAPAFNQQGSASFYNPSAPGLSMQQPVNLLGSLNQSFPSQSAVPFGHQTPQIAPT 1081
Query: 824 EMLTRLEDQSDSITT-------SSSLPGASVSGSAVPVPSTKDEVDDGAKTPISLSESTS 876
E D ++ ++ ++ P A+++ P+ ++ + L ++
Sbjct: 1082 ENHFGQMDVNELLSKLISNGIIKAAQPEATLTSGTGTAPAGSPLAEEDDEDDPELLDNDV 1141
Query: 877 TEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQ--FDRHLEWHATR 934
++R+F + ++ + SV++ L+ C +CS++ ++ + HL+WH
Sbjct: 1142 PDLRSFT-----IEAMKLRYESVVTKLYSGT--QCGLCSMRFSSRQPDLYGDHLDWHY-- 1192
Query: 935 EREHSGLIKAS-----RWYLKSSDWV----VGKVESLSENEFADSVDRYGKETDRNQEDA 985
+ H+G + + RWY DW+ + +E +++ F + + E +NQ A
Sbjct: 1193 RQNHNGKVPSKKVTHRRWYYGLRDWIEFEEIADLEERAKSLFFEKENEV--EVQKNQ--A 1248
Query: 986 MVLADENQCL----------CVLCGELFEEFYCQENGEWMFKGAVYV 1022
E QC+ C +C E FE ++ +E +W K A+ V
Sbjct: 1249 AAKEKEVQCVKATKDQVGESCEICQEQFETYWVEEEEDWFLKNAIRV 1295
>UniRef100_UPI000036EFCD UPI000036EFCD UniRef100 entry
Length = 1654
Score = 97.8 bits (242), Expect = 2e-18
Identities = 60/168 (35%), Positives = 86/168 (50%), Gaps = 8/168 (4%)
Query: 93 QELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLY 152
++ Y+S+L +LTFNSKP I LTI+A EN + I + + A + PS +KLP +Y
Sbjct: 115 EDACRDYQSSLEDLTFNSKPHINMLTILAEENLPFAKEIVSLIEAQTAKAPSSEKLPVMY 174
Query: 153 LLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIE 212
L+DSIVKN+GR Y+ F L F + VD S+ L TW +FP + L A++
Sbjct: 175 LMDSIVKNVGREYLTAFTKNLVATFICVFEKVDENTRKSLFKLRSTWDEIFPLKKLYALD 234
Query: 213 KELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLERQRLQQSS 260
V +S A SIHVNPK+L + + S+
Sbjct: 235 --------VRVNSLDPAWPIKPLPPNVNTSSIHVNPKFLNKSPEEPST 274
Score = 57.4 bits (137), Expect = 2e-06
Identities = 111/547 (20%), Positives = 196/547 (35%), Gaps = 128/547 (23%)
Query: 534 GNRSLLPKATIGLAGNLGQLHSVGDESPSRESSLRQQSPPPVSVHLPHLMQNLAEQDHPQ 593
G SLLP+ GL G G + +P + R PP V P D PQ
Sbjct: 1143 GQPSLLPRFD-GLHGQPGPRF---ERTPGQPGPQRFDGPPGQQVQ-PRFDGVPQRFDGPQ 1197
Query: 594 THKVSKFSGGLHSQNIKDISPALPPNIQVGKFHRSQLRDLQSGSFSSMTFQPSHKQQLGS 653
+ S+F +P +Q +F + L+S SF+ + G+
Sbjct: 1198 HQQASRFD--------------IPLGLQGTRFDNHPSQRLESVSFNQTG---PYNDPPGN 1240
Query: 654 SHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITN---------RLP 704
+ + + ++ + P F G G + + P+ L
Sbjct: 1241 AFNAPSQGLQFQRHEQI-----FDSPQGPNFNGPHGPGNQSFSNPLNRASGHYFDEKNLQ 1295
Query: 705 TSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRA 764
+S+ + GN+P+ + V Q+S +S +A P + G+ LP P
Sbjct: 1296 SSQFGNFGNIPAPMTVGNIQASQQV-------LSGVAQPVAFGQ---GQQFLPVHPQNPG 1345
Query: 765 AQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSE 824
+ ALP A + + V + L + L+ G++
