

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0358.5
(189 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
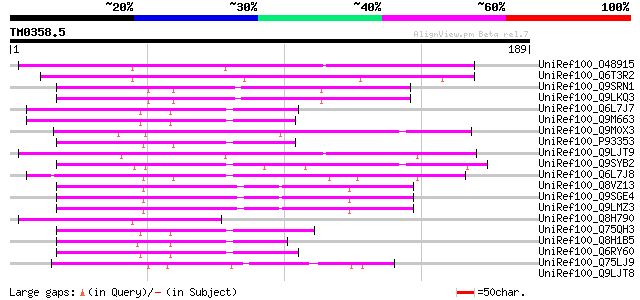
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O48915 Non-race specific disease resistance protein [A... 66 4e-10
UniRef100_Q6T3R2 NDR1-like protein [Nicotiana benthamiana] 63 3e-09
UniRef100_Q9SRN1 T19F11.5 protein [Arabidopsis thaliana] 57 2e-07
UniRef100_Q9LKQ3 NDR1/HIN1-Like protein 2 [Arabidopsis thaliana] 57 2e-07
UniRef100_Q6L7J7 Harpin inducing protein 1-like 18 [Nicotiana ta... 56 4e-07
UniRef100_Q9M663 Harpin inducing protein [Nicotiana tabacum] 56 5e-07
UniRef100_Q9M0X3 Hypothetical protein AT4g05220 [Arabidopsis tha... 54 3e-06
UniRef100_P93353 Hin1 protein [Nicotiana tabacum] 54 3e-06
UniRef100_Q9LJT9 Non-race specific disease resistance protein-li... 53 4e-06
UniRef100_Q9SYB2 T13M11.12 protein [Arabidopsis thaliana] 50 4e-05
UniRef100_Q6L7J8 Harpin inducing protein 1-like 9 [Nicotiana tab... 49 7e-05
UniRef100_Q8VZ13 Hypothetical protein At1g08150 [Arabidopsis tha... 48 1e-04
UniRef100_Q9SGE4 T23G18.2 [Arabidopsis thaliana] 48 1e-04
UniRef100_Q9LMZ3 T6D22.24 [Arabidopsis thaliana] 48 1e-04
UniRef100_Q8H790 Hypothetical protein [Arabidopsis thaliana] 48 1e-04
UniRef100_Q75QH3 Hin1 like protein [Capsicum chinense] 48 1e-04
UniRef100_Q8H1B5 Hin1-like protein [Solanum tuberosum] 48 1e-04
UniRef100_Q6RY60 Hin1-like protein [Solanum tuberosum] 47 2e-04
UniRef100_Q75LJ9 Putative harpin inducing protein [Oryza sativa] 46 6e-04
UniRef100_Q9LJT8 Non-race specific disease resistance protein-li... 45 0.001
>UniRef100_O48915 Non-race specific disease resistance protein [Arabidopsis thaliana]
Length = 219
Score = 66.2 bits (160), Expect = 4e-10
Identities = 48/178 (26%), Positives = 83/178 (45%), Gaps = 13/178 (7%)
Query: 4 GKRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSI-----EQHPGENGSIFYSLE 58
G+ + FI GL L LWL+LR P S+ I + + +N ++ + +
Sbjct: 11 GRNCCTCCLSFIFTAGLTSLFLWLSLRADKPKCSIQNFFIPALGKDPNSRDNTTLNFMVR 70
Query: 59 IENPNKDSSIYYDDIILSF-------LYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNAR 111
+NPNKD IYYDD+ L+F + VG T+ F+QG K + V
Sbjct: 71 CDNPNKDKGIYYDDVHLNFSTINTTKINSSALVLVGNYTVPKFYQGHKKKAKKWGQVKPL 130
Query: 112 HGAFKPLFKAISNATAELKVALTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKLSRKK 169
+ L + N +A ++ L T+ ++K K+K +G+ + + ++ DG ++KK
Sbjct: 131 NNQ-TVLRAVLPNGSAVFRLDLKTQVRFKIVFWKTKRYGVEVGADVEVNGDGVKAQKK 187
>UniRef100_Q6T3R2 NDR1-like protein [Nicotiana benthamiana]
Length = 205
Score = 63.2 bits (152), Expect = 3e-09
