

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
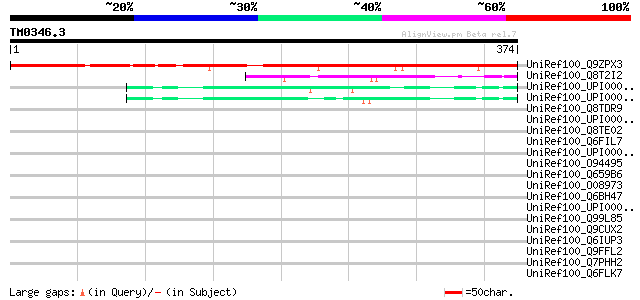
Query= TM0346.3
(374 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZPX3 Hypothetical protein At2g18410 [Arabidopsis tha... 321 2e-86
UniRef100_Q8T2I2 Putative ubiquitin carboxyl-terminal hydrolase ... 68 5e-10
UniRef100_UPI0000360BE7 UPI0000360BE7 UniRef100 entry 67 1e-09
UniRef100_UPI0000360BE6 UPI0000360BE6 UniRef100 entry 65 3e-09
UniRef100_Q8TDR9 S-phase 2 protein [Homo sapiens] 40 0.081
UniRef100_UPI00003C285F UPI00003C285F UniRef100 entry 40 0.11
UniRef100_Q8TE02 Dermal papilla derived protein 6 [Homo sapiens] 40 0.14
UniRef100_Q6FIL7 Candida glabrata strain CBS138 chromosome M com... 39 0.18
UniRef100_UPI0000260CBB UPI0000260CBB UniRef100 entry 39 0.23
UniRef100_O94495 SPBC18E5.05c protein [Schizosaccharomyces pombe] 39 0.31
UniRef100_Q659B6 Hypothetical protein DKFZp761D0721 [Homo sapiens] 38 0.40
UniRef100_O08973 Hypothetical protein [Mus musculus] 38 0.52
UniRef100_Q6BH47 Similar to CA3323|CaIKI1 Candida albicans CaIKI... 38 0.52
UniRef100_UPI00002BB5B1 UPI00002BB5B1 UniRef100 entry 37 0.68
UniRef100_Q99L85 S-phase 2 protein [Mus musculus] 37 0.68
UniRef100_Q9CUX2 Mus musculus adult male hippocampus cDNA, RIKEN... 37 0.68
UniRef100_Q6IUP3 DERP6 [Rattus norvegicus] 37 0.68
UniRef100_Q9FFL2 Nucleolar protein-like [Arabidopsis thaliana] 37 0.89
UniRef100_Q7PHH2 ENSANGP00000022479 [Anopheles gambiae str. PEST] 37 0.89
UniRef100_Q6FLK7 Similar to sp|P38630 Saccharomyces cerevisiae Y... 37 1.2
>UniRef100_Q9ZPX3 Hypothetical protein At2g18410 [Arabidopsis thaliana]
Length = 392
Score = 321 bits (822), Expect = 2e-86
Identities = 200/413 (48%), Positives = 252/413 (60%), Gaps = 60/413 (14%)
Query: 1 MAESICRTLRDGALEGELAPTLTITDSLTSPLASDVFSHLLLQLSSHLIASKSHSQFRSG 60
MAESI R LRDG EGELAP LTI +++ SP DV +LL LSS ++A KS SQ G
Sbjct: 1 MAESIFRKLRDGGEEGELAPALTIEETVASPFGLDVSGYLLTNLSSSILAGKSSSQ---G 57
Query: 61 IVIVALSRTPSSYAALFKTKGIHISSSNNNWIHILDCYTDPLGWKEKPRKSQNVTNPSHQ 120
+V++ SR+PS Y L K KGI +SSS+ WI ILDCYTDPLGW ++ S + + S
Sbjct: 58 LVLITFSRSPSFYLQLLKQKGIVVSSSSK-WIRILDCYTDPLGWIDQ--SSTSFSEGSSL 114
Query: 121 VSLATTSYKTVKDLDKLFSVITELGR------------------GLIGENKARFCVAIDS 162
+ L +K V DL KLFS I E GR L+G K RFCVAIDS
Sbjct: 115 IKL----HKCVSDLKKLFSSIIEAGRVRSLSKGLYEIKSDCFGAELVGTGKTRFCVAIDS 170
Query: 163 LSELLRHASLQSVAGLLSNLRSHDQISSIFGLLHSDLHEERAAAVLEYMSSMVASVAPLH 222
