

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0336.15
(501 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
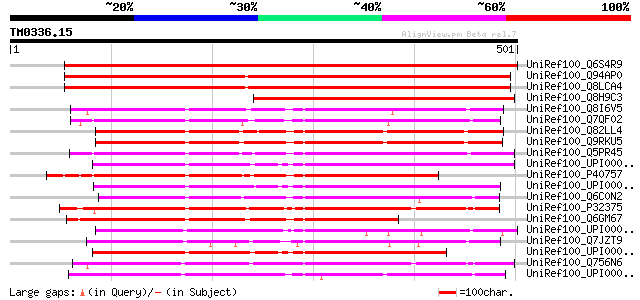
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6S4R9 Allantoinase [Robinia pseudoacacia] 832 0.0
UniRef100_Q94AP0 Hypothetical protein AT4g04955 [Arabidopsis tha... 646 0.0
UniRef100_Q8LCA4 Hypothetical protein [Arabidopsis thaliana] 643 0.0
UniRef100_Q8H9C3 Putative allantoinase [Solanum tuberosum] 389 e-107
UniRef100_Q8I6V5 Allantoinase [Ctenocephalides felis] 331 3e-89
UniRef100_Q7QF02 ENSANGP00000012435 [Anopheles gambiae str. PEST] 324 3e-87
UniRef100_Q82LL4 Putative allantoinase [Streptomyces avermitilis] 322 2e-86
UniRef100_Q9RKU5 Probable allantoinase [Streptomyces coelicolor] 320 8e-86
UniRef100_Q5PR45 Hypothetical protein [Brachydanio rerio] 319 1e-85
UniRef100_UPI0000360CF8 UPI0000360CF8 UniRef100 entry 317 7e-85
UniRef100_P40757 Allantoinase, mitochondrial precursor [Rana cat... 312 2e-83
UniRef100_UPI0000360CF9 UPI0000360CF9 UniRef100 entry 309 1e-82
UniRef100_Q6C0N2 Similar to sp|P32375 Saccharomyces cerevisiae A... 305 2e-81
UniRef100_P32375 Allantoinase [Saccharomyces cerevisiae] 301 4e-80
UniRef100_Q6GM67 MGC83388 protein [Xenopus laevis] 296 1e-78
UniRef100_UPI0000235FAE UPI0000235FAE UniRef100 entry 291 3e-77
UniRef100_Q7JZT9 RE13129p [Drosophila melanogaster] 283 6e-75
UniRef100_UPI0000360CF7 UPI0000360CF7 UniRef100 entry 282 1e-74
UniRef100_Q756N6 AER218Cp [Ashbya gossypii] 273 8e-72
UniRef100_UPI000042D9AE UPI000042D9AE UniRef100 entry 273 1e-71
>UniRef100_Q6S4R9 Allantoinase [Robinia pseudoacacia]
Length = 512
Score = 832 bits (2150), Expect = 0.0
Identities = 407/447 (91%), Positives = 427/447 (95%)
Query: 55 SEDLEINEGKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFV 114
S +EINEG+IVSI EGYGKQGNS QE VIDYGEAV+MPGLIDVHVHLDEPGRTEWEGF
Sbjct: 66 SGSVEINEGEIVSIIEGYGKQGNSMQEAVIDYGEAVVMPGLIDVHVHLDEPGRTEWEGFD 125
Query: 115 TGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNT 174
TGTRAAAAGGVTTVVDMPLNN+PTTVS+ETLQLKLEAAE +L+VDVGFWGGL+PENALNT
Sbjct: 126 TGTRAAAAGGVTTVVDMPLNNYPTTVSKETLQLKLEAAEKKLYVDVGFWGGLIPENALNT 185
Query: 175 SILEGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKN 234
SILEGLLSAGVLG+KSFMCPSGI+DFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKN
Sbjct: 186 SILEGLLSAGVLGVKSFMCPSGIDDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKN 245
Query: 235 NLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSSV 294
+LELKGN DPR Y TYLNTRPPSWE+AAIKELVDVTKDTI GGPLEGAHVHIVHLSDSS
Sbjct: 246 HLELKGNGDPRAYLTYLNTRPPSWEQAAIKELVDVTKDTIIGGPLEGAHVHIVHLSDSSA 305
Query: 295 SLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEG 354
SLDLIKEAK RGDSISVETC HYLAFSSEEIPD DTRFKCSPPIRDA NKEKLWEAVLEG
Sbjct: 306 SLDLIKEAKSRGDSISVETCPHYLAFSSEEIPDRDTRFKCSPPIRDALNKEKLWEAVLEG 365
Query: 355 HIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWS 414
HIDLL+SDHSPTVPELKLL+EGDFL+AWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWS
Sbjct: 366 HIDLLSSDHSPTVPELKLLEEGDFLRAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWS 425
Query: 415 QKPATLAGIASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKVL 474
+KPAT AG+ SKGAI VGNHADIVVW PELEF+LNDDYPVF+KHP+LSAYMGRRLSGKVL
Sbjct: 426 KKPATFAGLESKGAIAVGNHADIVVWQPELEFDLNDDYPVFIKHPSLSAYMGRRLSGKVL 485
Query: 475 DTFVRGNLVFKDGKHAPAACGVPILAK 501
DTFVRGNLVFKDGKHAPAACGVPILAK
Sbjct: 486 DTFVRGNLVFKDGKHAPAACGVPILAK 512
>UniRef100_Q94AP0 Hypothetical protein AT4g04955 [Arabidopsis thaliana]
Length = 506
Score = 646 bits (1666), Expect = 0.0
Identities = 309/442 (69%), Positives = 376/442 (84%), Gaps = 3/442 (0%)
Query: 55 SEDLEINEGKIVSITEGYGKQGNSKQEV-VIDYGEAVIMPGLIDVHVHLDEPGRTEWEGF 113
S +E+ G IVS+ + + + V VIDYGEAV+MPGLIDVHVHLD+PGR+EWEGF
Sbjct: 60 SGSVEVKGGIIVSVVKEVDWHKSQRSRVKVIDYGEAVLMPGLIDVHVHLDDPGRSEWEGF 119
Query: 114 VTGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALN 173
+GT+AAAAGG+TT+VDMPLN+ P+TVS ETL+LK+EAA+N +HVDVGFWGGLVP+NALN
Sbjct: 120 PSGTKAAAAGGITTLVDMPLNSFPSTVSPETLKLKIEAAKNRIHVDVGFWGGLVPDNALN 179
Query: 174 TSILEGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSK 233
+S LE LL AGVLGLKSFMCPSGINDFPMT I HIKEGLSVLAKY+RPLLVHAEI++D +
Sbjct: 180 SSALESLLDAGVLGLKSFMCPSGINDFPMTNITHIKEGLSVLAKYKRPLLVHAEIERDLE 239
Query: 234 NNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSS 293