Identities = 47/169 (27%), Positives = 75/169 (43%), Gaps = 11/169 (6%)
Query: 12 MEFIGFLGLLVLCLWLALRPKSPSYSVVFLSI--------EQHPGENGSIFYSLEIENPN 63
++FI GL L +WL+LR PS S+ + N ++ + L + N
Sbjct: 15 IQFILTAGLTALFIWLSLRTTKPSCSLDNFYLPALNNSDNSNSTRSNHTLSFQLNLNNKM 74
Query: 64 KDSSIYYDDIILSFLYGQQED-KVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAI 122
KD + YDDI LSF YG +G T+ F+QG K + + + K +
Sbjct: 75 KDKGVRYDDIKLSFYYGTNTSFPIGNYTVPGFYQGHDKKAHKKGILETQKMPWNDALKMV 134
Query: 123 SNAT-AELKVALTTRFQYKTWGIKSKFHGLHLQGI-LPIDSDGKLSRKK 169
SN + +V + TR +YK +K H ++ + +D GK S +K
Sbjct: 135 SNGSKVVFRVDVATRVRYKVILWYTKRHNFTVENSKVKVDGSGKSSAQK 183
>UniRef100_Q9SRN1 T19F11.5 protein [Arabidopsis thaliana]
Length = 240
Score = 57.4 bits (137), Expect = 2e-07
Identities = 39/140 (27%), Positives = 64/140 (44%), Gaps = 13/140 (9%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHPGE-NGSIFYSLE----IENPNKDSSIYYDD 72
LG+ L LWL RP + + V ++ + + N ++ YSL+ I NPN+ +YYD+
Sbjct: 66 LGVAALILWLIFRPNAVKFYVADANLNRFSFDPNNNLHYSLDLNFTIRNPNQRVGVYYDE 125
Query: 73 IILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARH------GAFKPLFKAISNAT 126
+S YG Q + G + SF+QG T + + ++ GA L +
Sbjct: 126 FSVSGYYGDQ--RFGSANVSSFYQGHKNTTVILTKIEGQNLVVLGDGARTDLKDDEKSGI 183
Query: 127 AELKVALTTRFQYKTWGIKS 146
+ L ++K W IKS
Sbjct: 184 YRINAKLRLSVRFKFWFIKS 203
>UniRef100_Q9LKQ3 NDR1/HIN1-Like protein 2 [Arabidopsis thaliana]
Length = 240
Score = 57.0 bits (136), Expect = 2e-07
Identities = 39/140 (27%), Positives = 64/140 (44%), Gaps = 13/140 (9%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHPGE-NGSIFYSLE----IENPNKDSSIYYDD 72
LG+ L LWL RP + + V ++ + + N ++ YSL+ I NPN+ +YYD+
Sbjct: 66 LGVAALILWLIFRPNAVKFYVADANLNRFSFDPNNNLHYSLDLNFTIRNPNQRVGVYYDE 125
Query: 73 IILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARH------GAFKPLFKAISNAT 126
+S YG Q + G + SF+QG T + + ++ GA L +
Sbjct: 126 FSVSGYYGDQ--RFGSANVSSFYQGHKNTTVILTKIEGQNLVVLGDGARTDLKDDEKSGI 183
Query: 127 AELKVALTTRFQYKTWGIKS 146
+ L ++K W IKS
Sbjct: 184 YRIDAKLRLSVRFKFWFIKS 203
>UniRef100_Q6L7J7 Harpin inducing protein 1-like 18 [Nicotiana tabacum]
Length = 229
Score = 56.2 bits (134), Expect = 4e-07
Identities = 35/105 (33%), Positives = 51/105 (48%), Gaps = 8/105 (7%)
Query: 7 FYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQH--PGENGSIFYSL----EIE 60
F+ + + LG++ L LWL LRP + V ++ Q N +IFY L I
Sbjct: 45 FFKIIFTLLIILGVIALVLWLVLRPNKVKFYVTDATLTQFDLSNTNNTIFYDLALNMTIR 104
Query: 61 NPNKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVY 105
NPNK IYYD I +Y Q ++ T + F+QG T S++
Sbjct: 105 NPNKRIGIYYDSIEARAMY--QGERFHSTNLKPFYQGHKNTSSLH 147