E++ + + +S QISS+F L++DLH+E+ LEY+S+M A++ PL
Sbjct: 171 KREVIIYLNFESA-----------QISSVFWSLNTDLHQEKVTNALEYISTMKANLEPLC 219
Query: 223 HSAE--RGNLENSLS-EQNFTKGRLNVRFKRRNGRVRVTCEEFKVEAGGISLTSVSSADG 279
S++ R LEN S Q+F KGR +VRFK R GRVRV EE+ V+ GI+ + +SS D
Sbjct: 220 PSSDGQRNALENLFSVHQDFGKGRFHVRFKLRKGRVRVMSEEYHVDQSGINFSPISSVDT 279
Query: 280 ATVA--GLLPK-----------VQFNLQLSEKEKTDRAKVVLPFEHQGTGRPIQIYDGRR 326
A LLPK VQFNLQLSEKE+ ++ KVVLPFEHQ G+ +IYDGRR
Sbjct: 280 VIAATKSLLPKASYLDLLFNYLVQFNLQLSEKERVEKEKVVLPFEHQDDGKSNEIYDGRR 339
Query: 327 SLDDNSSEAAPISSGKKE-----NSEMGEIIYFRDSDDEMPDSDEDPDDDLDI 374
SL D E P+SS + + + + GEIIYFRDSDDE PDSDEDPDDDLDI
Sbjct: 340 SLVDGKIETTPLSSMELQTDVVSSGKGGEIIYFRDSDDEHPDSDEDPDDDLDI 392
>UniRef100_Q8T2I2 Putative ubiquitin carboxyl-terminal hydrolase [Dictyostelium
discoideum]
Length = 253
Score = 67.8 bits (164), Expect = 5e-10
Identities = 63/208 (30%), Positives = 93/208 (44%), Gaps = 45/208 (21%)
Query: 175 VAGLLSN-LRSHDQISSIFGLLHSDLHE--ERAAAVLEYMSSMVASVAPLHHSAERGNLE 231
+ G +SN ++ S IF +LH+DLHE + L+Y+SS+ V PL L+
Sbjct: 83 IEGSVSNRIKMEKSFSCIFAILHTDLHEYEQSVQKQLQYISSVSIQVTPLS-----AKLK 137
Query: 232 NSLSEQNFTKGRLNVRFKRRNGRVRVTCEEFKVE--AGGI---SLTSVSSADGATVAGLL 286
S S + + + + K+R+GRV E + + G + S S+ +
Sbjct: 138 YSESLPHPYESTITLITKKRSGRVVRNVEYYYINNATGKVEFDSAESLQTKQEEQEPDPT 197
Query: 287 PKVQFNLQLSEKEKTDRAKVVLPFEHQGTGRPIQIYDGRRSLDDNSSEAAPISSGKKENS 346
+ FNL+L+E EK R VVLP+ HQG N++E
Sbjct: 198 QNLSFNLKLTEDEKQARDSVVLPYRHQG----------------NNNE------------ 229
Query: 347 EMGEIIYFRDSDDEMPDSDEDPDDDLDI 374
+ + D DDE D DEDPDDDLDI
Sbjct: 230 ---QTLLIEDPDDEDFD-DEDPDDDLDI 253
>UniRef100_UPI0000360BE7 UPI0000360BE7 UniRef100 entry
Length = 290
Score = 66.6 bits (161), Expect = 1e-09
Identities = 67/301 (22%), Positives = 114/301 (37%), Gaps = 69/301 (22%)
Query: 87 SNNNWIHILDCYTDPLGWKEKPRKSQNVTNPSHQVSLATTSYKTVKDLDKLFSVITELGR 146
++N +H + Y DPLGW + N H+ EL R
Sbjct: 46 THNQRLHFHNAYADPLGWND------NTPFTVHRFCAE------------------ELTR 81
Query: 147 GLIGENKARFCVAIDSLSELLRHASLQSVAGLLSNLRSHDQISSIFGLLHSDLHEERAAA 206
+ + + IDSLS +LRH +V L LR ++ GLLH+D+H+
Sbjct: 82 LMRTPHSKPTTLVIDSLSWVLRHVKCPTVCRTLHQLRKGGAARAVVGLLHTDMHQRGTVG 141
Query: 207 VLEYMSSMVASVAP----------LHHSAERGNLENSLSEQNFTKGRLNVRFKRR---NG 253
+ ++++ V +V+P + + G + + + G R R +
Sbjct: 142 SVGHLATCVITVSPGTKADEAVAKITKRLKSGKVIQDVGPNMLSLGSRRCRVTHRIYSHN 201
