

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0333b.4
(162 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
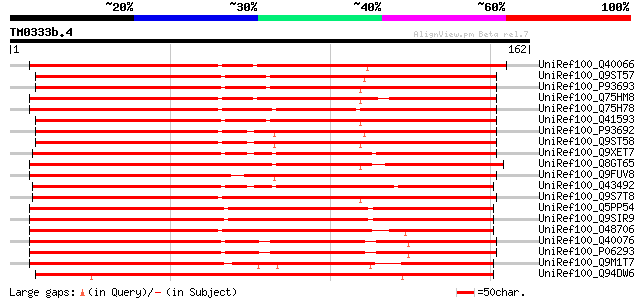
UniRef100_Q40066 Protein zx [Hordeum vulgare] 148 4e-35
UniRef100_Q9ST57 Serpin [Triticum aestivum] 144 1e-33
UniRef100_P93693 Serpin [Triticum aestivum] 143 1e-33
UniRef100_Q75HM8 Putative serine protease inhibitor [Oryza sativa] 143 2e-33
UniRef100_Q75H78 Putative serpin [Oryza sativa] 143 2e-33
UniRef100_Q41593 Serpin [Triticum aestivum] 142 2e-33
UniRef100_P93692 Serpin [Triticum aestivum] 140 9e-33
UniRef100_Q9ST58 Serpin [Triticum aestivum] 139 2e-32
UniRef100_Q9XET7 Barley protein Z homolog [Avena fatua] 137 1e-31
UniRef100_Q8GT65 Serpin-like protein [Citrus paradisi] 136 2e-31
UniRef100_Q9FUV8 Phloem serpin-1 [Cucurbita maxima] 134 8e-31
UniRef100_Q43492 Serpin [Hordeum vulgare] 133 1e-30
UniRef100_Q9S7T8 F16N3.3 protein [Arabidopsis thaliana] 132 4e-30
UniRef100_Q5PP54 At2g25240 [Arabidopsis thaliana] 127 8e-29
UniRef100_Q9SIR9 Putative serpin [Arabidopsis thaliana] 127 8e-29
UniRef100_O48706 Putative serpin [Arabidopsis thaliana] 127 8e-29
UniRef100_Q40076 Protein z-type serpin [Hordeum vulgare] 125 5e-28
UniRef100_P06293 Protein Z [Hordeum vulgare] 125 5e-28
UniRef100_Q9M1T7 Serpin-like protein [Arabidopsis thaliana] 123 1e-27
UniRef100_Q94DW6 Putative serpin [Oryza sativa] 113 2e-24
>UniRef100_Q40066 Protein zx [Hordeum vulgare]
Length = 398
Score = 148 bits (374), Expect = 4e-35
Identities = 82/151 (54%), Positives = 103/151 (67%), Gaps = 4/151 (2%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
QDGL L K+++EP F+E +P KV + FK+P+F ISF FEASD+LK LG+ LPFS
Sbjct: 250 QDGLWNLANKLSTEPEFMEKHMPMQKVPVGQFKLPKFKISFGFEASDMLKGLGLQLPFS- 308
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIR--ALRGGGGPPQGLKF 124
+AD ++MV + LYV S+F K F++V+EEGTEAAA T R LR P + F
Sbjct: 309 SEADLSEMVD-SPAARSLYVSSVFHKSFVEVNEEGTEAAARTARVVTLRSLPVEPVKVDF 367
Query: 125 VADHPFLFLIREDFSGTILFVGQVLNPLGGA 155
VADHPFLFLIRED +G +LFVG V NPL A
Sbjct: 368 VADHPFLFLIREDLTGVVLFVGHVFNPLVSA 398
>UniRef100_Q9ST57 Serpin [Triticum aestivum]
Length = 398
Score = 144 bits (362), Expect = 1e-33
Identities = 76/145 (52%), Positives = 101/145 (69%), Gaps = 3/145 (2%)
Query: 9 GLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGD 68
G+ L K+++EP LE +P+ KV L FK+P+F ISF EASD+LK LG+ LPFS +
Sbjct: 253 GIWSLAEKLSAEPELLERHIPRQKVALRQFKLPKFKISFGIEASDLLKHLGLQLPFSD-E 311
Query: 69 ADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATI-RALRGGGGPPQGLKFVAD 127
AD ++MV P L + S+F K F++V+E GTEAAAATI +A+ PP + F+AD
Sbjct: 312 ADLSEMVDSPMP-QGLRISSVFHKTFVEVNETGTEAAAATIAKAVLLSASPPSDMDFIAD 370
Query: 128 HPFLFLIREDFSGTILFVGQVLNPL 152
HPFLFLIRED SG +LF+G V+NPL
Sbjct: 371 HPFLFLIREDTSGVVLFIGHVVNPL 395
>UniRef100_P93693 Serpin [Triticum aestivum]
Length = 399
Score = 143 bits (361), Expect = 1e-33
Identities = 75/145 (51%), Positives = 104/145 (71%), Gaps = 3/145 (2%)
Query: 9 GLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGD 68
GLS L K+++EP FLE +P+ +V + FK+P+F ISF EASD+LK LG+ LPFS +
Sbjct: 254 GLSSLAEKLSAEPDFLERHIPRQRVAIRQFKLPKFKISFGMEASDLLKCLGLQLPFSD-E 312
Query: 69 ADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAAT-IRALRGGGGPPQGLKFVAD 127