>UniRef100_Q9M663 Harpin inducing protein [Nicotiana tabacum]
Length = 229
Score = 55.8 bits (133), Expect = 5e-07
Identities = 36/104 (34%), Positives = 50/104 (47%), Gaps = 8/104 (7%)
Query: 7 FYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQH--PGENGSIFYSL----EIE 60
F+ + + LG++ L LWL LRP + V ++ Q N +IFY L I
Sbjct: 45 FFKIIFTLLIILGVIALVLWLVLRPNKVKFYVTDATLTQFDLSTTNNTIFYDLALNMTIR 104
Query: 61 NPNKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSV 104
NPNK IYYD I LY Q ++ T + F+QG T S+
Sbjct: 105 NPNKRIGIYYDSIEARALY--QGERFDSTNLEPFYQGHKNTSSL 146
>UniRef100_Q9M0X3 Hypothetical protein AT4g05220 [Arabidopsis thaliana]
Length = 226
Score = 53.5 bits (127), Expect = 3e-06
Identities = 44/162 (27%), Positives = 68/162 (41%), Gaps = 12/162 (7%)
Query: 17 FLGLLVLCLWLALRPKSPSYSV---VFLSIEQHPG-ENGSIFYSLEIENPNKDSSIYYDD 72
F+G++ LWL+LRP P + + V ++Q G EN I +++ I NPN+ +Y+D
Sbjct: 56 FVGVIAFILWLSLRPHRPRFHIQDFVVQGLDQPTGVENARIAFNVTILNPNQHMGVYFDS 115
Query: 73 IILSFLYGQQEDKVGETTIGSFHQG------TGKTRSVYDTVNARHGAFKPLFKAISNAT 126
+ S Y Q + F Q TG TVN+ +A
Sbjct: 116 MEGSIYYKDQRVGLIPLLNPFFQQPTNTTIVTGTLTGASLTVNSNRWTEFSNDRAQGTVG 175
Query: 127 AELKVALTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKLSRK 168
L + T RF+ W SK H +H + + DG + K
Sbjct: 176 FRLDIVSTIRFKLHRW--ISKHHRMHANCNIVVGRDGLILPK 215
>UniRef100_P93353 Hin1 protein [Nicotiana tabacum]
Length = 221
Score = 53.5 bits (127), Expect = 3e-06
Identities = 35/93 (37%), Positives = 46/93 (48%), Gaps = 8/93 (8%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHP--GENGSIFYSLE----IENPNKDSSIYYD 71
LG++ L LWL LRP + V ++ Q N +IFY L I NPNK IYYD
Sbjct: 48 LGVIALVLWLVLRPNKVKFYVTDATLTQFDLSTTNNTIFYDLALNMTIRNPNKRIGIYYD 107
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKTRSV 104
I LY Q ++ T + F+QG T S+
Sbjct: 108 SIEARALY--QGERFDSTNLEPFYQGHKNTSSL 138
>UniRef100_Q9LJT9 Non-race specific disease resistance protein-like [Arabidopsis
thaliana]
Length = 255
Score = 53.1 bits (126), Expect = 4e-06
Identities = 46/180 (25%), Positives = 79/180 (43%), Gaps = 14/180 (7%)
Query: 4 GKRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVV-----FLSIEQHPGENGSIFYSLE 58
G++ +FI L L LWL+LR K P S+ LS +N ++ + +
Sbjct: 11 GRKCCTCFFKFIFTTRLGALILWLSLRAKKPKCSIQNFYIPALSKNLSSRDNTTLNFMVR 70
Query: 59 IENPNKDSSIYYDDIILSF-------LYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNAR 111
+NPNKD IYYDD+ L+F V T+ F+QG K + V
Sbjct: 71 CDNPNKDKGIYYDDVHLTFSTINTTTTNSSDLVLVANYTVPKFYQGHKKKAKKWGQVWPL 130
Query: 112 HGAFKPLFKAISNATAELKVALTTRFQYKTWGIKSK-FHGLHLQGILPIDSDGKLSRKKK 170
