

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.6
(408 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
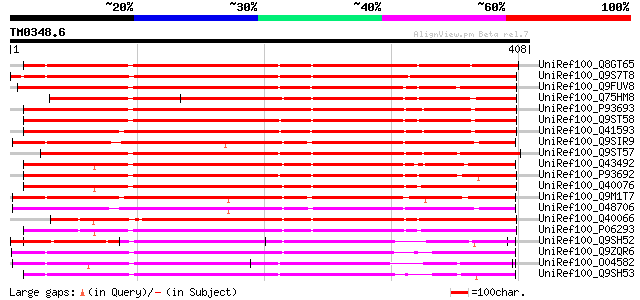
UniRef100_Q8GT65 Serpin-like protein [Citrus paradisi] 350 4e-95
UniRef100_Q9S7T8 F16N3.3 protein [Arabidopsis thaliana] 342 1e-92
UniRef100_Q9FUV8 Phloem serpin-1 [Cucurbita maxima] 331 3e-89
UniRef100_Q75HM8 Putative serine protease inhibitor [Oryza sativa] 303 4e-81
UniRef100_P93693 Serpin [Triticum aestivum] 297 4e-79
UniRef100_Q9ST58 Serpin [Triticum aestivum] 292 1e-77
UniRef100_Q41593 Serpin [Triticum aestivum] 291 2e-77
UniRef100_Q9SIR9 Putative serpin [Arabidopsis thaliana] 288 2e-76
UniRef100_Q9ST57 Serpin [Triticum aestivum] 286 6e-76
UniRef100_Q43492 Serpin [Hordeum vulgare] 285 1e-75
UniRef100_P93692 Serpin [Triticum aestivum] 283 5e-75
UniRef100_Q40076 Protein z-type serpin [Hordeum vulgare] 283 6e-75
UniRef100_Q9M1T7 Serpin-like protein [Arabidopsis thaliana] 281 2e-74
UniRef100_O48706 Putative serpin [Arabidopsis thaliana] 279 9e-74
UniRef100_Q40066 Protein zx [Hordeum vulgare] 278 3e-73
UniRef100_P06293 Protein Z [Hordeum vulgare] 277 4e-73
UniRef100_Q9SH52 F22C12.22 [Arabidopsis thaliana] 264 4e-69
UniRef100_Q9ZQR6 Putative serpin protein [Arabidopsis thaliana] 261 3e-68
UniRef100_O04582 F19K23.10 protein [Arabidopsis thaliana] 259 1e-67
UniRef100_Q9SH53 F22C12.21 [Arabidopsis thaliana] 256 6e-67
>UniRef100_Q8GT65 Serpin-like protein [Citrus paradisi]
Length = 389
Score = 350 bits (898), Expect = 4e-95
Identities = 192/390 (49%), Positives = 266/390 (67%), Gaps = 9/390 (2%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRFDSIDH 71
Q+DVALS TKH+ E ++ NL+FSP S++ LS+++AGS+G TLD+LLSFL+ S D
Sbjct: 8 QTDVALSLTKHVALTEA-KDSNLVFSPSSIHVLLSLISAGSKGPTLDQLLSFLKSKSDDQ 66
Query: 72 LNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASL 131
LNT+ S+ ++ VF+D P+ P LS NG++ID ++SL F++++ Y A+ +
Sbjct: 67 LNTFASELVAVVFADGSPSGGP---RLSVANGVWIDKSLSLKNTFKQVVDNVYKAASNQV 123
Query: 132 DFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDGL- 190
D + V +VN E+E+NGL+ ++LPPG+V N T+LIFANAL F+G W T D
Sbjct: 124 DSQTKAAEVSREVNMWAEKETNGLVKEVLPPGSVDNSTRLIFANALYFKGAWNETFDSSK 183
Query: 191 THVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDA 250
T F+LL+G S+KVP+MT+ KK Q+V AFDGFK+L LPYKQG D+ RRFSM FLPDA
Sbjct: 184 TKDYDFHLLNGGSIKVPFMTS-KKNQFVSAFDGFKVLGLPYKQGEDK-RRFSMYFFLPDA 241
Query: 251 QDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSP 310
+DGL L++K+ SE FL LP ++V V FR+P+F ISF E S VLK +G+V PFS
Sbjct: 242 KDGLPTLLEKMGSESKFLDHHLPSQRVEVGDFRIPRFKISFGIEVSKVLKGLGLVLPFSG 301
Query: 311 MDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDF 370
+MV+ + NL+V SIF K+FIEVNE+GT+A AA+ + + + IDF
Sbjct: 302 EGGGLAEMVD-SPVGKNLYVSSIFQKSFIEVNEEGTEAAAASAATVVLRSIL-LLDKIDF 359
Query: 371 IADHPFLFLIREDFTGTILFVGQVLNPLDG 400
+ADHPF+F+IRED TG ++F+G VLNPL G
Sbjct: 360 VADHPFVFMIREDMTGLVMFIGHVLNPLAG 389
>UniRef100_Q9S7T8 F16N3.3 protein [Arabidopsis thaliana]
Length = 391
Score = 342 bits (876), Expect = 1e-92
Identities = 186/399 (46%), Positives = 261/399 (64%), Gaps = 10/399 (2%)
Query: 1 MEIQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDEL 60
M++++S S Q+ V+++ KH+ + Q N+IFSP S+ LS++AAGS G T D++
Sbjct: 1 MDVRESISL--QNQVSMNLAKHVITTVS-QNSNVIFSPASINVVLSIIAAGSAGATKDQI 57
Query: 61 LSFLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLL 120
LSFL+F S D LN++ S+ +S V +D P LS NG +ID ++S F++LL
Sbjct: 58 LSFLKFSSTDQLNSFSSEIVSAVLADGSANGGPK---LSVANGAWIDKSLSFKPSFKQLL 114
Query: 121 STHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQ 180
Y A+ DF + V+ +VNS E+E+NGLIT++LP G+ ++TKLIFANAL F+
Sbjct: 115 EDSYKAASNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKLIFANALYFK 174
Query: 181 GMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQR 239
G W D LT F+LL G V P+MT+ KK QYV A+DGFK+L LPY QG+D+ R
Sbjct: 175 GTWNEKFDESLTQEGEFHLLDGNKVTAPFMTS-KKKQYVSAYDGFKVLGLPYLQGQDK-R 232
Query: 240 RFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVL 299
+FSM +LPDA +GLS L+ K+ S PGFL +P R+V+VR F++PKF SF F+ASNVL
Sbjct: 233 QFSMYFYLPDANNGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFSFGFDASNVL 292
Query: 300 KEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQ 359
K +G+ SPFS + T+MV NL V +IFHKA IEVNE+GT+A AA+ + +
Sbjct: 293 KGLGLTSPFSGEEG-LTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAAASAGVIKLR 351
Query: 360 CARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNPL 398
IDF+ADHPFL ++ E+ TG +LF+GQV++PL