+E +DPR Y TYL TRP SWEE AI+ L+ VT++T GG EGAH+HIVHLSD+S
Sbjct: 240 --IEDGSENDPRSYLTYLKTRPTSWEEGAIRNLLSVTENTRIGGSAEGAHLHIVHLSDAS 297
Query: 294 VSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLE 353
SLDLIKEAK +GDS++VETC HYLAFS+EEIP+GDTRFKCSPPIRDA N+EKLWEA++E
Sbjct: 298 SSLDLIKEAKGKGDSVTVETCPHYLAFSAEEIPEGDTRFKCSPPIRDAANREKLWEALME 357
Query: 354 GHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLW 413
G ID+L+SDHSPT PELKL+ +G+FLKAWGGISSLQF LP+TWSYGKK+G+TLEQ++ W
Sbjct: 358 GDIDMLSSDHSPTKPELKLMSDGNFLKAWGGISSLQFVLPITWSYGKKYGVTLEQVTSWW 417
Query: 414 SQKPATLAGIASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKV 473
S +P+ LAG+ SKGA+ VG HAD+VVW PE EF++++D+P+ KHP++SAY+GRRLSGKV
Sbjct: 418 SDRPSKLAGLHSKGAVTVGKHADLVVWEPEAEFDVDEDHPIHFKHPSISAYLGRRLSGKV 477
Query: 474 LDTFVRGNLVFKDGKHAPAACG 495
+ TFVRGNLVF +GKHA ACG
Sbjct: 478 VSTFVRGNLVFGEGKHASDACG 499
>UniRef100_Q8LCA4 Hypothetical protein [Arabidopsis thaliana]
Length = 506
Score = 643 bits (1658), Expect = 0.0
Identities = 308/442 (69%), Positives = 375/442 (84%), Gaps = 3/442 (0%)
Query: 55 SEDLEINEGKIVSITEGYGKQGNSKQEV-VIDYGEAVIMPGLIDVHVHLDEPGRTEWEGF 113
S +E+ G IVS+ + + + V VIDYGEAV+MPGLIDVHVHLD+PGR+EWEGF
Sbjct: 60 SGSVEVKGGIIVSVVKEVDWHKSQRSRVKVIDYGEAVLMPGLIDVHVHLDDPGRSEWEGF 119
Query: 114 VTGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALN 173
+GT+AAAAGG+TT+VDMPLN+ P+TVS ETL+LK+EAA+N +HVDVGFWGGLVP+NALN
Sbjct: 120 PSGTKAAAAGGITTLVDMPLNSFPSTVSPETLKLKIEAAKNRIHVDVGFWGGLVPDNALN 179
Query: 174 TSILEGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSK 233
+S LE LL AGVLGLKSFMCPSGINDFPMT I HIKEGLSVLAKY+RPLLVHAEI++D +
Sbjct: 180 SSALESLLDAGVLGLKSFMCPSGINDFPMTNITHIKEGLSVLAKYKRPLLVHAEIERDLE 239
Query: 234 NNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSS 293
+E +DPR Y TYL TRP SWEE AI+ L+ VT++T GG EGAH+HIVHLSD+S
Sbjct: 240 --IEDGSENDPRSYLTYLKTRPTSWEEGAIRNLLSVTENTRIGGSAEGAHLHIVHLSDAS 297
Query: 294 VSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLE 353
SLDLIKEAK +GDS++VETC HYLAFS+EEIP+GDTRFKCSPPIRDA N+EKLWEA++E
Sbjct: 298 SSLDLIKEAKGKGDSVTVETCPHYLAFSAEEIPEGDTRFKCSPPIRDAANREKLWEALME 357
Query: 354 GHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLW 413
G ID+L+SDHSPT PELKL+ +G+FLKAWGGISSLQF LP+TWSYGKK+G+TLEQ++ W
Sbjct: 358 GDIDMLSSDHSPTKPELKLMSDGNFLKAWGGISSLQFVLPITWSYGKKYGVTLEQVTSWW 417
Query: 414 SQKPATLAGIASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKV 473
S +P+ LA + SKGA+ VG HAD+VVW PE EF++++D+P+ KHP++SAY+GRRLSGKV
Sbjct: 418 SDRPSKLARLHSKGAVTVGKHADLVVWEPEAEFDVDEDHPIHFKHPSISAYLGRRLSGKV 477
Query: 474 LDTFVRGNLVFKDGKHAPAACG 495
+ TFVRGNLVF +GKHA ACG
Sbjct: 478 VSTFVRGNLVFGEGKHASDACG 499
>UniRef100_Q8H9C3 Putative allantoinase [Solanum tuberosum]
Length = 261
Score = 389 bits (1000), Expect = e-107
Identities = 181/258 (70%), Positives = 222/258 (85%)
Query: 242 DDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKE 301
++ R YSTYL TRP S EEAAI +L+ ++KDT GG EGAH+HIVHLSD+ SL+LIKE
Sbjct: 2 ENARSYSTYLKTRPASMEEAAINQLITLSKDTRAGGSAEGAHLHIVHLSDARTSLNLIKE 61
Query: 302 AKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLTS 361
AK RGDSI+VETC HYLAF++E+IPDGDTRFKC+PPIRDA NKEKLW+A+L+G ID+L+S
Sbjct: 62 AKQRGDSITVETCPHYLAFAAEDIPDGDTRFKCAPPIRDAANKEKLWDALLDGDIDMLSS 121
Query: 362 DHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPATLA 421
DHSP+VPE+KLL EGDFLKAWGGISSLQF LP TW++G+K+G+T EQL+ WS+ PA LA
Sbjct: 122 DHSPSVPEMKLLDEGDFLKAWGGISSLQFVLPATWTHGRKYGITFEQLASWWSENPAKLA 181
Query: 422 GIASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKVLDTFVRGN 481
G+ +KGAI VGN ADIVVW P++EF+L++DYPV +KHP++SAYMG RLSGKVL TFVRGN
Sbjct: 182 GLTTKGAIAVGNQADIVVWEPDMEFDLDNDYPVHIKHPSISAYMGSRLSGKVLATFVRGN 241
Query: 482 LVFKDGKHAPAACGVPIL 499
LV+K+G HA AC +PIL
Sbjct: 242 LVYKEGNHASQACALPIL 259
>UniRef100_Q8I6V5 Allantoinase [Ctenocephalides felis]
Length = 483
Score = 331 bits (848), Expect = 3e-89
Identities = 187/435 (42%), Positives = 264/435 (59%), Gaps = 25/435 (5%)
Query: 61 NEGKIVSITEGYGKQ--GNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTR 118
+ G+I SI G + N + V+DYG+ I PG+ID HVH++EPGR WEG+ T T+
Sbjct: 55 SSGRIKSIISGEEVERIANETKVEVLDYGQFSIWPGVIDSHVHVNEPGRESWEGYTTATK 114
Query: 119 AAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNTSILE 178
AAA GG+TT+VDMPLN+ P T + E L+ K+ +A + HVDV FWGG++P NA L
Sbjct: 115 AAAWGGITTIVDMPLNSIPPTTTVENLRTKVNSACGKTHVDVAFWGGVIPGNAHE---LL 171