+ L + N +A ++ L T ++K K+K + + + + ++ DG + +K+
Sbjct: 131 NNQ-TVLRAVLPNGSAVFRLDLKTHVRFKIVFWKTKWYRRIKVGADVEVNGDGVKANEKE 189
>UniRef100_Q9SYB2 T13M11.12 protein [Arabidopsis thaliana]
Length = 224
Score = 49.7 bits (117), Expect = 4e-05
Identities = 47/171 (27%), Positives = 72/171 (41%), Gaps = 18/171 (10%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIE--QHPG--ENGSIFYSLEIENPNKDSSIYYDDI 73
LG++ LW++L+P P + SI P E I + + NPN++ IYYD +
Sbjct: 55 LGIITFILWISLQPHRPRVHIRGFSISGLSRPDGFETSHISFKITAHNPNQNVGIYYDSM 114
Query: 74 ILSFLYGQQEDKVGETTI-GSFHQGTGKTRSVYD-------TVNARHGAFKPLFKAISNA 125
S Y +E ++G T + F+Q T S+ VN +
Sbjct: 115 EGSVYY--KEKRIGSTKLTNPFYQDPKNTSSIDGALSRPAMAVNKDRWMEMERDRNQGKI 172
Query: 126 TAELKVALTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKL--SRKKKKYPL 174
LKV RF+ TW SK H ++ + I DG L + K K+ P+
Sbjct: 173 MFRLKVRSMIRFKVYTW--HSKSHKMYASCYIEIGWDGMLLSATKDKRCPV 221
>UniRef100_Q6L7J8 Harpin inducing protein 1-like 9 [Nicotiana tabacum]
Length = 229
Score = 48.9 bits (115), Expect = 7e-05
Identities = 47/174 (27%), Positives = 76/174 (43%), Gaps = 17/174 (9%)
Query: 7 FYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHP--GENGSIFYSLE----IE 60
F +F + +G+ L LWL LRP + V ++ Q N +I Y L I
Sbjct: 46 FQIFFTILV-IIGVAALVLWLVLRPNKVKFYVTDATLTQFDLSTTNNTINYDLALNMTIR 104
Query: 61 NPNKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAF----- 115
NPNK IYYD I +Y Q ++ + F+QG T S++ +
Sbjct: 105 NPNKRIGIYYDSIEARAMY--QGERFASQNLEPFYQGHKNTSSLHPVFKGQSLVLLGDRE 162
Query: 116 KPLFKAISNA-TAELKVALTTRFQYKTWGIKSK--FHGLHLQGILPIDSDGKLS 166
K + NA E++V L+ R + K IK++ + +P++S+G+ S
Sbjct: 163 KTNYNNEKNAGVYEMEVKLSMRIRLKFGWIKTRKIKPKIECDFKVPLESNGRSS 216
>UniRef100_Q8VZ13 Hypothetical protein At1g08150 [Arabidopsis thaliana]
Length = 221
Score = 48.1 bits (113), Expect = 1e-04
Identities = 40/142 (28%), Positives = 60/142 (42%), Gaps = 15/142 (10%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHP------GENGSIFYSLEIENPNKDSSIYYD 71
+GL +L +L LRPK Y+V S+++ N Y ++ NP K S+ Y
Sbjct: 51 VGLAILITYLTLRPKRLIYTVEAASVQEFAIGNNDDHINAKFSYVIKSYNPEKHVSVRYH 110
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAI------SNA 125
+ +S + Q V I F Q K + +T H F A S
Sbjct: 111 SMRISTAHHNQS--VAHKNISPFKQRP-KNETRIETQLVSHNVALSKFNARDLRAEKSKG 167
Query: 126 TAELKVALTTRFQYKTWGIKSK 147
T E++V +T R YKTW +S+
Sbjct: 168 TIEMEVYITARVSYKTWIFRSR 189
>UniRef100_Q9SGE4 T23G18.2 [Arabidopsis thaliana]
Length = 2658
Score = 48.1 bits (113), Expect = 1e-04
Identities = 40/142 (28%), Positives = 60/142 (42%), Gaps = 15/142 (10%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHP------GENGSIFYSLEIENPNKDSSIYYD 71