Sbjct: 1346 FVQNPSGALPKAYPDNHLSQVDVNE-----------LFSKLLKTGILKLS---------- 1384
Query: 825 MLTRLEDQSDSITTSSSLPGASVSGSAVPVPSTKDEVDDGAKTPISLSESTSTEIRNFIG 884
Q+DS TT S +A P P +++ ++ P ++ NF
Sbjct: 1385 -------QTDSATTQVS------EVTAQPPPEEEEDQNEDQDVP---------DLTNFTV 1422
Query: 885 FEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQ--FDRHLEWHATR---EREHS 939
E K + + SVI+ L+ C C ++ + + HL+WH + E++ S
Sbjct: 1423 EELK-----QRYDSVINRLYTGI--QCYSCGMRFTTSQTDVYADHLDWHYRQNRTEKDVS 1475
Query: 940 GLIKASRWYLKSSDWV----VGKVESLSENEFADSVDRY----GKETDRNQEDAMVLADE 991
+ RWY +DW+ + +E ++++F + V +E + +E V A
Sbjct: 1476 RKVTHRRWYYSLTDWIEFEEIADLEERAKSQFFEKVHEEVVLKTQEAAKEKEFQSVPAGP 1535
Query: 992 NQCL--CVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCL 1049
+ C +C E FE+++ +E EW K A+ V G I H C
Sbjct: 1536 AGAVESCEICQEQFEQYWDEEEEEWHLKNAIRV-----------------DGKIYHPSCY 1578
Query: 1050 SENPVSS 1056
+ P +S
Sbjct: 1579 EDYPNTS 1585
>UniRef100_O94913 Pre-mRNA cleavage complex II protein Pcf11 [Homo sapiens]
Length = 1654
Score = 97.8 bits (242), Expect = 2e-18
Identities = 60/168 (35%), Positives = 86/168 (50%), Gaps = 8/168 (4%)
Query: 93 QELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLY 152
++ Y+S+L +LTFNSKP I LTI+A EN + I + + A + PS +KLP +Y
Sbjct: 115 EDACRDYQSSLEDLTFNSKPHINMLTILAEENLPFAKEIVSLIEAQTAKAPSSEKLPVMY 174
Query: 153 LLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIE 212
L+DSIVKN+GR Y+ F L F + VD S+ L TW +FP + L A++
Sbjct: 175 LMDSIVKNVGREYLTAFTKNLVATFICVFEKVDENTRKSLFKLRSTWDEIFPLKKLYALD 234
Query: 213 KELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLERQRLQQSS 260
V +S A SIHVNPK+L + + S+
Sbjct: 235 --------VRVNSLDPAWPIKPLPPNVNTSSIHVNPKFLNKSPEEPST 274
Score = 58.2 bits (139), Expect = 1e-06
Identities = 112/555 (20%), Positives = 206/555 (36%), Gaps = 113/555 (20%)
Query: 534 GNRSLLPKATIGLAGNLGQLHSVGDESPSRESSLRQQSPPPVSVHLPHLMQNLAEQDHPQ 593
G SLLP+ GL G G + +P + R PP V P D PQ
Sbjct: 1143 GQPSLLPRFD-GLHGQPGPRF---ERTPGQPGPQRFDGPPGQQVQ-PRFDGVPQRFDGPQ 1197
Query: 594 THKVSKFSGGLHSQNIKDISPALPPNIQVGKFHRSQLRDLQSGSFSSMTFQPSHKQQLGS 653
+ S+F +P +Q +F + L+S SF+ + G+
Sbjct: 1198 HQQASRFD--------------IPLGLQGTRFDNHPSQRLESVSFNQTG---PYNDPPGN 1240
Query: 654 SHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITN---------RLP 704
+ + + ++ + P F G G + + P+ L
Sbjct: 1241 AFNAPSQGLQFQRHEQI-----FDSPQGPNFNGPHGPGNQSFSNPLNRASGHYFDEKNLQ 1295
Query: 705 TSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRA 764
+S+ + GN+P+ + V Q+S +S +A P + G+ LP P
Sbjct: 1296 SSQFGNFGNIPAPMTVGNIQASQQV-------LSGVAQPVAFGQ---GQQFLPVHPQNPG 1345