Query: 179 GLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLEL 238
L++AGV G K F SG+++FP T + ++ L L K LL HAE+ +N
Sbjct: 172 PLINAGVRGFKCFTSESGVDEFPQVTKNDLEMALKELQKANSVLLYHAELPAPQEN---- 227
Query: 239 KGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDL 298
+++ Y TYL TRPPS E AI ++D+TK HIVHLS ++ +L
Sbjct: 228 VTSNETEKYMTYLKTRPPSMEVNAIDMIIDLTKK-------YKVRSHIVHLS-AAGALPQ 279
Query: 299 IKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDL 358
+K+A+ +S+ETC HYL F++E++PDG T +KC+PPIR+ N+EKLW+A+ ID+
Sbjct: 280 LKKARSENVPLSIETCHHYLTFAAEDVPDGHTEYKCAPPIREESNQEKLWQALENRDIDM 339
Query: 359 LTSDHSPTVPELKLLKEG---DFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQ 415
+ SDHSP+ LK L G DFLKAWGGI+ +QF L + + K G +S L S
Sbjct: 340 VVSDHSPSPAALKGLCNGCHPDFLKAWGGIAGMQFGLSLIRTGASKRGFKAHDVSRLLSA 399
Query: 416 KPATLAGI-ASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNL-SAYMGRRLSGKV 473
PA L G+ KG I G AD+V+W PE EF++ D ++H N + Y+G L GKV
Sbjct: 400 GPAKLTGLDGIKGQIKEGLDADLVIWDPEEEFKVTKD---IIQHKNKETPYLGMTLKGKV 456
Query: 474 LDTFVRGNLVFKDGK 488
T VRG+ V+++G+
Sbjct: 457 HATVVRGDFVYRNGQ 471
>UniRef100_Q7QF02 ENSANGP00000012435 [Anopheles gambiae str. PEST]
Length = 479
Score = 324 bits (831), Expect = 3e-87
Identities = 182/441 (41%), Positives = 261/441 (58%), Gaps = 36/441 (8%)
Query: 61 NEGKIVSI------TEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFV 114
++GK+ + E Y K E +D+G ++MPGLID HVH++EPGRTEWEGF
Sbjct: 31 HDGKVTRVLPSWAAVERYLKDNEGAYEH-LDFGNLMLMPGLIDTHVHINEPGRTEWEGFH 89
Query: 115 TGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNT 174
T T+AAAAGG TT+ DMPLN+ P T + L+ K+ AA ++ VDV FWGGLVP+N
Sbjct: 90 TATKAAAAGGFTTICDMPLNSIPPTTTVANLRAKVNAARGKIFVDVAFWGGLVPDNLPE- 148
Query: 175 SILEGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEI-----Q 229
L L++AGV G K F+CPSG+ +FP T + + E +L L HAE+ Q
Sbjct: 149 --LRRLIAAGVFGFKCFLCPSGVEEFPHVTEEQVMEAARLLEGTGAVLAFHAEVECQRLQ 206
Query: 230 QDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHL 289
D L DP Y ++L TRP + E+ AI+ + V + HIVHL
Sbjct: 207 SDCPRPL------DPHKYRSFLQTRPDTMEQRAIELVAKVAQ-------RHDVRAHIVHL 253
Query: 290 SDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWE 349
S ++ +L I+ A+ G ++VETC HYL+ ++E++PD T +KC PPIR N+E+LW
Sbjct: 254 S-TARALPTIRAARALGARLTVETCHHYLSLAAEDVPDARTEYKCCPPIRGRTNREQLWR 312
Query: 350 AVLEGHIDLLTSDHSPTVPELKLL----KEGDFLKAWGGISSLQFDLPVTWSYGKKHGLT 405
AV IDL+ SDHSP+ P+LKLL ++G+FL+AWGGISSLQ LP+ W+ +++GL+
Sbjct: 313 AVQARDIDLIVSDHSPSTPQLKLLTDGKRQGNFLEAWGGISSLQLGLPLFWTGCRRYGLS 372
Query: 406 LEQLSLLWSQKPATLAGI-ASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAY 464
L++ L +PA L+G+ K + VG D+ VW PE + +D F + Y
Sbjct: 373 LQETVRLLCTEPARLSGLDHRKSRLDVGYDGDMCVWDPEGTVTVTEDMLEF--QHKATPY 430
Query: 465 MGRRLSGKVLDTFVRGNLVFK 485
+ R L G V T +RG+L+++
Sbjct: 431 LNRTLQGVVHATILRGSLIYQ 451
>UniRef100_Q82LL4 Putative allantoinase [Streptomyces avermitilis]
Length = 445
Score = 322 bits (825), Expect = 2e-86
Identities = 175/405 (43%), Positives = 249/405 (61%), Gaps = 22/405 (5%)
Query: 85 DYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQET 144
D G+ V++PGL+D HVH+++PGRT WEGF T TRAAAAGG+TT+VDMPLN+ P T +
Sbjct: 49 DLGDDVLLPGLVDTHVHVNDPGRTHWEGFWTATRAAAAGGITTLVDMPLNSLPPTTTVGN 108
Query: 145 LQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPMTT 204
L+ K + A ++ H+DVGFWGG +P+N + L L AGV G K+F+ PSG+++FP
Sbjct: 109 LRTKRDVAADKAHIDVGFWGGALPDNVKD---LRPLHDAGVFGFKAFLSPSGVDEFPELD 165
Query: 205 IDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIK 264
+ + ++ +A + L+VHAE D + PR Y+ +L +RP E+ AI
Sbjct: 166 QERLARSMAEIAGFGGLLIVHAE---DPHHLAAAPQRGGPR-YTDFLASRPRDAEDTAIA 221
Query: 265 ELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEE 324
L+ K A VH++HLS SS +L LI AK G ++VETC HYL ++EE
Sbjct: 222 NLLAQAKRL-------NARVHVLHLS-SSDALPLIAGAKAEGVRVTVETCPHYLTLTAEE 273
Query: 325 IPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGG 384
+PDG + FKC PPIR+A N++ LW+A+ +G ID + +DHSP+ + LK DF AWGG
Sbjct: 274 VPDGASEFKCCPPIREAANQDLLWQALADGTIDCVVTDHSPSTAD---LKTDDFATAWGG 330
Query: 385 ISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPATLAGIASKGAIVVGNHADIVVWHPEL 444
IS LQ LP W+ ++ G +LE + S + A L G+A KGAI G AD V P+
Sbjct: 331 ISGLQLSLPAIWTEARRRGHSLEDVVRWMSARTARLVGLAQKGAIEAGRDADFAVLAPDE 390
Query: 445 EFELNDDYPVFLKHPN-LSAYMGRRLSGKVLDTFVRGNLVFKDGK 488