+GL +L +L LRPK Y+V S+++ N Y ++ NP K S+ Y
Sbjct: 51 VGLAILITYLTLRPKRLIYTVEAASVQEFAIGNNDDHINAKFSYVIKSYNPEKHVSVRYH 110
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAI------SNA 125
+ +S + Q V I F Q K + +T H F A S
Sbjct: 111 SMRISTAHHNQS--VAHKNISPFKQRP-KNETRIETQLVSHNVALSKFNARDLRAEKSKG 167
Query: 126 TAELKVALTTRFQYKTWGIKSK 147
T E++V +T R YKTW +S+
Sbjct: 168 TIEMEVYITARVSYKTWIFRSR 189
>UniRef100_Q9LMZ3 T6D22.24 [Arabidopsis thaliana]
Length = 2621
Score = 48.1 bits (113), Expect = 1e-04
Identities = 40/142 (28%), Positives = 60/142 (42%), Gaps = 15/142 (10%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHP------GENGSIFYSLEIENPNKDSSIYYD 71
+GL +L +L LRPK Y+V S+++ N Y ++ NP K S+ Y
Sbjct: 51 VGLAILITYLTLRPKRLIYTVEAASVQEFAIGNNDDHINAKFSYVIKSYNPEKHVSVRYH 110
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAI------SNA 125
+ +S + Q V I F Q K + +T H F A S
Sbjct: 111 SMRISTAHHNQS--VAHKNISPFKQRP-KNETRIETQLVSHNVALSKFNARDLRAEKSKG 167
Query: 126 TAELKVALTTRFQYKTWGIKSK 147
T E++V +T R YKTW +S+
Sbjct: 168 TIEMEVYITARVSYKTWIFRSR 189
>UniRef100_Q8H790 Hypothetical protein [Arabidopsis thaliana]
Length = 95
Score = 48.1 bits (113), Expect = 1e-04
Identities = 26/79 (32%), Positives = 41/79 (50%), Gaps = 5/79 (6%)
Query: 4 GKRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSI-----EQHPGENGSIFYSLE 58
G+ + FI GL L LWL+LR P S+ I + + +N ++ + +
Sbjct: 11 GRNCCTCCLSFIFTAGLTSLFLWLSLRADKPKCSIQNFFIPALGKDPNSRDNTTLNFMVR 70
Query: 59 IENPNKDSSIYYDDIILSF 77
+NPN+D IYYDD+ L+F
Sbjct: 71 CDNPNRDQGIYYDDVHLNF 89
>UniRef100_Q75QH3 Hin1 like protein [Capsicum chinense]
Length = 228
Score = 48.1 bits (113), Expect = 1e-04
Identities = 30/100 (30%), Positives = 48/100 (48%), Gaps = 8/100 (8%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQH--PGENGSIFYSL----EIENPNKDSSIYYD 71
LG++ L LWL LRP + V ++ Q N +++Y L I NPNK IYYD
Sbjct: 56 LGVIALVLWLVLRPNKVKFYVTDATLTQFDLSTANNTLYYDLALNMTIRNPNKRIGIYYD 115
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNAR 111
I +Y + ++ + F+QG T S++ + +
Sbjct: 116 SIEARGMY--KGERFASQNLEPFYQGHKNTSSLHPVLKGQ 153
>UniRef100_Q8H1B5 Hin1-like protein [Solanum tuberosum]
Length = 225
Score = 47.8 bits (112), Expect = 1e-04
Identities = 30/90 (33%), Positives = 44/90 (48%), Gaps = 8/90 (8%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQ--HPGENGSIFYSL----EIENPNKDSSIYYD 71
LG++ L LWL LRP ++ V ++ Q + N +++Y L I NPNK IYYD
Sbjct: 54 LGVIALVLWLVLRPNKVNFYVTDATLTQFDYSTTNNTLYYDLALNMTIRNPNKRIGIYYD 113
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKT 101
I +Y Q + + F+QG T
Sbjct: 114 SIEARGMY--QGQRFASQNLERFYQGHKNT 141