Query: 254 RVRVTCEEFKVEAGGISLTSVSSADGATVAGLLPKVQFNLQLSEKEKTDRAKVVLPFEHQ 313
++ V + A T FNL+LS+ E+ + K+ LPF
Sbjct: 202 TLKPNVARLNVPLWLFTFLQTDPAANLT---------FNLRLSDSEREAKEKLALPF--- 249
Query: 314 GTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLD 373
+ + A + SG G+I+Y D++D+ +EDPDDDLD
Sbjct: 250 --------------MFSKEKKTALLCSGPGS----GQIVYEPDANDDY--DEEDPDDDLD 289
Query: 374 I 374
+
Sbjct: 290 V 290
>UniRef100_UPI0000360BE6 UPI0000360BE6 UniRef100 entry
Length = 278
Score = 65.1 bits (157), Expect = 3e-09
Identities = 67/295 (22%), Positives = 118/295 (39%), Gaps = 69/295 (23%)
Query: 87 SNNNWIHILDCYTDPLGWKEKPRKSQNVTNPSHQVSLATTSYKTVKDLDKLFSVITELGR 146
++N +H + Y DPLGW + N H+ EL R
Sbjct: 46 THNQRLHFHNAYADPLGWND------NTPFTVHRFCAE------------------ELTR 81
Query: 147 GLIGENKARFCVAIDSLSELLRHASLQSVAGLLSNLRSHDQISSIFGLLHSDLHEERAAA 206
+ + + IDSLS +LRH +V L LR ++ GLLH+D+H+
Sbjct: 82 LMRTPHSKPTTLVIDSLSWVLRHVKCPTVCRTLHQLRKGGAARAVVGLLHTDMHQRGTVG 141
Query: 207 VLEYMSSMVASVAPLHHSAERGNLENSLSEQNFTKGRLNVRFKRRNGRVRVTC-----EE 261
+ ++++ V +V+P + +++ K + + ++G+V E+
Sbjct: 142 SVGHLATCVITVSP-----------GTKADEAVAK----ITKRLKSGKVIQDVVFSIQED 186
Query: 262 FKV--EAGGISLTSVSSADGATVAGLLPKVQFNLQLSEKEKTDRAKVVLPFEHQGTGRPI 319
F V + + G + FNL+LS+ E+ + K+ LPF
Sbjct: 187 FTVVLQCRANQARPKHAEPGEQETDPAANLTFNLRLSDSEREAKEKLALPF--------- 237
Query: 320 QIYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLDI 374
+ + A + SG G+I+Y D++D+ +EDPDDDLD+
Sbjct: 238 --------MFSKEKKTALLCSGPGS----GQIVYEPDANDDY--DEEDPDDDLDV 278
>UniRef100_Q8TDR9 S-phase 2 protein [Homo sapiens]
Length = 307
Score = 40.4 bits (93), Expect = 0.081
Identities = 57/241 (23%), Positives = 88/241 (35%), Gaps = 69/241 (28%)
Query: 158 VAIDSLSELLRHASLQSVAGLLSNLRSHDQIS---------SIFGLLHSDLHEERAAAVL 208
+A+DSLS LL ++ +L + D S+ GLLH +LH L
Sbjct: 112 IALDSLSWLLLRLPCTTLCQVLHAVSHQDSRPGDSSSVGKVSVLGLLHEELHGPSPVGAL 171
Query: 209 EYMSSMVASVAPLHHSAERGNLENSLSEQNFTKGRLNVRFKRRNGRVRVTCE-------- 260
++ ++ T G+ + R R R T +
Sbjct: 172 NSLAQTEVTLGG-------------------TMGQASAHILCRRPRQRPTDQTQWFSILP 212
Query: 261 EFKVEAG-GISLTSVSSADGATVAGLLPKVQ------FNLQLSEKEKTDRAKVVLPFEHQ 313
+F ++ G S+ S +D +P+V FNL LS+KE+ R ++LPF+
Sbjct: 213 DFSLDLKEGPSVESQPYSDPH-----IPRVDPTTHLTFNLHLSKKEREARDSLILPFQF- 266
Query: 314 GTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLD 373
SSE + I Y D+ D++ EDPDDDLD