F ++ P L+H N ++AY G+ LSG V T++RG + DG+
Sbjct: 391 TFTVD---PAALQHRNRVTAYAGKTLSGVVKSTWLRGERIMADGE 432
>UniRef100_Q9RKU5 Probable allantoinase [Streptomyces coelicolor]
Length = 445
Score = 320 bits (819), Expect = 8e-86
Identities = 175/405 (43%), Positives = 250/405 (61%), Gaps = 23/405 (5%)
Query: 85 DYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQET 144
D G+ V++PGL+D HVH+++PGRTEWEGF T TRAAAAGG+TT+VDMPLN+ P T + +
Sbjct: 49 DVGDHVVLPGLVDTHVHVNDPGRTEWEGFWTATRAAAAGGITTLVDMPLNSIPPTTTVDN 108
Query: 145 LQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPMTT 204
L+ K E A ++ H+DVGFWGG +P+N + L L AGV G K+F+ PSG+++FP
Sbjct: 109 LRTKREVAADKAHIDVGFWGGALPDNVKD---LRPLHEAGVFGFKAFLSPSGVDEFPHLD 165
Query: 205 IDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIK 264
+ + L+ +A + L+VHAE ++L Y+ +L +RP E+ AI
Sbjct: 166 QEQLARSLAEIAAFDGLLIVHAE----DPHHLAAAPQQGGPKYTHFLASRPRDAEDTAIA 221
Query: 265 ELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEE 324
L+ K A VH++HLS SS +L LI EA+ G ++VETC HYL ++EE
Sbjct: 222 TLLAQAKRF-------NARVHVLHLS-SSDALPLIAEARADGVRVTVETCPHYLTLTAEE 273
Query: 325 IPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGG 384
+PDG + FKC PPIR+A N++ LW+A+ +G ID + +DHSP+ + LK DF AWGG
Sbjct: 274 VPDGASEFKCCPPIREAANQDLLWQALADGTIDCVVTDHSPSTAD---LKTDDFATAWGG 330
Query: 385 ISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPATLAGI-ASKGAIVVGNHADIVVWHPE 443
I+ LQ LP W+ + GL LE + S++ A L G+ A KGAI G+ AD V P+
Sbjct: 331 IAGLQLSLPAMWTAARGRGLGLEDVVRWMSERTAALVGLDARKGAIAPGHDADFAVLAPD 390
Query: 444 LEFELNDDYPVFLKHPN-LSAYMGRRLSGKVLDTFVRGNLVFKDG 487
F ++ P L+H N ++AY G+ L G V T++RG + DG
Sbjct: 391 ETFTVD---PAALQHRNRVTAYAGKTLYGVVKSTWLRGERIVADG 432
>UniRef100_Q5PR45 Hypothetical protein [Brachydanio rerio]
Length = 459
Score = 319 bits (817), Expect = 1e-85
Identities = 183/442 (41%), Positives = 256/442 (57%), Gaps = 21/442 (4%)
Query: 60 INEGKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRA 119
I EGKI S+ + +V +D G+++IMPG++D HVH++EPGRT+WEG+ T TRA
Sbjct: 28 IKEGKIHSVLPDSEASAHVAGKV-LDVGDSLIMPGIVDSHVHVNEPGRTDWEGYWTATRA 86
Query: 120 AAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNTSILEG 179
AAAGG+TT+ DMPLN P T + KL AA + VD FWGG++P N + L+
Sbjct: 87 AAAGGITTIADMPLNTIPPTTTLRNFNEKLCAATGQCFVDTAFWGGVIPGNQME---LKP 143
Query: 180 LLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLELK 239
+ AGV G K F+ SG+ +FP T + + L LL HAE Q+ + + K
Sbjct: 144 MCQAGVAGFKCFLIHSGVEEFPHVTDADLHAAMKQLQGTNSVLLFHAE--QELSHPVAEK 201
Query: 240 GNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLI 299
G DP+ YST+L +RP E AI +++ HIVHLS S+ L+LI
Sbjct: 202 G--DPQEYSTFLKSRPDIMELEAIHTVIEFCLQY-------QVRCHIVHLS-SAEPLELI 251
Query: 300 KEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLL 359
+ AK G ++VET HYL S+E+IP T+FKC PPIR N+E LW A+ G ID++
Sbjct: 252 RAAKQAGAPLTVETTHHYLNLSAEDIPGRATQFKCCPPIRGTANQELLWSALKAGDIDMV 311
Query: 360 TSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPAT 419
SDHSP P+LK L GDF +AWGGISSLQF L + W+ K G + + L ++PA
Sbjct: 312 VSDHSPCTPDLKCLDSGDFTQAWGGISSLQFGLSLFWTSASKRGFSFSDAARLLREEPAR 371
Query: 420 LAGIAS-KGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPN-LSAYMGRRLSGKVLDTF 477
L + + KG+++ G+ AD+V+W PE EF + +D + H N L+ Y+ R L G V
Sbjct: 372 LCRLDNQKGSLIPGHDADLVIWDPEKEFTVKEDN---IHHKNKLTPYLDRVLRGMVRFPI 428
Query: 478 VRGNLVFKDGKHAPAACGVPIL 499
VRG +V+ G + G +L
Sbjct: 429 VRGQVVYSHGSFSSQPLGKHLL 450
>UniRef100_UPI0000360CF8 UPI0000360CF8 UniRef100 entry
Length = 403
Score = 317 bits (811), Expect = 7e-85
Identities = 178/418 (42%), Positives = 246/418 (58%), Gaps = 16/418 (3%)
Query: 83 VIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQ 142
V+D G++V+MPG++D HVH++EPGR+ WEGF T TRAAAAGGVTT+VDMPLN+ P T +
Sbjct: 1 VLDVGDSVVMPGIVDCHVHVNEPGRSSWEGFWTATRAAAAGGVTTIVDMPLNSIPPTTTL 60
Query: 143 ETLQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPM 202
+ KL A+ + VD FWGG++P N L + ++ AGV G K F+ SG+ +FP
Sbjct: 61 DHFNEKLREAKGKCFVDTAFWGGVIPGNELE---IRPMIQAGVAGFKCFLIHSGVEEFPP 117
Query: 203 TTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAA 262
T + + L LLV+ +L L DP YST+L +RP E A
Sbjct: 118 VTDLDLHRAMKQLQGTGSVLLVNTCTLYLFPEHLHL----DPTEYSTFLQSRPDLMEIEA 173
Query: 263 IKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSS 322
I+ VT+ +H HIVHLS S+ L I+ A+ G ++VET HYL+ +