>UniRef100_Q6RY60 Hin1-like protein [Solanum tuberosum]
Length = 187
Score = 47.4 bits (111), Expect = 2e-04
Identities = 30/94 (31%), Positives = 45/94 (46%), Gaps = 8/94 (8%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQH--PGENGSIFYSL----EIENPNKDSSIYYD 71
LG++ L LWL LRP + V ++ Q N +++Y L I NPNK IYYD
Sbjct: 56 LGVIALVLWLVLRPNKVKFYVTDATLTQFDLSTANNTLYYDLALNMTIRNPNKRVGIYYD 115
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVY 105
I +Y Q + + F+QG T +++
Sbjct: 116 SIEARAMY--QGQRFASHNLEPFYQGHKNTSNLH 147
>UniRef100_Q75LJ9 Putative harpin inducing protein [Oryza sativa]
Length = 212
Score = 45.8 bits (107), Expect = 6e-04
Identities = 44/144 (30%), Positives = 67/144 (45%), Gaps = 24/144 (16%)
Query: 16 GFLGLLVLCLWLALRPKSPSYSVVFLSIEQHPGE---NGSIFYS----LEIENPNKDSSI 68
G + LLV L PK+ + V L + PG N ++ Y+ L + NPN I
Sbjct: 31 GLIVLLVFAFALVFPPKATADDAVLLRLALSPGSPPSNSTVSYNATVTLSLRNPNLYRGI 90
Query: 69 YYDDIILSFLY-GQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAIS---- 123
YD + ++F + G + D+ T+ +F+ KT + + TV GA KP+ K +
Sbjct: 91 SYDPVAVAFSFNGTRFDE--SATVPAFYHRPRKTATFHVTVG---GAGKPVPKLTAAGVA 145
Query: 124 -----NATA--ELKVALTTRFQYK 140
NAT E++V L T QYK
Sbjct: 146 AFRAENATGRFEVEVRLDTVMQYK 169
>UniRef100_Q9LJT8 Non-race specific disease resistance protein-like [Arabidopsis
thaliana]
Length = 222
Score = 44.7 bits (104), Expect = 0.001
Identities = 43/169 (25%), Positives = 71/169 (41%), Gaps = 9/169 (5%)
Query: 11 LMEFIGFLGLLVLCLWLA-LRPKSPSYSVVFLSIEQH-----PGENGSIFYSLEIENPNK 64
LM + LL LCLWL+ L P S+ + I +N ++ + + ++N N
Sbjct: 17 LMTLLYVSFLLALCLWLSTLVHHIPRCSIHYFYIPALNKSLISSDNTTLNFMVRLKNINA 76
Query: 65 DSSIYYDDIILSF--LYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAI 122
IYY+D+ LSF V T+ F+QG K + + + +
Sbjct: 77 KQGIYYEDLHLSFSTRINNSSLLVANYTVPRFYQGHEKKAKKWGQALPFNNQ-TVIQAVL 135
Query: 123 SNATAELKVALTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKLSRKKKK 171
N +A +V L + +YK K+K + L L ++ DG K K+
Sbjct: 136 PNGSAIFRVDLKMQVKYKVMSWKTKRYKLKASVNLEVNEDGATKVKDKE 184
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 313,735,568
Number of Sequences: 2790947
Number of extensions: 12796016
Number of successful extensions: 29692
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 29645
Number of HSP's gapped (non-prelim): 70
length of query: 189
length of database: 848,049,833
effective HSP length: 120
effective length of query: 69
effective length of database: 513,136,193
effective search space: 35406397317
effective search space used: 35406397317
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0358.5