Sbjct: 352 GLLMEEDEIDFVADHPFLLVVTENITGVVLFIGQVVDPL 390
>UniRef100_Q9FUV8 Phloem serpin-1 [Cucurbita maxima]
Length = 389
Score = 331 bits (848), Expect = 3e-89
Identities = 183/393 (46%), Positives = 263/393 (66%), Gaps = 11/393 (2%)
Query: 7 KSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRF 66
++ R DVA++ TK + ++ + N++ SPLS+Y LS++AAGS+GR LD+LLSFL+
Sbjct: 5 EAIRNHGDVAMAITKRILQHDEAKGSNVVISPLSIYVLLSLVAAGSKGRPLDQLLSFLKS 64
Query: 67 DSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNA 126
+SID+LN + S I VF+D P L+F NG++ID ++SL F++++ +Y A
Sbjct: 65 NSIDNLNAFASHIIDKVFADASSCGGP---RLAFVNGVWIDQSLSLKSSFQQVVDKYYKA 121
Query: 127 SLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHT 186
L +DF + + V+ +VNS +E+ + GLI ++LP G+V + T+L+ ANAL F+ W+
Sbjct: 122 ELRQVDFLTKANEVISEVNSWVEKNTYGLIREILPAGSVGSSTQLVLANALYFKAAWQQA 181
Query: 187 LDG-LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCI 245
D +T F L+ G+SVK P+M+ +K QYV FDGFK+L LPY QG D RRFSM
Sbjct: 182 FDASITMKRDFYLIDGSSVKAPFMSG-EKDQYVAVFDGFKVLALPYSQGPD-PRRFSMYF 239
Query: 246 FLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVV 305
FLPD +DGL++LI+KL SEPGF+ +P +K + F +PKF ISF E S+VLK++G+V
Sbjct: 240 FLPDRKDGLASLIEKLDSEPGFIDRHIPCKKQELGGFLIPKFKISFGIEVSDVLKKLGLV 299
Query: 306 SPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHH 365
PF+ + MV + + NL V +IFHKAFIEV+E+GTKA A+ SA+
Sbjct: 300 LPFT--EGGLLGMVE-SPVAQNLRVSNIFHKAFIEVDEEGTKAAAS--SAVTVGIVSLPI 354
Query: 366 PSIDFIADHPFLFLIREDFTGTILFVGQVLNPL 398
IDFIA+ PFL+LIRED +GT+LF+GQVLNPL
Sbjct: 355 NRIDFIANRPFLYLIREDKSGTLLFIGQVLNPL 387
>UniRef100_Q75HM8 Putative serine protease inhibitor [Oryza sativa]
Length = 653
Score = 303 bits (777), Expect = 4e-81
Identities = 171/369 (46%), Positives = 245/369 (66%), Gaps = 12/369 (3%)
Query: 32 KNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRFD-SIDHLNTYFSQGISPVFSDEDPA 90
+N+ FSPLSL+ ALS++AAG+ G T D+L S L S + L+ + Q + V +D A
Sbjct: 35 RNVAFSPLSLHVALSLVAAGAGGATRDQLASALGGPGSAEGLHAFAEQLVQLVLADASGA 94
Query: 91 SSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASLDFNLRGDSVLHDVNSLIEQ 150
P ++F +G+F+D ++SL F + Y A S+DF + V VNS +E+
Sbjct: 95 GGP---RVAFADGVFVDASLSLKKTFGDVAVGKYKAETHSVDFQTKAAEVASQVNSWVEK 151
Query: 151 ESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDGL-THVSSFNLLSGTSVKVPYM 209
++GLI ++LPPG+V + T+L+ NAL F+G W D T F+LL G SV+ P+M
Sbjct: 152 VTSGLIKEILPPGSVDHTTRLVLGNALYFKGAWTEKFDASKTKDGEFHLLDGKSVQAPFM 211
Query: 210 TTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDGLSALIQKLSSEPGFLK 269
+T KK QY+ ++D K+L+LPY+QG D+ R+FSM I LP+AQDGL +L +KL+SEP FL+
Sbjct: 212 STSKK-QYILSYDNLKVLKLPYQQGGDK-RQFSMYILLPEAQDGLWSLAEKLNSEPEFLE 269
Query: 270 GKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSPMDAHFTKMVNVNSPSDNLF 329
+P R+V V F++PKF ISF FEAS++LK +G+ PFS +A T+MV+ + NLF
Sbjct: 270 KHIPTRQVTVGQFKLPKFKISFGFEASDLLKSLGLHLPFS-SEADLTEMVD-SPEGKNLF 327
Query: 330 VQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDFIADHPFLFLIREDFTGTIL 389
V S+FHK+F+EVNE+GT+A AAT + + R + DF+ADHPFLFLI+ED TG +L
Sbjct: 328 VSSVFHKSFVEVNEEGTEAAAATAAVIT---LRSAPIAEDFVADHPFLFLIQEDMTGVVL 384
Query: 390 FVGQVLNPL 398
FVG V+NPL
Sbjct: 385 FVGHVVNPL 393
Score = 227 bits (579), Expect = 4e-58
Identities = 126/265 (47%), Positives = 177/265 (66%), Gaps = 9/265 (3%)
Query: 135 LRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLD-GLTHV 193
L VL VNS +++ ++GLI + P ++ + TKL+ ANAL F+G W D T
Sbjct: 394 LAAAEVLGQVNSWVDRVTSGLIKNIATPRSINHNTKLVLANALYFKGAWAEKFDVSKTED 453
Query: 194 SSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDG 253
F+LL G SV+ P+M+T KK QY+ ++D K+L+LPY QG D+ R+FSM I LP+AQDG
Sbjct: 454 GEFHLLDGESVQAPFMSTRKK-QYLSSYDSLKVLKLPYLQGGDK-RQFSMYILLPEAQDG 511
Query: 254 LSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSPMDA 313
L +L +KL+SEP F++ +P R V V F++PKF ISF F AS +LK +G+ F +
Sbjct: 512 LWSLAEKLNSEPEFMENHIPMRPVHVGQFKLPKFKISFGFGASGLLKGLGLPLLFG-SEV 570
Query: 314 HFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDFIAD 373
+MV+ + + NLFV S+FHK+FIEVNE+GT+ATAA ++ R ++F+AD
Sbjct: 571 DLIEMVD-SPGAQNLFVSSVFHKSFIEVNEEGTEATAAVMVSMEHSRPR----RLNFVAD 625
Query: 374 HPFLFLIREDFTGTILFVGQVLNPL 398
HPF+FLIRED TG ILF+G V+NPL
Sbjct: 626 HPFMFLIREDVTGVILFIGHVVNPL 650
>UniRef100_P93693 Serpin [Triticum aestivum]
Length = 399
Score = 297 bits (760), Expect = 4e-79
Identities = 169/389 (43%), Positives = 244/389 (62%), Gaps = 10/389 (2%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRFDSIDH 71
Q+ AL + S N FSP+SL++ALS++AAG+ T D+L++ L ++