Sbjct: 174 IQT---VTELCLH----YQVRCHIVHLS-SAKPLKQIQAARQAGAPLTVETTHHYLSLCA 225
Query: 323 EEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAW 382
E+IP G T+FKC PPIRD+ N+ +LW A+ GHID++ SDHSP P+LK L GDF KAW
Sbjct: 226 EKIPAGATQFKCCPPIRDSNNQAQLWSALKAGHIDMVVSDHSPCTPDLKSLDSGDFTKAW 285
Query: 383 GGISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPATLAGIAS-KGAIVVGNHADIVVWH 441
GGISSLQF LP+ WS K G L + L S++ A L G+ KG++ AD+V+W
Sbjct: 286 GGISSLQFALPLFWSSASKRGFELPDVVRLLSKRTAQLCGLDHLKGSLAPSYDADLVIWD 345
Query: 442 PELEFELNDDYPVFLKHPNLSAYMGRRLSGKVLDTFVRGNLVFKDGKHAPAACGVPIL 499
P+ +F+++ + Y+G L G V T VRG LV+++G P G +L
Sbjct: 346 PDSQFQVSKKNHLQYSFSYGFPYLGITLQGVVRATIVRGRLVYQEGSFCPEPLGKHLL 403
>UniRef100_P40757 Allantoinase, mitochondrial precursor [Rana catesbeiana]
Length = 484
Score = 312 bits (799), Expect = 2e-83
Identities = 172/387 (44%), Positives = 245/387 (62%), Gaps = 18/387 (4%)
Query: 37 RVGIDFEKRKADPYTLNHSEDLEINEGKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLI 96
++ + KR T++ S D+ I++GKI S+ +GK S ++ +D G+ V+M G+I
Sbjct: 17 KISVIRSKRVIQANTIS-SCDIIISDGKISSVL-AWGKHVTSGAKL-LDVGDLVVMAGII 73
Query: 97 DVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENEL 156
D HVH++EPGRT+WEG+ T T AAAAGG+T +VDMPLN+ P T S KL+AA+ +
Sbjct: 74 DPHVHVNEPGRTDWEGYRTATLAAAAGGITAIVDMPLNSLPPTTSVTNFHTKLQAAKRQC 133
Query: 157 HVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLA 216
+VDV FWGG++P+N + L +L AGV G K F+ SG+ +FP ++ + +S L
Sbjct: 134 YVDVAFWGGVIPDNQVE---LIPMLQAGVAGFKCFLINSGVPEFPHVSVTDLHTAMSELQ 190
Query: 217 KYRRPLLVHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHG 276
LL HAE++ +K E+ D +Y T+L++RP E AA++ + D+ +
Sbjct: 191 GTNSVLLFHAELEI-AKPAPEI---GDSTLYQTFLDSRPDDMEIAAVQLVADLCQQY--- 243
Query: 277 GPLEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSP 336
HIVHLS S+ SL +I++AK G ++VET HYL+ SSE IP G T FKC P
Sbjct: 244 ----KVRCHIVHLS-SAQSLTIIRKAKEAGAPLTVETTHHYLSLSSEHIPPGATYFKCCP 298
Query: 337 PIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTW 396
P+R NKE LW A+L+GHID++ SDHSP P+LKLLKEGD++KAWGGISSLQF LP+ W
Sbjct: 299 PVRGHRNKEALWNALLQGHIDMVVSDHSPCTPDLKLLKEGDYMKAWGGISSLQFGLPLFW 358
Query: 397 SYGKKHGLTLEQLSLLWSQKPATLAGI 423
+ + G +L +S L S A L G+
Sbjct: 359 TSARTRGFSLTDVSQLLSSNTAKLCGL 385
>UniRef100_UPI0000360CF9 UPI0000360CF9 UniRef100 entry
Length = 391
Score = 309 bits (792), Expect = 1e-82
Identities = 172/403 (42%), Positives = 240/403 (58%), Gaps = 13/403 (3%)
Query: 84 IDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQE 143
+D G++V+MPG++D HVH++EPGR+ WEGF T TRAAAAGGVTT+VDMPLN+ P T + +
Sbjct: 1 LDVGDSVVMPGIVDCHVHVNEPGRSSWEGFWTATRAAAAGGVTTIVDMPLNSIPPTTTLD 60
Query: 144 TLQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPMT 203
KL A+ + VD FWGG++P N L + ++ AGV G K F+ SG+ +FP
Sbjct: 61 HFNEKLREAKGKCFVDTAFWGGVIPGNELE---IRPMIQAGVAGFKCFLIHSGVEEFPPV 117
Query: 204 TIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAI 263
T + + L LLV+ + +N + D P YST+L +RP E AI
Sbjct: 118 TDLDLHRAMKQLQGTGSVLLVNTFVITVQMHNTQTPALD-PTEYSTFLQSRPDLMEIEAI 176
Query: 264 KELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSE 323
+ VT+ +H HIVHLS S+ L I+ A+ G ++VET HYL+ +E
Sbjct: 177 QT---VTELCLH----YQVRCHIVHLS-SAKPLKQIQAARQAGAPLTVETTHHYLSLCAE 228
Query: 324 EIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWG 383
+IP G T+FKC PPIRD+ N+ +LW A+ GHID++ SDHSP P+LK L GDF KAWG
Sbjct: 229 KIPAGATQFKCCPPIRDSNNQAQLWSALKAGHIDMVVSDHSPCTPDLKSLDSGDFTKAWG 288
Query: 384 GISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPATLAGIAS-KGAIVVGNHADIVVWHP 442
GISSLQF LP+ WS K G L + L S++ A L G+ KG++ AD+V+W P
Sbjct: 289 GISSLQFALPLFWSSASKRGFELPDVVRLLSKRTAQLCGLDHLKGSLAPSYDADLVIWDP 348
Query: 443 ELEFELNDDYPVFLKHPNLSAYMGRRLSGKVLDTFVRGNLVFK 485
+ +F+++ + Y+G L G V T VRG LV++
Sbjct: 349 DSQFQVSKKNHLQYSFSYGFPYLGITLQGVVRATIVRGRLVYQ 391
>UniRef100_Q6C0N2 Similar to sp|P32375 Saccharomyces cerevisiae Allantoinase
[Yarrowia lipolytica]
Length = 457
Score = 305 bits (782), Expect = 2e-81
Identities = 167/400 (41%), Positives = 241/400 (59%), Gaps = 17/400 (4%)
Query: 88 EAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQL 147
E VI+PGL+D HVHL+EPGRTEWEGF TGT+AAAAGGVTTV+DMPLN P T +
Sbjct: 51 EQVILPGLVDAHVHLNEPGRTEWEGFATGTKAAAAGGVTTVIDMPLNAIPPTTTVANFDT 110
Query: 148 KLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPMTTIDH 207