Sbjct: 267 ------------------SSEKQQALLRPRPGQATSHIFYEPDAYDDL--DQEDPDDDLD 306
Query: 374 I 374
I
Sbjct: 307 I 307
>UniRef100_UPI00003C285F UPI00003C285F UniRef100 entry
Length = 1327
Score = 40.0 bits (92), Expect = 0.11
Identities = 28/102 (27%), Positives = 46/102 (44%), Gaps = 21/102 (20%)
Query: 273 SVSSADGATVAGLLPKVQFNLQLSEKEKTDRAKVVLPFEHQGTGRPIQIYDGRRSLDDNS 332
S + + GA + ++ + FNL + ++ R V LP+ + N+
Sbjct: 1247 SAACSGGAAHSSMISTLPFNLSETVDQRQRRETVALPYAYH----------------QNA 1290
Query: 333 SEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLDI 374
+A G N++ I+F DD+ D DEDPDDDLD+
Sbjct: 1291 QQAERALRGTTGNAK----IFFEPEDDDDED-DEDPDDDLDL 1327
>UniRef100_Q8TE02 Dermal papilla derived protein 6 [Homo sapiens]
Length = 316
Score = 39.7 bits (91), Expect = 0.14
Identities = 57/241 (23%), Positives = 87/241 (35%), Gaps = 69/241 (28%)
Query: 158 VAIDSLSELLRHASLQSVAGLLSNLRSHDQIS---------SIFGLLHSDLHEERAAAVL 208
+A+DSLS LL ++ +L + D S+ GLLH +LH L
Sbjct: 121 IALDSLSWLLLRLPCTTLCQVLHAVSHQDSCPGDSSSVGKVSVLGLLHEELHGPGPVGAL 180
Query: 209 EYMSSMVASVAPLHHSAERGNLENSLSEQNFTKGRLNVRFKRRNGRVRVTCE-------- 260
++ ++ T G+ + R R R T +
Sbjct: 181 SSLAQTEVTLGG-------------------TMGQASAHILCRRPRQRPTDQTQWFSILP 221
Query: 261 EFKVEAG-GISLTSVSSADGATVAGLLPKVQ------FNLQLSEKEKTDRAKVVLPFEHQ 313
+F ++ G S+ S +D +P V FNL LS+KE+ R ++LPF+
Sbjct: 222 DFSLDLQEGPSVESQPYSDPH-----IPPVDPTTHLTFNLHLSKKEREARDSLILPFQF- 275
Query: 314 GTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLD 373
SSE + I Y D+ D++ EDPDDDLD
Sbjct: 276 ------------------SSEKQQALLRPRPGQATSHIFYEPDAYDDL--DQEDPDDDLD 315
Query: 374 I 374
I
Sbjct: 316 I 316
>UniRef100_Q6FIL7 Candida glabrata strain CBS138 chromosome M complete sequence
[Candida glabrata]
Length = 307
Score = 39.3 bits (90), Expect = 0.18
Identities = 54/238 (22%), Positives = 91/238 (37%), Gaps = 61/238 (25%)
Query: 155 RFCVAIDSLSELLRHASLQSVAGLLSNLRSHDQISSIFGLLHSDLHEE----------RA 204
R + IDSL+ + R A L G ++ + +I LH D+H+ A
Sbjct: 106 RNVIVIDSLNHVPR-AQLTKFIGSIATPHA-----TILATLHRDMHDSPYEHGLDNYPSA 159
Query: 205 AAVLEYMSSMVASVAPLHHSAERGNLENSLSEQNFTKG------RLNVRFKRRNGRVRVT 258
+L +M++ V P ++ L LS+ +G ++++ +KR++GR
Sbjct: 160 IELLHFMATTSLDVTPASTISDDDELNQQLSKLQIPRGLNNTTFKVSLTYKRKSGRELTY 219
Query: 259 CEEFKVEAGGISLTSVSSADGA------TVAGLLPKVQFNLQLSEKEKTDRAKVVLPFEH 312
+FKV+ + T L FNL S K+K + +V LPF
Sbjct: 220 --DFKVDYNTHQYYQIMDKKDVDENALETPEMLQGLTTFNLSTSAKQKAAKDQVELPF-- 275