Sbjct: 16 QTRFALRLASTISSNPKSAASNAAFSPVSLHSALSLLAAGAGSATRDQLVATLGTGEVEG 75
Query: 72 LNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASL 131
+ Q + V +D A P ++F NG+F+D ++ L F+ L Y A S+
Sbjct: 76 GHALAEQVVQFVLADASSAGGP---RVAFANGVFVDASLLLKPSFQELAVCKYKAETQSV 132
Query: 132 DFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDGL- 190
DF + V VNS +E+ ++G I +LP G+V N TKL+ ANAL F+G W D
Sbjct: 133 DFQTKAAEVTTQVNSWVEKVTSGRIKNILPSGSVDNTTKLVLANALYFKGAWTDQFDSYG 192
Query: 191 THVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDA 250
T F LL G+SV+ P+M++ QY+ + DG K+L+LPYKQG D R+FSM I LP+A
Sbjct: 193 TKNDYFYLLDGSSVQTPFMSSMDDDQYISSSDGLKVLKLPYKQGGDN-RQFSMYILLPEA 251
Query: 251 QDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSP 310
GLS+L +KLS+EP FL+ +P ++V +R F++PKF ISF EAS++LK +G+ PFS
Sbjct: 252 PGGLSSLAEKLSAEPDFLERHIPRQRVAIRQFKLPKFKISFGMEASDLLKCLGLQLPFSD 311
Query: 311 MDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSI-D 369
+A F++MV+ P L V S+FH+AF+EVNE+GT+A A+T + Q AR PS+ D
Sbjct: 312 -EADFSEMVDSPMP-QGLRVSSVFHQAFVEVNEQGTEAAASTAIKMVPQQARP--PSVMD 367
Query: 370 FIADHPFLFLIREDFTGTILFVGQVLNPL 398
FIADHPFLFL+RED +G +LF+G V+NPL
Sbjct: 368 FIADHPFLFLLREDISGVVLFMGHVVNPL 396
>UniRef100_Q9ST58 Serpin [Triticum aestivum]
Length = 398
Score = 292 bits (747), Expect = 1e-77
Identities = 169/389 (43%), Positives = 246/389 (62%), Gaps = 11/389 (2%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRFDSIDH 71
Q+ AL + S N +FSP+SL+ ALS++AAG+ T D+L++ L ++
Sbjct: 16 QTRFALRLASTISSNPKSAASNAVFSPVSLHVALSLLAAGAGSATRDQLVATLGTGEVEG 75
Query: 72 LNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASL 131
L+ Q + V +D A P H++F NG+F+D ++ L F+ L Y A S+
Sbjct: 76 LHALAEQVVQFVLADASSAGGP---HVAFANGVFVDASLPLKPSFQELAVCKYKADTQSV 132
Query: 132 DFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDGL- 190
DF + V VNS +E+ ++G I +LP G+V N TKL+ ANAL F+G W D
Sbjct: 133 DFQTKAAEVATQVNSWVEKVTSGRIKDILPSGSVDNTTKLVLANALYFKGAWTDQFDSSG 192
Query: 191 THVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDA 250
T F L G+SV+ P+M++ QY+ + DG K+L+LPYKQG D+ R+FSM I LP+A
Sbjct: 193 TKNDYFYLPDGSSVQTPFMSSM-DDQYLSSSDGLKVLKLPYKQGGDK-RQFSMYILLPEA 250
Query: 251 QDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSP 310
GLS L +KLS+EP FL+ +P ++V +R F++PKF ISF EAS++LK +G+ PFS
Sbjct: 251 PGGLSNLAEKLSAEPDFLERHIPRQRVALRQFKLPKFKISFETEASDLLKCLGLQLPFS- 309
Query: 311 MDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSI-D 369
+A F++MV+ + + L V S+FH+AF+EVNE+GT+A A+T +A AR PS+ D
Sbjct: 310 NEADFSEMVD-SPMAHGLRVSSVFHQAFVEVNEQGTEAAASTAIKMALLQARP--PSVMD 366
Query: 370 FIADHPFLFLIREDFTGTILFVGQVLNPL 398
FIADHPFLFL+RED +G +LF+G V+NPL
Sbjct: 367 FIADHPFLFLLREDISGVVLFMGHVVNPL 395
>UniRef100_Q41593 Serpin [Triticum aestivum]
Length = 398
Score = 291 bits (746), Expect = 2e-77
Identities = 169/389 (43%), Positives = 243/389 (62%), Gaps = 11/389 (2%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRFDSIDH 71
Q+ AL + S N FSP+SLY+ALS++AAG+ T D+L++ L ++
Sbjct: 16 QTRFALRLASTISSNPKSAASNAAFSPVSLYSALSLLAAGAGSATRDQLVATLGTGKVEG 75
Query: 72 LNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASL 131
L+ Q + V +D ASS F NG+F+D ++ L F+ + Y A S+
Sbjct: 76 LHALAEQVVQFVLAD---ASSTGGSACRFANGVFVDASLLLKPSFQEIAVCKYKAETQSV 132
Query: 132 DFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDGL- 190
DF + V VNS +E+ ++G I +LPPG++ N TKL+ ANAL F+G W D
Sbjct: 133 DFQTKAAEVTTQVNSWVEKVTSGRIKDILPPGSIDNTTKLVLANALYFKGAWTEQFDSYG 192
Query: 191 THVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDA 250
T F LL G+SV+ P+M++ QY+ + DG K+L+LPYKQG D R+F M I LP+A
Sbjct: 193 TKNDYFYLLDGSSVQTPFMSSM-DDQYLLSSDGLKVLKLPYKQGGD-NRQFFMYILLPEA 250
Query: 251 QDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSP 310
GLS+L +KLS+EP FL+ +P ++V +R F++PKF ISF EAS++LK +G+ PF
Sbjct: 251 PGGLSSLAEKLSAEPDFLERHIPRQRVALRQFKLPKFKISFGIEASDLLKCLGLQLPFGD 310
Query: 311 MDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSI-D 369
+A F++MV+ P L V S+FH+AF+EVNE+GT+A A+T + Q AR PS+ D
Sbjct: 311 -EADFSEMVDSLMP-QGLRVSSVFHQAFVEVNEQGTEAAASTAIKMVLQQARP--PSVMD 366
Query: 370 FIADHPFLFLIREDFTGTILFVGQVLNPL 398
FIADHPFLFL+RED +G +LF+G V+NPL
Sbjct: 367 FIADHPFLFLVREDISGVVLFMGHVVNPL 395
>UniRef100_Q9SIR9 Putative serpin [Arabidopsis thaliana]
Length = 385
Score = 288 bits (737), Expect = 2e-76
Identities = 167/398 (41%), Positives = 245/398 (60%), Gaps = 19/398 (4%)
Query: 3 IQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLS 62
++ KS +DV + TKH+ + NL+FSP+S+ LS++AAGS T +++LS