KL AA + VDVGFWGG +P N+ + L L+ GV G K F+ SG+++FP +
Sbjct: 111 KLAAARGQCWVDVGFWGGAIPGNSED---LVPLIEKGVRGFKCFLIESGVDEFPCLEVPD 167
Query: 208 IKEGLSVLAKYRRPLLVHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELV 267
+K+ L + + + HAE+ + + +L N DP+ Y ++L++RP E AI ++
Sbjct: 168 VKKALKAIEGHPTIFMFHAEMA--NGESTDLPPNADPKSYMSFLHSRPDKLELTAINSII 225
Query: 268 DVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPD 327
+ +K + +HIVHL+ ++ ++ +IK+AK G +SVETC HYL F++E IP+
Sbjct: 226 EASKSS------PDVKLHIVHLA-TAEAVPIIKKAKEEGVPLSVETCFHYLHFAAESIPE 278
Query: 328 GDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISS 387
T+ KC PPIR N+ LW+A+ +G + + SDHSP P LK L+ GDF KAWGGI+S
Sbjct: 279 KSTQHKCCPPIRTEDNRSLLWKALEDGVVTSVVSDHSPCTPNLKDLQTGDFFKAWGGIAS 338
Query: 388 LQFDLPVTWSYGKKHG--LTLEQLSLLWSQKPATLAG-IASKGAIVVGNHADIVVWHPEL 444
+ LP+ W+ +K G +T+ +S S A G + SKG+I G AD ++ PE
Sbjct: 339 VGLGLPILWTTARKDGRNVTVADISKWCSYNTAEQVGLLKSKGSIKEGKDADFCIFDPEA 398
Query: 445 EFELNDDYPVFLKHPNLSAYMGRRLSGKVLDTFVRGNLVF 484
+ ++ D F +S Y L G+V T VRG +V+
Sbjct: 399 KVKITTDKLHFKN--KISPYKDGTLVGEVRRTIVRGQVVY 436
>UniRef100_P32375 Allantoinase [Saccharomyces cerevisiae]
Length = 460
Score = 301 bits (770), Expect = 4e-80
Identities = 177/438 (40%), Positives = 266/438 (60%), Gaps = 24/438 (5%)
Query: 50 YTLNHSEDLEINEGKIVSITEGYGKQGNSKQEV--VIDYGEAVIMPGLIDVHVHLDEPGR 107
Y+ L++ EG +V K +K E+ + + I+PGL+D HVHL+EPGR
Sbjct: 24 YSTESGTILDVLEGSVVM-----EKTEITKYEIHTLENVSPCTILPGLVDSHVHLNEPGR 78
Query: 108 TEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLV 167
T WEGF TGT+AA +GGVTTVVDMPLN P T + E ++KLEAAE ++ DVGFWGGLV
Sbjct: 79 TSWEGFETGTQAAISGGVTTVVDMPLNAIPPTTNVENFRIKLEAAEGQMWCDVGFWGGLV 138
Query: 168 PENALNTSILEGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAE 227
P N + L L+ AGV G K F+ SG+ +FP ++I+E L VLA+ ++ HAE
Sbjct: 139 PHNLPD---LIPLVKAGVRGFKGFLLDSGVEEFPPIGKEYIEEALKVLAEEDTMMMFHAE 195
Query: 228 IQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIV 287
+ + ++ + + R YS++L++RP S+E AI +++ + GP+ VHIV
Sbjct: 196 LPKAHED--QQQPEQSHREYSSFLSSRPDSFEIDAINLILECLR--ARNGPV--PPVHIV 249
Query: 288 HLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKL 347
HL+ S ++ LI++A+ G ++ ETC HYL ++E+IPDG T FKC PPIR N++ L
Sbjct: 250 HLA-SMKAIPLIRKARASGLPVTTETCFHYLCIAAEQIPDGATYFKCCPPIRSESNRQGL 308
Query: 348 WEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLE 407
W+A+ EG I + SDHSP PELK L++GDF +WGGI+S+ LP+ ++ G +L
Sbjct: 309 WDALREGVIGSVVSDHSPCTPELKNLQKGDFFDSWGGIASVGLGLPLMFT----QGCSLV 364
Query: 408 QLSLLWSQKPATLAGIA-SKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMG 466
+ + + G++ KG I G AD+VV+ + ++++ V+ K+ L+AY G
Sbjct: 365 DIVTWCCKNTSHQVGLSHQKGTIAPGYDADLVVFDTASKHKISNS-SVYFKN-KLTAYNG 422
Query: 467 RRLSGKVLDTFVRGNLVF 484
+ G VL T +RG +V+
Sbjct: 423 MTVKGTVLKTILRGQVVY 440
>UniRef100_Q6GM67 MGC83388 protein [Xenopus laevis]
Length = 369
Score = 296 bits (757), Expect = 1e-78
Identities = 155/328 (47%), Positives = 212/328 (64%), Gaps = 16/328 (4%)
Query: 57 DLEINEGKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTG 116
D+ I +GKI+ I + +GKQ + ++D G+ V+MPG+ID HVH++EPGRT+WEG+ T
Sbjct: 27 DIIIKDGKIMGI-QPWGKQTVTMGSQLLDAGDRVVMPGVIDPHVHVNEPGRTDWEGYFTA 85
Query: 117 TRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNTSI 176
TRAAAAGG+TT+VDMPLN P T S KL+AA+ + VDV FWGG++P N L
Sbjct: 86 TRAAAAGGITTIVDMPLNCLPPTTSVSNFHTKLKAAKGQCFVDVAFWGGVIPGNQLE--- 142
Query: 177 LEGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNL 236
L +L AGV G K F+ SG+ +FP ++ + +S L LL HAE + S+
Sbjct: 143 LVPMLHAGVAGFKCFLISSGVPEFPHVSLMDLHSAMSELQGTNSVLLFHAEFDEPSR--- 199
Query: 237 ELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSL 296
N DP +Y T+L +RPP E AA++ + ++ HIVHLS S+++L
Sbjct: 200 -APTNGDPMLYETFLASRPPCMEIAAVEAVANLCLKY-------KVRCHIVHLS-SAMAL 250
Query: 297 DLIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHI 356
+I++AK G +SVET HYL S+E+IP G T FKC PP+R+ NKE LW A+L+GHI
Sbjct: 251 PIIRKAKKAGAPLSVETTHHYLTLSAEKIPPGATYFKCCPPVREQSNKEALWNALLQGHI 310
Query: 357 DLLTSDHSPTVPELKLLKEGDFLKAWGG 384
D++ SDHSP PELKLL++GDFL AWGG
Sbjct: 311 DMVVSDHSPCTPELKLLQDGDFLNAWGG 338
>UniRef100_UPI0000235FAE UPI0000235FAE UniRef100 entry
Length = 506
Score = 291 bits (745), Expect = 3e-77
Identities = 180/460 (39%), Positives = 258/460 (55%), Gaps = 55/460 (11%)
Query: 85 DYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQET 144