Query: 313 QGTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSD--EDP 368
EA +++G G I+Y + DD+ + D EDP
Sbjct: 276 --------------------LEAQSLTAG-------GAIVYEYEKDDDYDEEDPYEDP 306
>UniRef100_UPI0000260CBB UPI0000260CBB UniRef100 entry
Length = 179
Score = 38.9 bits (89), Expect = 0.23
Identities = 46/177 (25%), Positives = 76/177 (41%), Gaps = 40/177 (22%)
Query: 124 ATTSYKTVKDLDKLFSVITELG---RGLIGENKA-RFCVAID---SLSELLRHASLQSVA 176
AT + D+ +T +G G GE A R CV +D S S ++ SL+ V
Sbjct: 30 ATVTMTRTTHQDESGPAVTAVGLQATGTSGETAATRICVLVDTSASQSGAIQQQSLEVVR 89
Query: 177 GLLSNLRSHDQI-----SSIFGLLHSDLHEERAAAVLEYMSSMVASVAPLHHSAERGNLE 231
GLL N+R HD+I F L +D + ++ AV++ +++ A+ PL ++ NL
Sbjct: 90 GLLGNVRPHDRILLAAADITFASLMTDFAQAQSNAVIDAQNTL-ANRTPLGNT----NLL 144
Query: 232 NSLSEQNFTKGRLNVRFKRRNGRVRVTCEEFKVEAGGISLTSVSSADGATVAGLLPK 288
L R E + E+G ++ + DG + G+LP+
Sbjct: 145 EVL---------------------RAALESLQAESGPAAILYI--GDGPGLTGILPE 178
>UniRef100_O94495 SPBC18E5.05c protein [Schizosaccharomyces pombe]
Length = 314
Score = 38.5 bits (88), Expect = 0.31
Identities = 30/85 (35%), Positives = 37/85 (43%), Gaps = 24/85 (28%)
Query: 289 VQFNLQLSEKEKTDRAKVVLP-FEHQGTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSE 347
V FNL +SEKE+ +R KV LP F Q G S K +
Sbjct: 251 VSFNLNVSEKERKERDKVFLPYFSAQMVG----------------------SQHKSSFVD 288
Query: 348 MGEIIYFRDSDDEMPDSDEDPDDDL 372
G IIY D D+ D +ED D+DL
Sbjct: 289 EGTIIYHADEADDF-DEEEDADEDL 312
>UniRef100_Q659B6 Hypothetical protein DKFZp761D0721 [Homo sapiens]
Length = 175
Score = 38.1 bits (87), Expect = 0.40
Identities = 27/84 (32%), Positives = 36/84 (42%), Gaps = 21/84 (25%)
Query: 291 FNLQLSEKEKTDRAKVVLPFEHQGTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSEMGE 350
FNL LS+KE+ R ++LPF+ SSE +
Sbjct: 113 FNLHLSKKEREARDSLILPFQF-------------------SSEKQQALLRPRPGQATSH 153
Query: 351 IIYFRDSDDEMPDSDEDPDDDLDI 374
I Y D+ D++ EDPDDDLDI
Sbjct: 154 IFYEPDAYDDL--DQEDPDDDLDI 175
>UniRef100_O08973 Hypothetical protein [Mus musculus]
Length = 300
Score = 37.7 bits (86), Expect = 0.52
Identities = 55/234 (23%), Positives = 86/234 (36%), Gaps = 55/234 (23%)
Query: 158 VAIDSLSELLRHASLQSVAGLLSNLRSHD---------QISSIFGLLHSDLHEERAAAVL 208
+A+DSLS LL H ++ L L + + + GLLH +LH + L
Sbjct: 105 IALDSLSWLLCHIPCVTLCQALHALSQQNGDPGDNSLVEQVHVLGLLHEELHGPGSMGAL 164
Query: 209 EYMSSMVASVAPLHHSAERGNLENSLSEQNFTKGRLNVRFKRRNGRVRVTCEEFKVEAG- 267