Sbjct: 1 MELGKSIENHNDVVVRLTKHVIATVA-NGSNLVFSPISINVLLSLIAAGSCSVTKEQILS 59
Query: 63 FLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLST 122
FL S DHLN +Q I D + S LS NG++ID SL F+ LL
Sbjct: 60 FLMLPSTDHLNLVLAQII-------DGGTEKSDLRLSIANGVWIDKFFSLKLSFKDLLEN 112
Query: 123 HYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNL--TKLIFANALRFQ 180
Y A+ + +DF + V+ +VN+ E +NGLI Q+L ++ + + L+ ANA+ F+
Sbjct: 113 SYKATCSQVDFASKPSEVIDEVNTWAEVHTNGLIKQILSRDSIDTIRSSTLVLANAVYFK 172
Query: 181 GMWKHTLDG-LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQR 239
G W D +T + F+LL GTSVKVP+MT ++ QY+R++DGFK+LRLPY + QR
Sbjct: 173 GAWSSKFDANMTKKNDFHLLDGTSVKVPFMTNYED-QYLRSYDGFKVLRLPYIED---QR 228
Query: 240 RFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVL 299
+FSM I+LP+ ++GL+ L++K+ SEP F +P + V FR+PKF SF F AS VL
Sbjct: 229 QFSMYIYLPNDKEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVL 288
Query: 300 KEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQ 359
K++G+ SPF+ T+MV+ S D+L+V SI HKA IEV+E+GT+A A + ++
Sbjct: 289 KDMGLTSPFN-NGGGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEAAAVSVGVVSCT 347
Query: 360 CARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
R + DF+AD PFLF +RED +G ILF+GQVL+P
Sbjct: 348 SFRRNP---DFVADRPFLFTVREDKSGVILFMGQVLDP 382
>UniRef100_Q9ST57 Serpin [Triticum aestivum]
Length = 398
Score = 286 bits (733), Expect = 6e-76
Identities = 161/378 (42%), Positives = 236/378 (61%), Gaps = 9/378 (2%)
Query: 25 SKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRFDSIDHLNTYFSQGISPVF 84
S + N+ FSP+SL+ ALS++ AG+ G T D+L++ L + L+ Q + V
Sbjct: 29 SNPESTANNVAFSPVSLHVALSLITAGAGGATRDQLVATLGEGEAERLHALAEQVVQFVL 88
Query: 85 SDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASLDFNLRGDSVLHDV 144
+D A SP ++F NG+F+D ++ L F+ L Y A S+DF + V V
Sbjct: 89 ADASYADSP---RVTFANGVFVDASLPLKPSFQELAVCKYKAEAQSVDFQTKAAEVTAQV 145
Query: 145 NSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDG-LTHVSSFNLLSGTS 203
NS +E+ + GLI +LP G+++N T+L+ NAL F+G W D +T F LL G+S
Sbjct: 146 NSWVEKVTTGLIKDILPAGSISNTTRLVLGNALYFKGAWTDQFDSRVTKSDYFYLLDGSS 205
Query: 204 VKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDGLSALIQKLSS 263
++ P+M + + QY+ + DG K+L+LPYKQG D+ R+FSM I LP+A G+ +L +KLS+
Sbjct: 206 IQTPFMYS-SEEQYISSSDGLKVLKLPYKQGGDK-RQFSMYILLPEAPSGIWSLAEKLSA 263
Query: 264 EPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSPMDAHFTKMVNVNS 323
EP L+ +P +KV +R F++PKF ISF EAS++LK +G+ PFS +A ++MV+
Sbjct: 264 EPELLERHIPRQKVALRQFKLPKFKISFGIEASDLLKHLGLQLPFSD-EADLSEMVDSPM 322
Query: 324 PSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDFIADHPFLFLIRED 383
P L + S+FHK F+EVNE GT+A AAT A A + +DFIADHPFLFLIRED
Sbjct: 323 P-QGLRISSVFHKTFVEVNETGTEAAAAT-IAKAVLLSASPPSDMDFIADHPFLFLIRED 380
Query: 384 FTGTILFVGQVLNPLDGL 401
+G +LF+G V+NPL L
Sbjct: 381 TSGVVLFIGHVVNPLRSL 398
>UniRef100_Q43492 Serpin [Hordeum vulgare]
Length = 397
Score = 285 bits (730), Expect = 1e-75
Identities = 161/390 (41%), Positives = 242/390 (61%), Gaps = 16/390 (4%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLR---FDS 68
Q+ L + S + N+ FSP+SL+ ALS++AAG+ G T D+L++ L
Sbjct: 16 QTRFGLRLASAISSDPESAATNVAFSPVSLHVALSLVAAGARGATRDQLVAVLGGGGAGE 75
Query: 69 IDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASL 128
+ L + Q + V +D S P ++F NG+F+D ++SL F+ L +Y + +
Sbjct: 76 AEALQSLAEQVVQFVLADASINSGP---RIAFANGVFVDASLSLKPSFQELAVCNYKSEV 132
Query: 129 ASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLD 188
S+DF + VNS ++ + GLI ++LP G++ N T+L+ NAL F+G+W D
Sbjct: 133 QSVDFKTKAPEAASQVNSWVKNVTAGLIEEILPAGSIDNTTRLVLGNALYFKGLWTKKFD 192
Query: 189 -GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFL 247
T F+LL+G++V+ P+M++ K QY+ + DG K+L+LPY+ G D R+FSM I L
Sbjct: 193 ESKTKYDDFHLLNGSTVQTPFMSSTNK-QYLSSSDGLKVLKLPYQHGGDN-RQFSMYILL 250
Query: 248 PDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSP 307
P+A DGLS L QKLS+EP FL+ ++P +V V F +PKF ISF FEA+ +LK +G+ P
Sbjct: 251 PEAHDGLSRLAQKLSTEPDFLENRIPTEEVEVGQFMLPKFKISFGFEANKLLKTLGLQLP 310
Query: 308 FSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPS 367
FS ++A+ ++M VNSP L++ S+FHK F+EV+E+GTKA AAT + R
Sbjct: 311 FS-LEANLSEM--VNSPM-GLYISSVFHKTFVEVDEEGTKAGAATGDVIV---DRSLPIR 363
Query: 368 IDFIADHPFLFLIREDFTGTILFVGQVLNP 397
+DF+A+HPFLFLIRED G +LF+G V NP
Sbjct: 364 MDFVANHPFLFLIREDIAGVVLFIGHVANP 393
>UniRef100_P93692 Serpin [Triticum aestivum]
Length = 398
Score = 283 bits (725), Expect = 5e-75