D+ V++PGL+D HVHL+EPGRTEWEGF TGT+AAA GGVTTV+DMPLN P T + E
Sbjct: 57 DFSPYVLLPGLVDAHVHLNEPGRTEWEGFYTGTQAAAFGGVTTVIDMPLNAIPPTTTVEN 116
Query: 145 LQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPMTT 204
+LKL+AAE + VDVGF+GG++P NA L+ L+ GV G K F+ SG+++FP +
Sbjct: 117 FKLKLKAAEGKCWVDVGFYGGIIPGNA---GELKALVREGVRGFKGFLIDSGVDEFPAVS 173
Query: 205 IDHIKEGLSVLAKYRRPLLVHAE-IQQDSKNNL-ELKGNDDPRVYSTYLNTRPPSWEEAA 262
+ +++ ++ LA L+ HAE + + + L E+ P YST+L +RP +E A
Sbjct: 174 SEDVRKAMAELADEPTTLMFHAEMVPPKTPSELPEVMPEGAPEAYSTFLASRPSEYELCA 233
Query: 263 IKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSS 322
++E++ ++ H P +HIVHLS + ++ L+++A+ G I+ ETC HYL+ ++
Sbjct: 234 VEEILSLS----HLAP--KLPLHIVHLS-AMEAIPLLRKARAEGVPITAETCYHYLSLAA 286
Query: 323 EEIPDGDTRFKCSPPIRDAFNKEKLWEAV----LEGHIDLLTSDHSPTVPELKLL----- 373
EEI DGDTR KC PPIR N++ LW + +G I + SDHSP P+LKLL
Sbjct: 287 EEIRDGDTRHKCCPPIRSKSNQDALWAELDRHAEDGVIKTIVSDHSPCTPDLKLLPSHIP 346
Query: 374 -----------------KEGDFLKAWGGISSLQFDLPVTWS-YGKKHGLT---------- 405
EG FL AWGGISS+ LP+ W+ ++ GLT
Sbjct: 347 GNCSHGSSKHANTTPVVNEGSFLSAWGGISSVGLGLPILWTELSRRKGLTSSPDDTTTKQ 406
Query: 406 -LEQLSLLWSQKPATLAGI-ASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSA 463
L+ + L A G+ SKG +V G AD V+ E+E+ +F S
Sbjct: 407 ALQDIVHLCCANTAAQVGLHKSKGDLVPGYDADFCVFDDTAEWEVEPSTMLF--RNKCSP 464
Query: 464 YMGRRLSGKVLDTFVRGNLVFK--DGKHAPAACGVPILAK 501
Y GR + G V +T++RG +F DG A G +L K
Sbjct: 465 YQGRTMRGMVRETWLRGEKIFNRDDGFTVKAPSGSLLLEK 504
>UniRef100_Q7JZT9 RE13129p [Drosophila melanogaster]
Length = 492
Score = 283 bits (725), Expect = 6e-75
Identities = 174/430 (40%), Positives = 245/430 (56%), Gaps = 40/430 (9%)
Query: 77 NSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNH 136
N++ E V D+G+ V+MPGLID +VH++EPGR +WEGF T T+AAAAGG TT++D P N
Sbjct: 55 NTESESVYDFGDLVLMPGLIDPNVHINEPGRKDWEGFATATKAAAAGGFTTIIDRPTNAQ 114
Query: 137 PTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSG 196
P TVS L+ K A +++VDVGFWGGLVP N L LL AGV+GL+ +C
Sbjct: 115 PPTVSVAHLKAKTSTARGKIYVDVGFWGGLVPGNG---DQLAALLGAGVMGLQCTLCDPA 171
Query: 197 I---NDFPMTTIDHIKEGLSVLAKYRRP----LLVHAEIQQDSKNNLELKGNDDPRVYST 249
+FP ++E LS L K + + VHAE+ ++ + + + PR Y T
Sbjct: 172 APVSQEFPAVNESQLEEALSQLDKDQAEGEAIVAVHAELPLTTEIHPD---EEAPREYGT 228
Query: 250 YLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAH----VHIVHLSDSSVSLDLIKEAKLR 305
+L TRPP E +A + L L H +HI++ S S SL L++E + +
Sbjct: 229 FLVTRPPQMEISATQLLC----------RLANRHPRRCIHILNCS-SGESLPLVEECRRQ 277
Query: 306 GDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLE-GHIDLLTSDHS 364
G +++V+TC HYLA ++E++PD T FK PPIR+ N+E+LW+A+ G I ++ SDHS
Sbjct: 278 GGNLTVDTCPHYLALAAEDVPDCGTEFKTWPPIRERRNQEQLWQALRPGGAIRMIGSDHS 337
Query: 365 PTVPELKLLK----EGDFLKAWGGISSLQFDLPVTWSYGKKH----GLTLEQLSLLWSQK 416
P P + L G+FLKAW GI+SLQ L V W+ LT+ + L Q+
Sbjct: 338 PATPGARALTCGRGRGNFLKAWPGINSLQLSLSVVWTAAANRRGSANLTIADIHRLMCQE 397
Query: 417 PATLAGI-ASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKVLD 475
PA L G+ ASKG I G AD VW PE EF + + + + Y G++L G V
Sbjct: 398 PANLCGLSASKGRIAEGYDADFCVWSPEEEFTVGPE--LLYTATKATPYAGQKLRGVVHA 455
Query: 476 TFVRGNLVFK 485
T VRG V++
Sbjct: 456 TVVRGLHVYQ 465
>UniRef100_UPI0000360CF7 UPI0000360CF7 UniRef100 entry
Length = 406
Score = 282 bits (722), Expect = 1e-74
Identities = 158/350 (45%), Positives = 213/350 (60%), Gaps = 16/350 (4%)
Query: 83 VIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQ 142
V+D G++V+MPG++D HVH++EPGR+ WEGF T TRAAAAGGVTT+VDMPLN+ P T +
Sbjct: 51 VLDVGDSVVMPGIVDCHVHVNEPGRSSWEGFWTATRAAAAGGVTTIVDMPLNSIPPTTTL 110
Query: 143 ETLQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPM 202
+ KL A+ + VD FWGG++P N L + ++ AGV G K F+ SG+ +FP
Sbjct: 111 DHFNEKLREAKGKCFVDTAFWGGVIPGNELE---IRPMIQAGVAGFKCFLIHSGVEEFPP 167
Query: 203 TTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAA 262
T + + L LLV+ +L L DP YST+L +RP E A
Sbjct: 168 VTDLDLHRAMKQLQGTGSVLLVNTCTLYLFPEHLHL----DPTEYSTFLQSRPDLMEIEA 223
Query: 263 IKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSS 322
I+ VT+ +H HIVHLS S+ L I+ A+ G ++VET HYL+ +
Sbjct: 224 IQT---VTELCLH----YQVRCHIVHLS-SAKPLKQIQAARQAGAPLTVETTHHYLSLCA 275
Query: 323 EEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAW 382
E+IP G T+FKC PPIRD+ N+ +LW A+ GHID++ SDHSP P+LK L GDF KAW
Sbjct: 276 EKIPAGATQFKCCPPIRDSNNQAQLWSALKAGHIDMVVSDHSPCTPDLKSLDSGDFTKAW 335