++ +++ G ++ + + ++ +R R F V
Sbjct: 165 NTLAHTEVTLS--------GKVDQTSA---------SILCRRPQQRATYQTWWFSVLPDF 207
Query: 268 ------GISLTSVSSADGATV-AGLLPKVQFNLQLSEKEKTDRAKVVLPFEHQGTGRPIQ 320
G+ L S D T + FNL LS+KE+ R + LPF+
Sbjct: 208 SLTLHEGLPLRSELHPDHHTTQVDPTAHLTFNLHLSKKEREARDSLTLPFQF-------- 259
Query: 321 IYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLDI 374
SSE + G I Y D+ D++ EDPDDDLDI
Sbjct: 260 -----------SSEKQKALLHPVPSRTTGRIFYEPDAFDDV--DQEDPDDDLDI 300
>UniRef100_Q6BH47 Similar to CA3323|CaIKI1 Candida albicans CaIKI1 [Debaryomyces
hansenii]
Length = 298
Score = 37.7 bits (86), Expect = 0.52
Identities = 70/324 (21%), Positives = 128/324 (38%), Gaps = 69/324 (21%)
Query: 44 LSSHLIASKSHSQFRSGIVIVALSRTPSSYAALFKTKGIHISSSNNNWIHILDCYTDPLG 103
L + L++ K +S F ++V S SSY L + SNN I+I
Sbjct: 9 LLNRLLSLKENSPF----ILVLDSLIQSSYNIL---REFAYKCSNNKIIYI--------- 52
Query: 104 WKEKPRKSQNVTNPSHQVSLATTSYKTVKDLDK-LFSVITELGRGLIGENKARFCVAIDS 162
+ + PS S S ++ K + S I E+G ++ V +DS
Sbjct: 53 ------SFETLNKPSFASSFIQCSGLPAAEVSKKIKSEILEVGE----TTSSKCLVIVDS 102
Query: 163 LSELLRHASLQSVAG-LLSNLRSHDQISSIFGLLHSDLHEERAA--------AVLEYMSS 213
L+ + ++ +L N+ S+ G+ H++L + ++ ++L Y++S
Sbjct: 103 LNYIANDELAHFISSTILPNI-------SLVGVFHTNLPQPHSSVMNYPSSLSLLSYIAS 155
Query: 214 MVASVAPLHHSA-ERGNLENSLSEQNF--------TKGRLNVRFKRRNGRVRVTCEEFKV 264
+ + P + + L+N +S F T +L + +R++GR + ++
Sbjct: 156 SIFEIEPFYQDKIDEETLDNKISRLRFPINCSLNSTTYKLTLTNRRKSGRSLI----YQF 211
Query: 265 EAGGISLT-------SVSSADGATVAGLLPKVQFNLQLSEKEKTDRAKVVLPF----EHQ 313
I+ T + D + L FNL S K+K R +V LPF E
Sbjct: 212 MVNSITHTYDVFKEAKEEAIDPEDESMLKDLTTFNLTTSSKQKLAREQVELPFMEAQEAM 271
Query: 314 GTGRPIQIYDGRRSLDDNSSEAAP 337
G+ +Y+ + DD+ E P
Sbjct: 272 GSSGGAIVYEFEK--DDDYDEEDP 293
>UniRef100_UPI00002BB5B1 UPI00002BB5B1 UniRef100 entry
Length = 99
Score = 37.4 bits (85), Expect = 0.68
Identities = 26/84 (30%), Positives = 40/84 (46%), Gaps = 23/84 (27%)
Query: 291 FNLQLSEKEKTDRAKVVLPFEHQGTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSEMGE 350
FNL+LS+ E + K+ LPF + + A + SG G+
Sbjct: 39 FNLRLSDSELEAKEKLALPF-----------------VFSKEKKTALLCSGPGS----GQ 77
Query: 351 IIYFRDSDDEMPDSDEDPDDDLDI 374
I+Y D++D+ +EDPDDDLD+
Sbjct: 78 ILYEPDANDDY--DEEDPDDDLDV 99
>UniRef100_Q99L85 S-phase 2 protein [Mus musculus]
Length = 300
Score = 37.4 bits (85), Expect = 0.68
Identities = 55/234 (23%), Positives = 86/234 (36%), Gaps = 55/234 (23%)