Identities = 160/390 (41%), Positives = 238/390 (61%), Gaps = 13/390 (3%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRFDSIDH 71
Q+ A + S + N FSP+SL+ ALS++ AG+ G T ++L + L ++
Sbjct: 16 QTRFAFRLASAISSNPESTVNNAAFSPVSLHVALSLITAGAGGATRNQLAATLGEGEVEG 75
Query: 72 LNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASL 131
L+ Q + V +D P ++F NG+F+D ++ L F+ L Y A S+
Sbjct: 76 LHALAEQVVQFVLADASNIGGP---RVAFANGVFVDASLQLKPSFQELAVCKYKAEAQSV 132
Query: 132 DFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLD-GL 190
DF + V VNS +E+ + GLI +LP G++ N T+L+ NAL F+G W D
Sbjct: 133 DFQTKAAEVTAQVNSWVEKVTTGLIKDILPAGSIDNTTRLVLGNALYFKGAWTDQFDPRA 192
Query: 191 THVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDA 250
T F LL G+S++ P+M + + QY+ + DG K+L+LPYKQG D+ R+FSM I LP+A
Sbjct: 193 TQSDDFYLLDGSSIQTPFMYS-SEEQYISSSDGLKVLKLPYKQGGDK-RQFSMYILLPEA 250
Query: 251 QDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSP 310
GL +L +KLS+EP FL+ +P +KV +R F++PKF IS EAS++LK +G++ PF
Sbjct: 251 LSGLWSLAEKLSAEPEFLEQHIPRQKVALRQFKLPKFKISLGIEASDLLKGLGLLLPFG- 309
Query: 311 MDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPS--I 368
+A ++MV+ + + NL++ SIFHKAF+EVNE GT+A A T +A+ R P +
Sbjct: 310 AEADLSEMVD-SPMAQNLYISSIFHKAFVEVNETGTEAAATT---IAKVVLRQAPPPSVL 365
Query: 369 DFIADHPFLFLIREDFTGTILFVGQVLNPL 398
DFI DHPFLFLIRED +G +LF+G V+NPL
Sbjct: 366 DFIVDHPFLFLIREDTSGVVLFIGHVVNPL 395
>UniRef100_Q40076 Protein z-type serpin [Hordeum vulgare]
Length = 400
Score = 283 bits (724), Expect = 6e-75
Identities = 162/391 (41%), Positives = 237/391 (60%), Gaps = 13/391 (3%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLR---FDS 68
Q+ AL + S + N+ FSPLSL+ ALS++ AG+ G T D+L++ L
Sbjct: 16 QTRFALRLASAISSNPERAAGNVAFSPLSLHVALSLITAGAGGATRDQLVAILGDGGAGD 75
Query: 69 IDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASL 128
LN Q + V ++E P ++F NG+F+D ++SL F L Y A
Sbjct: 76 AKELNALAEQVVQFVLANESSTGGP---RIAFANGIFVDASLSLKPSFEELAVCQYKAKT 132
Query: 129 ASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLD 188
S+DF + + VNS +EQ + GLI Q+LPPG+V N TKL+ NAL F+G W D
Sbjct: 133 QSVDFQHKTLEAVGQVNSWVEQVTTGLIKQILPPGSVDNTTKLVLGNALYFKGAWDQKFD 192
Query: 189 -GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFL 247
T SF+LL G+S++ +M++ KK QY+ + D K+L+LPY +G D+ R+FSM I L
Sbjct: 193 ESNTKCDSFHLLDGSSIQTQFMSSTKK-QYISSSDNLKVLKLPYAKGHDK-RQFSMYILL 250
Query: 248 PDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSP 307
P AQDGL +L ++LS+EP F++ +P + V V F++PKF IS+ FEAS++L+ +G+ P
Sbjct: 251 PGAQDGLWSLAKRLSTEPEFIENHIPKQTVEVGRFQLPKFKISYQFEASSLLRALGLQLP 310
Query: 308 FSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPS 367
FS +A ++MV+ S L + +FHK+F+EVNE+GT+A AAT +
Sbjct: 311 FSE-EADLSEMVD---SSQGLEISHVFHKSFVEVNEEGTEAGAATVAMGVAMSMPLKVDL 366
Query: 368 IDFIADHPFLFLIREDFTGTILFVGQVLNPL 398
+DF+A+HPFLFLIRED G ++FVG V NPL
Sbjct: 367 VDFVANHPFLFLIREDIAGVVVFVGHVTNPL 397
>UniRef100_Q9M1T7 Serpin-like protein [Arabidopsis thaliana]
Length = 393
Score = 281 bits (719), Expect = 2e-74
Identities = 163/406 (40%), Positives = 249/406 (61%), Gaps = 27/406 (6%)
Query: 3 IQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLS 62
++ KS Q+DV + KH+ NL+FSP+S+ L ++AAGS T +++LS
Sbjct: 1 MELGKSMENQTDVMVLLAKHVIPTVA-NGSNLVFSPMSINVLLCLIAAGSNCVTKEQILS 59
Query: 63 FLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLST 122
F+ S D+LN ++ +S +D S HLS G++ID ++S F+ LL
Sbjct: 60 FIMLPSSDYLNAVLAKTVSVALND---GMERSDLHLSTAYGVWIDKSLSFKPSFKDLLEN 116
Query: 123 HYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTK--LIFANALRFQ 180
YNA+ +DF + V+++VN+ E +NGLI ++L ++ + + LI ANA+ F+
Sbjct: 117 SYNATCNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFK 176
Query: 181 GMWKHTLDG-LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQR 239
G W D LT F+LL GT VKVP+MT +KK QY+ +DGFK+LRLPY + QR
Sbjct: 177 GAWSKKFDAKLTKSYDFHLLDGTMVKVPFMTNYKK-QYLEYYDGFKVLRLPYVED---QR 232
Query: 240 RFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVL 299
+F+M I+LP+ +DGL L++++SS+P FL +P +++ F++PKF SF F+AS+VL
Sbjct: 233 QFAMYIYLPNDRDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVL 292
Query: 300 KEVGVVSPFSPMDAHFTKMVNVNSPS--------DNLFVQSIFHKAFIEVNEKGTKATAA 351
KE+G+ PF+ H + V SPS +NLFV ++FHKA IEV+E+GT+A A
Sbjct: 293 KEMGLTLPFT----HGSLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAV 348
Query: 352 TWSALARQCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
+ +++ +D DF+ADHPFLF +RE+ +G ILF+GQVL+P
Sbjct: 349 SVASM----TKDMLLMGDFVADHPFLFTVREEKSGVILFMGQVLDP 390
>UniRef100_O48706 Putative serpin [Arabidopsis thaliana]