Query: 765 AQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSE 824
+ ALP A + + V + L + L+ G++
Sbjct: 1346 FVQNPSGALPKAYPDNHLSQVDVNE-----------LFSKLLKTGILKLS---------- 1384
Query: 825 MLTRLEDQSDSITTSSSLPGASVSGSAVPVPSTKDEVDDGAKTPISLSESTSTEIRNFIG 884
Q+DS TT S +A P P +++ ++ P ++ NF
Sbjct: 1385 -------QTDSATTQVS------EVTAQPPPEEEEDQNEDQDVP---------DLTNFTV 1422
Query: 885 FEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQ--FDRHLEWHATR---EREHS 939
E K + + SVI+ L+ C C ++ + + HL+WH + E++ S
Sbjct: 1423 EELK-----QRYDSVINRLYTGI--QCYSCGMRFTTSQTDVYADHLDWHYRQNRTEKDVS 1475
Query: 940 GLIKASRWYLKSSDWV----VGKVESLSENEFADSVDRY----GKETDRNQEDAMVLADE 991
+ RWY +DW+ + +E ++++F + V +E + +E V A
Sbjct: 1476 RKVTHRRWYYSLTDWIEFEEIADLEERAKSQFFEKVHEEVVLKTQEAAKEKEFQSVPAGP 1535
Query: 992 NQCL--CVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDM--GIRDASTGGGPIIHAR 1047
+ C +C E FE+++ +E EW K A+ V + ++ S+ ++
Sbjct: 1536 AGAVESCEICQEQFEQYWDEEEEEWHLKNAIRVDGKIYHPSCYEDYQNTSSFDCTPSPSK 1595
Query: 1048 CLSENPVSSVLNMVR 1062
ENP++ +LN+V+
Sbjct: 1596 TPVENPLNIMLNIVK 1610
>UniRef100_UPI00001CE95F UPI00001CE95F UniRef100 entry
Length = 1635
Score = 97.4 bits (241), Expect = 2e-18
Identities = 59/161 (36%), Positives = 83/161 (50%), Gaps = 8/161 (4%)
Query: 93 QELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLY 152
++ Y+S+L +LTFNSKP I LTI+A EN + I + + A + PS +KLP +Y
Sbjct: 100 EDACRDYQSSLEDLTFNSKPHINMLTILAEENLPFAKEIVSLIEAQTAKAPSSEKLPVMY 159
Query: 153 LLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIE 212
L+DSIVKN+GR Y+ F L F + VD S+ L TW +FP + L A++
Sbjct: 160 LMDSIVKNVGREYLTAFTKNLVATFICVFEKVDENTRKSLFKLRSTWDEIFPLKKLYALD 219
Query: 213 KELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLER 253
V +S A SIHVNPK+L +
Sbjct: 220 --------VRVNSLDPAWPIKPLPPNVNTSSIHVNPKFLNK 252
Score = 53.1 bits (126), Expect = 4e-05
Identities = 43/193 (22%), Positives = 89/193 (45%), Gaps = 19/193 (9%)
Query: 887 FKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQ--FDRHLEWHATR---EREHSGL 941
F + +++ + SVI+ L+ C C ++ + + HL+WH + E++ S
Sbjct: 1401 FTIEELKQRYDSVINRLYTGI--QCYSCGMRFTTSQTDVYADHLDWHYRQNRTEKDVSRK 1458
Query: 942 IKASRWYLKSSDWV----VGKVESLSENEFADSVDRY----GKETDRNQEDAMVLADENQ 993
+ RWY +DW+ + +E ++++F + V +E + +E V A
Sbjct: 1459 VTHRRWYYSLTDWIEFEEIADLEERAKSQFFEKVHEEVVLKTQEAAKEKEFQSVPAGPAG 1518
Query: 994 CL--CVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDM--GIRDASTGGGPIIHARCL 1049
+ C +C E FE+++ +E EW K A+ V + ++ S+ ++
Sbjct: 1519 AVESCEICQEQFEQYWDEEEEEWHLKNAIRVDGKIYHPSCYEDYQNTSSFDCTPSPSKTP 1578