Query: 383 GGISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPATLAGIAS-KGAIVV 431
GGISSLQF LP+ WS K G L + L S++ A L G+ KG++ V
Sbjct: 336 GGISSLQFALPLFWSSASKRGFELPDVVRLLSKRTAQLCGLDHLKGSLAV 385
>UniRef100_Q756N6 AER218Cp [Ashbya gossypii]
Length = 468
Score = 273 bits (698), Expect = 8e-72
Identities = 171/444 (38%), Positives = 239/444 (53%), Gaps = 20/444 (4%)
Query: 63 GKIVSITEGYGKQ-----GNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGT 117
GKI +I EG +Q G + D V+MPGL+D HVHLDEPGR WEGF +GT
Sbjct: 34 GKIEAIIEGPCEQYLDVCGPEAAKEFRDVTPLVVMPGLVDTHVHLDEPGRVHWEGFQSGT 93
Query: 118 RAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNTSIL 177
++A +GGVTTVVDMPLN+ P T + K +AA+ DV FWGGLVP N L
Sbjct: 94 QSAVSGGVTTVVDMPLNSIPPTTTMANFDAKKKAAQGRCWCDVAFWGGLVPG---NLEEL 150
Query: 178 EGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLE 237
L AGV G K F+ S +F I+ ++ L YR LL HAE+ +
Sbjct: 151 RPLAEAGVRGFKGFLIDSATPEFQAIDRPTIEAAMATLQGYRTVLLFHAELDEVPALPTP 210
Query: 238 LKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLD 297
+DP +YS++L TRP E A+ +V + P VHIVH+S S +L
Sbjct: 211 ---PEDPTLYSSFLETRPDRLEVDALNLIVKCLERATCRLPGPPPPVHIVHMS-SKHALP 266
Query: 298 LIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHID 357
+++ A+ G IS ETC HYL +E IP G T KC PPIR N+ LW A+ G +
Sbjct: 267 VLERARAAGLPISAETCFHYLTMDAETIPKGSTLHKCCPPIRPHENQAHLWNALRNGVLS 326
Query: 358 LLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKP 417
+ SDHSP PE++ GDFL+AWGGISSL L + +S G TL +++ ++
Sbjct: 327 SVVSDHSPCEPEIRCCDSGDFLQAWGGISSLGLGLSLIYS----KGATLPEIARWCAENT 382
Query: 418 ATLAGIA-SKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKVLDT 476
A AG+ KG I G AD+V++ P +++ + +H ++++ G + G+V T
Sbjct: 383 ALQAGLGHRKGFIRAGYDADLVLFDPRERHTISNS-QLHFRH-KMTSFHGTEVVGRVRQT 440
Query: 477 FVRGNLVFKDGK-HAPAACGVPIL 499
+RG +V+ K +PA G +L
Sbjct: 441 LLRGRVVYDAAKGPSPAPAGTLLL 464
>UniRef100_UPI000042D9AE UPI000042D9AE UniRef100 entry
Length = 481
Score = 273 bits (697), Expect = 1e-71
Identities = 170/461 (36%), Positives = 245/461 (52%), Gaps = 44/461 (9%)
Query: 59 EINEGKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFVTGTR 118
+ + G I S+T G + + K +V +++PGLID HVHL++PGRT WEGF TGT
Sbjct: 27 DTDSGTITSVTLGIHQPEHGKADVWEIEDGKILLPGLIDTHVHLNQPGRTAWEGFRTGTL 86
Query: 119 AAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAA-ENELHVDVGFWGGLVPENALNTSIL 177
AA +GG+TT++DMPLN+ P T + L K A E + DVGFWGG+VP N L
Sbjct: 87 AAISGGITTLIDMPLNSIPPTTTLSALATKQACAKEVGVSCDVGFWGGIVPG---NEGEL 143
Query: 178 EGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLE 237
GL AGV G K F+ SG+ +FP I + L ++ HAE+ S +
Sbjct: 144 RGLWEAGVKGFKCFLIESGVEEFPCVGEQDIIKACDALKGTNALIMFHAELDAPSSDAGH 203
Query: 238 LKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLD 297
G+ DP YST+L +RP WE +A++ ++ + + HIVHLS +S ++
Sbjct: 204 ATGHADPSEYSTFLQSRPQQWEISALQLILRIARS------YPDLRFHIVHLS-ASDAIP 256
Query: 298 LIKEAKLRG--------------------------DSISVETCSHYLAFSSEEIPDGDTR 331
+++ A G ++++VETC HYL +IP T+
Sbjct: 257 ILQSALGTGKGKGKDTGAGTGKDTGKDTDTENPTLENLTVETCFHYLCLCDTDIPPNGTQ 316
Query: 332 FKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFD 391
FKC PPIRD N+ KL EA+L+G I + SDHSP VPE LK+GDF+ AWGG+S L
Sbjct: 317 FKCCPPIRDESNRVKLIEALLDGTIHYVVSDHSPCVPE---LKKGDFMSAWGGVSGLGLG 373
Query: 392 LPVTWS-YGKKHGLTLEQLSLLWSQKPATLAGI-ASKGAIVVGNHADIVVWHPELEFELN 449
L + W+ G + L ++ A G+ KG I G AD VV+ P+ FE+
Sbjct: 374 LSLLWTELGAGGRVELGRIVEWMGPVQARQVGLQGKKGVIQTGAAADFVVFDPDATFEVT 433
Query: 450 DDYPVFLKHPNLSAYMGRRLSGKVLDTFVRGNLVFKDGKHA 490
+ F +S Y+G+RL+G+V T++ G LV++ G A
Sbjct: 434 LETLRFKN--KVSPYIGKRLTGRVEKTYLGGALVWEHGAGA 472
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 892,604,749
Number of Sequences: 2790947
Number of extensions: 39576195
Number of successful extensions: 92512
Number of sequences better than 10.0: 1592
Number of HSP's better than 10.0 without gapping: 1068
Number of HSP's successfully gapped in prelim test: 524
Number of HSP's that attempted gapping in prelim test: 89390
Number of HSP's gapped (non-prelim): 1872
length of query: 501
length of database: 848,049,833
effective HSP length: 132
effective length of query: 369
effective length of database: 479,644,829
effective search space: 176988941901
effective search space used: 176988941901
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0336.15