Length = 389
Score = 279 bits (714), Expect = 9e-74
Identities = 163/401 (40%), Positives = 240/401 (59%), Gaps = 21/401 (5%)
Query: 3 IQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLS 62
++ KS Q++V K + + N++FSP+S+ LS++AAGS T +E+LS
Sbjct: 1 MELGKSIENQNNVVARLAKKVIETDVANGSNVVFSPMSINVLLSLIAAGSNPVTKEEILS 60
Query: 63 FLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLST 122
FL S DHLN ++ D + S LS +G++ID + L F+ LL
Sbjct: 61 FLMSPSTDHLNAVLAK-------IADGGTERSDLCLSTAHGVWIDKSSYLKPSFKELLEN 113
Query: 123 HYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTK-----LIFANAL 177
Y AS + +DF + V+ +VN + +NGLI Q+L + + LI ANA+
Sbjct: 114 SYKASCSQVDFATKPVEVIDEVNIWADVHTNGLIKQILSRDCTDTIKEIRNSTLILANAV 173
Query: 178 RFQGMWKHTLDG-LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRD 236
F+ W D LT + F+LL G +VKVP+M ++K QY+R +DGF++LRLPY + +
Sbjct: 174 YFKAAWSRKFDAKLTKDNDFHLLDGNTVKVPFMMSYKD-QYLRGYDGFQVLRLPYVEDK- 231
Query: 237 RQRRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEAS 296
R FSM I+LP+ +DGL+AL++K+S+EPGFL +P + V R+PK N SF F+AS
Sbjct: 232 --RHFSMYIYLPNDKDGLAALLEKISTEPGFLDSHIPLHRTPVDALRIPKLNFSFEFKAS 289
Query: 297 NVLKEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSAL 356
VLK++G+ SPF+ + T+MV+ S D L V SI HKA IEV+E+GT+A A + + +
Sbjct: 290 EVLKDMGLTSPFT-SKGNLTEMVDSPSNGDKLHVSSIIHKACIEVDEEGTEAAAVSVAIM 348
Query: 357 ARQCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
QC + DF+ADHPFLF +RED +G ILF+GQVL+P
Sbjct: 349 MPQCLMRNP---DFVADHPFLFTVREDNSGVILFIGQVLDP 386
>UniRef100_Q40066 Protein zx [Hordeum vulgare]
Length = 398
Score = 278 bits (710), Expect = 3e-73
Identities = 161/370 (43%), Positives = 234/370 (62%), Gaps = 14/370 (3%)
Query: 33 NLIFSPLSLYAALSVMAAGSEGRTLDELLSFL---RFDSIDHLNTYFSQGISPVFSDEDP 89
N FSPLSL+ ALS++AAG+ T D+L + L + L+ Q + V +D
Sbjct: 36 NAAFSPLSLHVALSLVAAGAAA-TRDQLAATLGAAEKGDAEGLHALAEQVVQVVLADASG 94
Query: 90 ASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASLDFNLRGDSVLHDVNSLIE 149
A P SF N +F+D+++ L F+ L+ Y S+DF + V VNS +E
Sbjct: 95 AGGPR----SFAN-VFVDSSLKLKPSFKDLVVGKYKGETQSVDFQTKAPEVAGQVNSWVE 149
Query: 150 QESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDGL-THVSSFNLLSGTSVKVPY 208
+ + GLI ++LP G+V + T+L+ NAL F+G W D T F+LL G+SV+ P+
Sbjct: 150 KITTGLIKEILPAGSVDSTTRLVLGNALYFKGSWTEKFDASKTKDEKFHLLDGSSVQTPF 209
Query: 209 MTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDGLSALIQKLSSEPGFL 268
M++ KK QY+ ++D K+L+LPY+QG D+ R+FSM I LP+AQDGL L KLS+EP F+
Sbjct: 210 MSSTKK-QYISSYDSLKVLKLPYQQGGDK-RQFSMYILLPEAQDGLWNLANKLSTEPEFM 267
Query: 269 KGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSPMDAHFTKMVNVNSPSDNL 328
+ +P +KV V F++PKF ISF FEAS++LK +G+ PFS +A ++MV+ + + +L
Sbjct: 268 EKHMPMQKVPVGQFKLPKFKISFGFEASDMLKGLGLQLPFSS-EADLSEMVD-SPAARSL 325
Query: 329 FVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDFIADHPFLFLIREDFTGTI 388
+V S+FHK+F+EVNE+GT+A A T + + +DF+ADHPFLFLIRED TG +
Sbjct: 326 YVSSVFHKSFVEVNEEGTEAAARTARVVTLRSLPVEPVKVDFVADHPFLFLIREDLTGVV 385
Query: 389 LFVGQVLNPL 398
LFVG V NPL
Sbjct: 386 LFVGHVFNPL 395
>UniRef100_P06293 Protein Z [Hordeum vulgare]
Length = 399
Score = 277 bits (708), Expect = 4e-73
Identities = 162/391 (41%), Positives = 236/391 (59%), Gaps = 14/391 (3%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLR---FDS 68
Q+ AL + S + N+ FSPLSL+ ALS++ AG+ T D+L++ L
Sbjct: 16 QTRFALRLRSAISSNPERAAGNVAFSPLSLHVALSLITAGAAA-TRDQLVAILGDGGAGD 74
Query: 69 IDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASL 128
LN Q + V ++E P ++F NG+F+D ++SL F L Y A
Sbjct: 75 AKELNALAEQVVQFVLANESSTGGP---RIAFANGIFVDASLSLKPSFEELAVCQYKAKT 131
Query: 129 ASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLD 188
S+DF + + VNS +EQ + GLI Q+LPPG+V N TKLI NAL F+G W D
Sbjct: 132 QSVDFQHKTLEAVGQVNSWVEQVTTGLIKQILPPGSVDNTTKLILGNALYFKGAWDQKFD 191
Query: 189 -GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFL 247
T SF+LL G+S++ +M++ KK QY+ + D K+L+LPY +G D+ R+FSM I L
Sbjct: 192 ESNTKCDSFHLLDGSSIQTQFMSSTKK-QYISSSDNLKVLKLPYAKGHDK-RQFSMYILL 249
Query: 248 PDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSP 307
P AQDGL +L ++LS+EP F++ +P + V V F++PKF IS+ FEAS++L+ +G+ P
Sbjct: 250 PGAQDGLWSLAKRLSTEPEFIENHIPKQTVEVGRFQLPKFKISYQFEASSLLRALGLQLP 309
Query: 308 FSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPS 367
FS +A ++MV+ S L + +FHK+F+EVNE+GT+A AAT +
Sbjct: 310 FSE-EADLSEMVD---SSQGLEISHVFHKSFVEVNEEGTEAGAATVAMGVAMSMPLKVDL 365
Query: 368 IDFIADHPFLFLIREDFTGTILFVGQVLNPL 398