Query: 158 VAIDSLSELLRHASLQSVAGLLSNLRSHD---------QISSIFGLLHSDLHEERAAAVL 208
+A+DSLS LL H ++ L L + + + GLLH +LH + L
Sbjct: 105 IALDSLSWLLCHIPCVTLCQALHALSQQNGDPGDNSLVEQVRVLGLLHEELHGPGSMGAL 164
Query: 209 EYMSSMVASVAPLHHSAERGNLENSLSEQNFTKGRLNVRFKRRNGRVRVTCEEFKVEAG- 267
++ +++ G ++ + + ++ +R R F V
Sbjct: 165 NTLAHTEVTLS--------GKVDQTSA---------SILCRRPQQRATYQTWWFSVLPDF 207
Query: 268 ------GISLTSVSSADGATV-AGLLPKVQFNLQLSEKEKTDRAKVVLPFEHQGTGRPIQ 320
G+ L S D T + FNL LS+KE+ R + LPF+
Sbjct: 208 SLTLHEGLPLRSELHPDHHTTQVDPTAHLTFNLHLSKKEREARDSLTLPFQF-------- 259
Query: 321 IYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLDI 374
SSE + G I Y D+ D++ EDPDDDLDI
Sbjct: 260 -----------SSEKQKALLHPVPSRTTGHIFYEPDAFDDV--DPEDPDDDLDI 300
>UniRef100_Q9CUX2 Mus musculus adult male hippocampus cDNA, RIKEN full-length
enriched library, clone:2900052B08 product:retinoic acid
induced 12, full insert sequence [Mus musculus]
Length = 277
Score = 37.4 bits (85), Expect = 0.68
Identities = 55/234 (23%), Positives = 86/234 (36%), Gaps = 55/234 (23%)
Query: 158 VAIDSLSELLRHASLQSVAGLLSNLRSHD---------QISSIFGLLHSDLHEERAAAVL 208
+A+DSLS LL H ++ L L + + + GLLH +LH + L
Sbjct: 82 IALDSLSWLLCHIPCVTLCQALHALSQQNGDPGDNSLVEQVRVLGLLHEELHGPGSMGAL 141
Query: 209 EYMSSMVASVAPLHHSAERGNLENSLSEQNFTKGRLNVRFKRRNGRVRVTCEEFKVEAG- 267
++ +++ G ++ + + ++ +R R F V
Sbjct: 142 NTLAHTEVTLS--------GKVDQTSA---------SILCRRPQQRATYQTWWFSVLPDF 184
Query: 268 ------GISLTSVSSADGATV-AGLLPKVQFNLQLSEKEKTDRAKVVLPFEHQGTGRPIQ 320
G+ L S D T + FNL LS+KE+ R + LPF+
Sbjct: 185 SLTLHEGLPLRSELHPDHHTTQVDPTAHLTFNLHLSKKEREARDSLTLPFQF-------- 236
Query: 321 IYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLDI 374
SSE + G I Y D+ D++ EDPDDDLDI
Sbjct: 237 -----------SSEKQKALLHPVPSRTTGHIFYEPDAFDDV--DPEDPDDDLDI 277
>UniRef100_Q6IUP3 DERP6 [Rattus norvegicus]
Length = 318
Score = 37.4 bits (85), Expect = 0.68
Identities = 58/233 (24%), Positives = 89/233 (37%), Gaps = 53/233 (22%)
Query: 158 VAIDSLSELLRHASLQSVAGLLSNLRSHD---------QISSIFGLLHSDLHEERAAAVL 208
+A+DSLS LL H ++ L L + + + GLLH +LH
Sbjct: 123 IALDSLSWLLCHIPCVTLCQALHALSQRNVDPGDNPLIEQVRVLGLLHEELHGP------ 176
Query: 209 EYMSSMVASVAPLHHSAERGNLENSLS-EQNFTKGRLNVRFKRRNGRVRV----TCEEFK 263
V +V+ L H+ E +LS + + T + R ++ + +F
Sbjct: 177 ----GPVGAVSSLAHT------EVTLSGKMDQTSASILCRRPQQRATYQTWWFSILPDFS 226
Query: 264 VEAG-GISLTSVSSADGATV-AGLLPKVQFNLQLSEKEKTDRAKVVLPFEHQGTGRPIQI 321