Query: 1050 SENPVSSVLNMVR 1062
ENP++ +LN+V+
Sbjct: 1579 VENPLNIMLNIVK 1591
>UniRef100_Q69ZY3 MKIAA0824 protein [Mus musculus]
Length = 1641
Score = 97.4 bits (241), Expect = 2e-18
Identities = 59/161 (36%), Positives = 83/161 (50%), Gaps = 8/161 (4%)
Query: 93 QELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLY 152
++ Y+S+L +LTFNSKP I LTI+A EN + I + + A + PS +KLP +Y
Sbjct: 104 EDACRDYQSSLEDLTFNSKPHINMLTILAEENLPFAKEIVSLIEAQTAKAPSSEKLPVMY 163
Query: 153 LLDSIVKNIGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIE 212
L+DSIVKN+GR Y+ F L F + VD S+ L TW +FP + L A++
Sbjct: 164 LMDSIVKNVGREYLTAFTKNLVATFICVFEKVDENTRKSLFKLRSTWDEIFPLKKLYALD 223
Query: 213 KELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVNPKYLER 253
V +S A SIHVNPK+L +
Sbjct: 224 --------VRVNSLDPAWPIKPLPPNVNTSSIHVNPKFLNK 256
Score = 53.1 bits (126), Expect = 4e-05
Identities = 43/193 (22%), Positives = 89/193 (45%), Gaps = 19/193 (9%)
Query: 887 FKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQ--FDRHLEWHATR---EREHSGL 941
F + +++ + SVI+ L+ C C ++ + + HL+WH + E++ S
Sbjct: 1407 FTIEELKQRYDSVINRLYTGI--QCYSCGMRFTTSQTDVYADHLDWHYRQNRTEKDVSRK 1464
Query: 942 IKASRWYLKSSDWV----VGKVESLSENEFADSVDRY----GKETDRNQEDAMVLADENQ 993
+ RWY +DW+ + +E ++++F + V +E + +E V A
Sbjct: 1465 VTHRRWYYSLTDWIEFEEIADLEERAKSQFFEKVHEEVVLKTQEAAKEKEFQSVPAGPAG 1524
Query: 994 CL--CVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDM--GIRDASTGGGPIIHARCL 1049
+ C +C E FE+++ +E EW K A+ V + ++ S+ ++
Sbjct: 1525 AVESCEICQEQFEQYWDEEEEEWHLKNAIRVDGKIYHPSCYEDYQNTSSFDCTPSPSKTP 1584
Query: 1050 SENPVSSVLNMVR 1062
ENP++ +LN+V+
Sbjct: 1585 VENPLNIMLNIVK 1597
>UniRef100_Q9ST17 S-locus protein 4 [Brassica campestris]
Length = 413
Score = 97.1 bits (240), Expect = 3e-18
Identities = 55/193 (28%), Positives = 85/193 (43%), Gaps = 25/193 (12%)
Query: 871 LSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEW 930
LS + E + IG +F + H SV+ L+ D P CS C ++ K QE+ +H++W
Sbjct: 215 LSVLNNNEKESDIGLDFDKINLNVRHESVVKSLYSDMPRQCSSCGVRFKCQEEHSKHMDW 274
Query: 931 HATRER-----------EHSGLIKASR-WYLKSSDWVVGKVESLSENEFADSVDRYGKET 978
H + R S +K SR W+ S W+ + E + +
Sbjct: 275 HVRKNRLAKDATKAMTTRASQKLKKSRDWFASLSLWLSAATGAAIEGAKPAFGETQKMKE 334
Query: 979 DRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDAST 1038
+ Q+ V ADENQ +C LC E EEF+ E +WM++ A Y+
Sbjct: 335 EEKQQQRNVPADENQKMCALCLESLEEFFSHEEDDWMYRDAAYL-------------HMN 381
Query: 1039 GGGPIIHARCLSE 1051
G GP++H C+ E
Sbjct: 382 GTGPVVHVNCMPE 394
>UniRef100_Q9C710 Hypothetical protein F28G11.6 [Arabidopsis thaliana]