+DF+A+HPFLFLIRED G ++FVG V NPL
Sbjct: 366 VDFVANHPFLFLIREDIAGVVVFVGHVTNPL 396
>UniRef100_Q9SH52 F22C12.22 [Arabidopsis thaliana]
Length = 651
Score = 264 bits (674), Expect = 4e-69
Identities = 154/386 (39%), Positives = 228/386 (58%), Gaps = 36/386 (9%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLD-ELLSFLRFDSID 70
Q+ VA+ + H+ S ++ N+IFSP S+ +A+++ AAG G + ++LSFLR SID
Sbjct: 10 QTHVAMILSGHVLSSAP-KDSNVIFSPASINSAITMHAAGPGGDLVSGQILSFLRSSSID 68
Query: 71 HLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLAS 130
L T F + S V++D P ++ NG++ID ++ F+ L + A
Sbjct: 69 ELKTVFRELASVVYADRSATGGPK---ITAANGLWIDKSLPTDPKFKDLFENFFKAVYVP 125
Query: 131 LDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDG- 189
+DF + V +VNS +E +N LI LLP G+VT+LT I+ANAL F+G WK +
Sbjct: 126 VDFRSEAEEVRKEVNSWVEHHTNNLIKDLLPDGSVTSLTNKIYANALSFKGAWKRPFEKY 185
Query: 190 LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGR-DRQRRFSMCIFLP 248
T + F L++GTSV VP+M+++ + QYVRA+DGFK+LRLPY++G D R+FSM +LP
Sbjct: 186 YTRDNDFYLVNGTSVSVPFMSSY-ENQYVRAYDGFKVLRLPYQRGSDDTNRKFSMYFYLP 244
Query: 249 DAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPF 308
D +DGL L++K++S PGFL +P + + FR+PKF I F F ++VL +G+ S
Sbjct: 245 DKKDGLDDLLEKMASTPGFLDSHIPTYRDELEKFRIPKFKIEFGFSVTSVLDRLGLRS-- 302
Query: 309 SPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDH---H 365
S++HKA +E++E+G +A AAT C+ D
Sbjct: 303 ----------------------MSMYHKACVEIDEEGAEAAAATADGDC-GCSLDFVEPP 339
Query: 366 PSIDFIADHPFLFLIREDFTGTILFV 391
IDF+ADHPFLFLIRE+ TGT+LFV
Sbjct: 340 KKIDFVADHPFLFLIREEKTGTVLFV 365
Score = 155 bits (391), Expect = 3e-36
Identities = 83/196 (42%), Positives = 117/196 (59%), Gaps = 27/196 (13%)
Query: 202 TSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDGLSALIQKL 261
T+V V M+++K QY+ A+DGFK+L+LP++QG D R FSM +LPD +DGL L++K+
Sbjct: 479 TNVSVSLMSSYKD-QYIEAYDGFKVLKLPFRQGNDTSRNFSMHFYLPDEKDGLDNLVEKM 537
Query: 262 SSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSPMDAHFTKMVNV 321
+S GFL +P +KV+V F +PKF I F F AS +G
Sbjct: 538 ASSVGFLDSHIPSQKVKVGEFGIPKFKIEFGFSASRAFNRLG------------------ 579
Query: 322 NSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDFIADHPFLFLIR 381
L +++ KA +E++E+G +A AAT CA IDF+ADHPFLF+IR
Sbjct: 580 ------LDEMALYQKACVEIDEEGAEAIAATAVVGGFGCA--FVKRIDFVADHPFLFMIR 631
Query: 382 EDFTGTILFVGQVLNP 397
ED TGT+LFVGQ+ +P
Sbjct: 632 EDKTGTVLFVGQIFDP 647
Score = 50.4 bits (119), Expect = 9e-05
Identities = 32/87 (36%), Positives = 53/87 (60%), Gaps = 3/87 (3%)
Query: 1 MEIQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTL-DE 59
+E++ + + Q++VA+ + HLFS N +FSP S+ A ++MA+G + D+
Sbjct: 365 VEMEDEGAMKKQNEVAMILSWHLFSTVAKHSNN-VFSPASITAVFTMMASGPGSSLISDQ 423
Query: 60 LLSFLRFDSIDHLNTYFSQGISPVFSD 86
+LSFL SID LN+ F + I+ VF+D
Sbjct: 424 ILSFLGSSSIDELNSVF-RVITTVFAD 449
>UniRef100_Q9ZQR6 Putative serpin protein [Arabidopsis thaliana]
Length = 407
Score = 261 bits (666), Expect = 3e-68
Identities = 153/400 (38%), Positives = 233/400 (58%), Gaps = 34/400 (8%)
Query: 2 EIQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDE-L 60
+I ++ + Q++V+L + S + N +FSP S+ A L+V AA ++ +TL +
Sbjct: 28 KIDMQEAMKNQNEVSLLLVGKVISAVA-KNSNCVFSPASINAVLTVTAANTDNKTLRSFI 86
Query: 61 LSFLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLL 120
LSFL+ S + N F + S VF D P ++ NG++++ ++S + + L
Sbjct: 87 LSFLKSSSTEETNAIFHELASVVFKDGSETGGPK---IAAVNGVWMEQSLSCNPDWEDLF 143
Query: 121 STHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQ 180
+ AS A +DF + + V DVN+ + +N LI ++LP G+VT+LT I+ NAL F+
Sbjct: 144 LNFFKASFAKVDFRHKAEEVRLDVNTWASRHTNDLIKEILPRGSVTSLTNWIYGNALYFK 203
Query: 181 GMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRD-RQ 238
G W+ D +T F+LL+G SV VP+M +++K Q++ A+DGFK+LRLPY+QGRD
Sbjct: 204 GAWEKAFDKSMTRDKPFHLLNGKSVSVPFMRSYEK-QFIEAYDGFKVLRLPYRQGRDDTN 262
Query: 239 RRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNV 298
R FSM ++LPD + L L+++++S PGFL +P +V V FR+PKF I F FEAS+V
Sbjct: 263 REFSMYLYLPDKKGELDNLLERITSNPGFLDSHIPEYRVDVGDFRIPKFKIEFGFEASSV 322
Query: 299 LKEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALAR 358
+ +NV S+ KA IE++E+GT+A AAT +
Sbjct: 323 FNDF---------------ELNV----------SLHQKALIEIDEEGTEAAAATTVVVVT 357
Query: 359 -QCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
C + IDF+ADHPFLFLIRED TGT+LF GQ+ +P
Sbjct: 358 GSCLWEPKKKIDFVADHPFLFLIREDKTGTLLFAGQIFDP 397
>UniRef100_O04582 F19K23.10 protein [Arabidopsis thaliana]
Length = 559
Score = 259 bits (662), Expect = 1e-67