++ G+ L S D T + FNL LS+KE+ + + LPF+
Sbjct: 227 LDLHEGLPLHSELHRDPHTTQVDPASHLTFNLHLSKKEREAKDSLTLPFQF--------- 277
Query: 322 YDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDDDLDI 374
SSE G I Y D+ D++ EDPDDDLDI
Sbjct: 278 ----------SSEKQQALLHPVPGQTTGRIFYEPDAFDDV--DQEDPDDDLDI 318
>UniRef100_Q9FFL2 Nucleolar protein-like [Arabidopsis thaliana]
Length = 834
Score = 37.0 bits (84), Expect = 0.89
Identities = 19/53 (35%), Positives = 31/53 (57%), Gaps = 7/53 (13%)
Query: 323 DGRRSLDDNSSEAAPI-------SSGKKENSEMGEIIYFRDSDDEMPDSDEDP 368
DG + L D+S EA+ I S K +SEMG+II + D++ D++++P
Sbjct: 687 DGVKELKDDSEEASEIVGTSGRVSESKVSSSEMGDIILLENGDEKKVDAEDEP 739
>UniRef100_Q7PHH2 ENSANGP00000022479 [Anopheles gambiae str. PEST]
Length = 223
Score = 37.0 bits (84), Expect = 0.89
Identities = 27/131 (20%), Positives = 57/131 (42%), Gaps = 4/131 (3%)
Query: 247 RFKRRNGRVRVTCEEFKVEAGGISLTSVSSA----DGATVAGLLPKVQFNLQLSEKEKTD 302
R K+R + EE K + + V++ D A + + Q QL +K KT
Sbjct: 47 RLKKREMLKQKQKEEKKTKKATKNAAKVAAVPNTFDPAEIFNIEKSRQKYQQLGQKSKTP 106
Query: 303 RAKVVLPFEHQGTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMP 362
+ ++ + ++D +D++ +AA + + S+M + + +S+DE
Sbjct: 107 QPVATNGHQNGSAKKGRNLFDSDNEMDEDEQKAATGKKVQVKKSQMKKFLQRAESEDESD 166
Query: 363 DSDEDPDDDLD 373
+E+ D D++
Sbjct: 167 SEEEEMDSDVE 177
>UniRef100_Q6FLK7 Similar to sp|P38630 Saccharomyces cerevisiae YOR217w RFC1 DNA
replication factor C [Candida glabrata]
Length = 854
Score = 36.6 bits (83), Expect = 1.2
Identities = 17/64 (26%), Positives = 33/64 (51%), Gaps = 11/64 (17%)
Query: 311 EHQGTGRPIQIYDGRRSLDDNSSEAAPISSGKKENSEMGEIIYFRDSDDEMPDSDEDPDD 370
++ G P+ IY + + P+S G +E + +++ D+DD +P ++ED D
Sbjct: 772 KYNGMTHPVAIY--------RTGSSVPVSGGSRETPDFEDVV---DADDAVPAAEEDTQD 820
Query: 371 DLDI 374
D D+
Sbjct: 821 DNDL 824
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 585,737,526
Number of Sequences: 2790947
Number of extensions: 23252052
Number of successful extensions: 75674
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 75491
Number of HSP's gapped (non-prelim): 113
length of query: 374
length of database: 848,049,833
effective HSP length: 129
effective length of query: 245
effective length of database: 488,017,670
effective search space: 119564329150
effective search space used: 119564329150
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0346.3