Length = 416
Score = 96.7 bits (239), Expect = 3e-18
Identities = 62/213 (29%), Positives = 95/213 (44%), Gaps = 31/213 (14%)
Query: 853 PVPSTKDEVD-----DGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDF 907
P+ +K+ D + K +L S S + + F+ P + H SVI L+ D
Sbjct: 194 PIVLSKELTDLLSLLNNEKEKKTLEASNSDSLPVGLSFD-NPSSLNVRHESVIKSLYSDM 252
Query: 908 PHHCSICSLKLKFQEQFDRHLEWHATREREHSGLI-------KASRWYLKSSDWVVGKV- 959
P CS C L+ K QE+ +H++WH + R K+ W +S W+
Sbjct: 253 PRQCSSCGLRFKCQEEHSKHMDWHVRKNRSVKTTTRLGQQPKKSRGWLASASLWLCAATG 312
Query: 960 -ESLSENEFADSVDRYGKETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKG 1018
E++ F + + K D + MV ADE+Q C LC E FEEF+ E+ +WM+K
Sbjct: 313 GETVEVASFGGEMQK-KKGKDEEPKQLMVPADEDQKNCALCVEPFEEFFSHEDDDWMYKD 371
Query: 1019 AVYVPNSDINDDMGIRDASTGGGPIIHARCLSE 1051
AVY+ T G I+H +C+ E
Sbjct: 372 AVYL---------------TKNGRIVHVKCMPE 389
>UniRef100_Q8VZG9 At1g66500/F28G11_6 [Arabidopsis thaliana]
Length = 416
Score = 96.7 bits (239), Expect = 3e-18
Identities = 62/213 (29%), Positives = 95/213 (44%), Gaps = 31/213 (14%)
Query: 853 PVPSTKDEVD-----DGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDF 907
P+ +K+ D + K +L S S + + F+ P + H SVI L+ D
Sbjct: 194 PIVLSKELTDLLSLLNNEKEKKTLEASNSDSLPVGLSFD-NPSSLNVRHESVIKSLYSDM 252
Query: 908 PHHCSICSLKLKFQEQFDRHLEWHATREREHSGLI-------KASRWYLKSSDWVVGKV- 959
P CS C L+ K QE+ +H++WH + R K+ W +S W+
Sbjct: 253 PRQCSSCGLRFKCQEEHSKHMDWHVRKNRSVKTTTRLGQQPKKSRGWLASASLWLCAATG 312
Query: 960 -ESLSENEFADSVDRYGKETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKG 1018
E++ F + + K D + MV ADE+Q C LC E FEEF+ E+ +WM+K
Sbjct: 313 GETVEVASFGGEMQK-KKGKDEEPKQLMVPADEDQKNCALCVEPFEEFFSHEDDDWMYKD 371
Query: 1019 AVYVPNSDINDDMGIRDASTGGGPIIHARCLSE 1051
AVY+ T G I+H +C+ E
Sbjct: 372 AVYL---------------TKNGRIVHVKCMPE 389
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,829,958,329
Number of Sequences: 2790947
Number of extensions: 81878680
Number of successful extensions: 266357
Number of sequences better than 10.0: 2022
Number of HSP's better than 10.0 without gapping: 119
Number of HSP's successfully gapped in prelim test: 2018
Number of HSP's that attempted gapping in prelim test: 255884
Number of HSP's gapped (non-prelim): 7642
length of query: 1065
length of database: 848,049,833
effective HSP length: 138
effective length of query: 927
effective length of database: 462,899,147
effective search space: 429107509269
effective search space used: 429107509269
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0359.12