Identities = 158/402 (39%), Positives = 231/402 (57%), Gaps = 39/402 (9%)
Query: 3 IQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDEL-- 60
I ++ + Q+DVA+ T + S + N +FSP S+ AAL+++AA S G +EL
Sbjct: 188 IDVGEAMKKQNDVAIFLTGIVISSVA-KNSNFVFSPASINAALTMVAASSGGEQGEELRS 246
Query: 61 --LSFLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRR 118
LSFL+ S D LN F + S V D P ++ NGM++D ++S++ +
Sbjct: 247 FILSFLKSSSTDELNAIFREIASVVLVDGSKKGGPK---IAVVNGMWMDQSLSVNPLSKD 303
Query: 119 LLSTHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALR 178
L ++A+ A +DF + + V +VN+ +NGLI LLP G+VT+LT ++ +AL
Sbjct: 304 LFKNFFSAAFAQVDFRSKAEEVRTEVNAWASSHTNGLIKDLLPRGSVTSLTDRVYGSALY 363
Query: 179 FQGMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDR 237
F+G W+ +T F LL+GTSV VP+M++F+K QY+ A+DGFK+LRLPY+QGRD
Sbjct: 364 FKGTWEEKYSKSMTKCKPFYLLNGTSVSVPFMSSFEK-QYIAAYDGFKVLRLPYRQGRDN 422
Query: 238 -QRRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEAS 296
R F+M I+LPD + L L+++++S PGFL P R+V+V FR+PKF I F FEAS
Sbjct: 423 TNRNFAMYIYLPDKKGELDDLLERMTSTPGFLDSHNPERRVKVGKFRIPKFKIEFGFEAS 482
Query: 297 NVLKEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKA-TAATWSA 355
+ SD S + K IE++EKGT+A T + +
Sbjct: 483 SAF-------------------------SDFELDVSFYQKTLIEIDEKGTEAVTFTAFRS 517
Query: 356 LARQCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
CA IDF+ADHPFLFLIRE+ TGT+LF GQ+ +P
Sbjct: 518 AYLGCAL--VKPIDFVADHPFLFLIREEQTGTVLFAGQIFDP 557
Score = 153 bits (386), Expect = 1e-35
Identities = 88/207 (42%), Positives = 122/207 (58%), Gaps = 28/207 (13%)
Query: 190 LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDR-QRRFSMCIFLP 248
+T F L++G SV VP+M++ K QY+ A+DGFK+LRLPY+QGRD R FSM +LP
Sbjct: 1 MTKRKPFYLVNGISVSVPFMSS-SKDQYIEAYDGFKVLRLPYRQGRDNTNRNFSMYFYLP 59
Query: 249 DAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPF 308
D + L L+++++S PGFL P +V V FR+PKF I F FEAS+V
Sbjct: 60 DKKGELDDLLKRMTSTPGFLDSHTPRERVEVDEFRIPKFKIEFGFEASSVF--------- 110
Query: 309 SPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSI 368
SD S + KA IE++E+GT+A AAT + + + ++
Sbjct: 111 ----------------SDFEIDVSFYQKALIEIDEEGTEAAAAT-AFVDNEDGCGFVETL 153
Query: 369 DFIADHPFLFLIREDFTGTILFVGQVL 395
DF+ADHPFLFLIRE+ TGT+LF V+
Sbjct: 154 DFVADHPFLFLIREEQTGTVLFADLVI 180
>UniRef100_Q9SH53 F22C12.21 [Arabidopsis thaliana]
Length = 543
Score = 256 bits (655), Expect = 6e-67
Identities = 154/392 (39%), Positives = 223/392 (56%), Gaps = 38/392 (9%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDE-LLSFLRFDSID 70
Q+DVAL + S D + N++FSP S+ + L++ AA S+ L +LSFLR S D
Sbjct: 178 QNDVALFLAGEVISAAD-KNSNVVFSPASINSVLTMAAATSDSEALKSCILSFLRSSSTD 236
Query: 71 HLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLAS 130
LN F + S V D P ++ NG++ + ++ S + L Y ++ A
Sbjct: 237 ELNDIFREIASVVLVDGSKTGGPK---ITVVNGVWREQSLPCSPESKDLFENFYKSAFAQ 293
Query: 131 LDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTL-DG 189
+DF + + V +VNS + +NG+I LLPPG+VT+ T I+ NAL F+G W+
Sbjct: 294 VDFRSKFEEVREEVNSWALRHTNGIIKDLLPPGSVTSETLWIYGNALYFKGAWEDKFYKS 353
Query: 190 LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQG-RDRQRRFSMCIFLP 248
+T F L++G V VP+M + ++QYV+A+DGFK+LR PY+QG D R+FSMC +LP
Sbjct: 354 MTKHKPFYLVNGKQVHVPFMQS-SQSQYVKAYDGFKVLRQPYRQGVNDTSRQFSMCTYLP 412
Query: 249 DAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPF 308
D +DGL L++K++S GFL +P +V V FR+PKF I F FEAS+V +
Sbjct: 413 DEKDGLDNLVEKMTSTDGFLDSHIPSWRVEVGEFRIPKFKIEFGFEASSVFNDFA----- 467
Query: 309 SPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHH--- 365
+D S++ KA IE++E+GT+A AAT AL C +
Sbjct: 468 --LDV------------------SLYQKAMIEIDEEGTEAAAAT--ALVGACGCSLYRPP 505
Query: 366 PSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
P IDF+ADHPF F IRED TGT+LF GQ+ +P
Sbjct: 506 PPIDFVADHPFFFFIREDKTGTVLFAGQIFDP 537
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 654,405,697
Number of Sequences: 2790947
Number of extensions: 26560368
Number of successful extensions: 58713
Number of sequences better than 10.0: 995
Number of HSP's better than 10.0 without gapping: 635
Number of HSP's successfully gapped in prelim test: 360
Number of HSP's that attempted gapping in prelim test: 55433
Number of HSP's gapped (non-prelim): 1111
length of query: 408
length of database: 848,049,833
effective HSP length: 130
effective length of query: 278
effective length of database: 485,226,723
effective search space: 134893028994
effective search space used: 134893028994
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0348.6