ADF++MV P L V S+F + F++V+E+GTEAAA+T I+ + PP + F+AD
Sbjct: 313 ADFSEMVDSPMP-QGLRVSSVFHQAFVEVNEQGTEAAASTAIKMVPQQARPPSVMDFIAD 371
Query: 128 HPFLFLIREDFSGTILFVGQVLNPL 152
HPFLFL+RED SG +LF+G V+NPL
Sbjct: 372 HPFLFLLREDISGVVLFMGHVVNPL 396
>UniRef100_Q75HM8 Putative serine protease inhibitor [Oryza sativa]
Length = 653
Score = 143 bits (360), Expect = 2e-33
Identities = 81/148 (54%), Positives = 102/148 (68%), Gaps = 7/148 (4%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
QDGL L K+ SEP FLE +P +V + FK+P+F ISF FEASD+LK LG+ LPFS
Sbjct: 251 QDGLWSLAEKLNSEPEFLEKHIPTRQVTVGQFKLPKFKISFGFEASDLLKSLGLHLPFS- 309
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAAT--IRALRGGGGPPQGLKF 124
+AD T+MV + L+V S+F K F++V+EEGTEAAAAT + LR P F
Sbjct: 310 SEADLTEMVD-SPEGKNLFVSSVFHKSFVEVNEEGTEAAAATAAVITLRSA---PIAEDF 365
Query: 125 VADHPFLFLIREDFSGTILFVGQVLNPL 152
VADHPFLFLI+ED +G +LFVG V+NPL
Sbjct: 366 VADHPFLFLIQEDMTGVVLFVGHVVNPL 393
Score = 129 bits (325), Expect = 2e-29
Identities = 72/146 (49%), Positives = 95/146 (64%), Gaps = 4/146 (2%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
QDGL L K+ SEP F+E +P V + FK+P+F ISF F AS +LK LG+ L F
Sbjct: 509 QDGLWSLAEKLNSEPEFMENHIPMRPVHVGQFKLPKFKISFGFGASGLLKGLGLPLLFG- 567
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFVA 126
+ D +MV + L+V S+F K FI+V+EEGTEA AA + ++ P+ L FVA
Sbjct: 568 SEVDLIEMVD-SPGAQNLFVSSVFHKSFIEVNEEGTEATAAVMVSME--HSRPRRLNFVA 624
Query: 127 DHPFLFLIREDFSGTILFVGQVLNPL 152
DHPF+FLIRED +G ILF+G V+NPL
Sbjct: 625 DHPFMFLIREDVTGVILFIGHVVNPL 650
>UniRef100_Q75H78 Putative serpin [Oryza sativa]
Length = 442
Score = 143 bits (360), Expect = 2e-33
Identities = 81/146 (55%), Positives = 100/146 (68%), Gaps = 3/146 (2%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
QDGL L K+ SEP FLE R+P +V + FK+P+F ISF FEASD+LK LG+ LPFS
Sbjct: 299 QDGLWSLAAKLNSEPEFLEKRIPTRQVTVGKFKLPKFKISFGFEASDLLKILGLQLPFS- 357
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFVA 126
AD T MV N L+V S+F K F++V EEGTEAAAA+ A+ P + FVA
Sbjct: 358 SKADLTGMVGSPERHN-LFVSSLFHKSFVQVDEEGTEAAAAS-AAVVSFRSAPVTVDFVA 415
Query: 127 DHPFLFLIREDFSGTILFVGQVLNPL 152
DHPFLFLIRED +G +LF+G V+NPL
Sbjct: 416 DHPFLFLIREDMTGVVLFIGHVVNPL 441
>UniRef100_Q41593 Serpin [Triticum aestivum]
Length = 398
Score = 142 bits (359), Expect = 2e-33
Identities = 75/145 (51%), Positives = 103/145 (70%), Gaps = 3/145 (2%)
Query: 9 GLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGD 68
GLS L K+++EP FLE +P+ +V L FK+P+F ISF EASD+LK LG+ LPF +
Sbjct: 253 GLSSLAEKLSAEPDFLERHIPRQRVALRQFKLPKFKISFGIEASDLLKCLGLQLPFGD-E 311
Query: 69 ADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAAT-IRALRGGGGPPQGLKFVAD 127
ADF++MV P L V S+F + F++V+E+GTEAAA+T I+ + PP + F+AD
Sbjct: 312 ADFSEMVDSLMP-QGLRVSSVFHQAFVEVNEQGTEAAASTAIKMVLQQARPPSVMDFIAD 370
Query: 128 HPFLFLIREDFSGTILFVGQVLNPL 152
HPFLFL+RED SG +LF+G V+NPL
Sbjct: 371 HPFLFLVREDISGVVLFMGHVVNPL 395
>UniRef100_P93692 Serpin [Triticum aestivum]
Length = 398
Score = 140 bits (354), Expect = 9e-33
Identities = 76/146 (52%), Positives = 101/146 (69%), Gaps = 5/146 (3%)
Query: 9 GLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGD 68
GL L K+++EP FLE +P+ KV L FK+P+F IS EASD+LK LG++LPF +
Sbjct: 253 GLWSLAEKLSAEPEFLEQHIPRQKVALRQFKLPKFKISLGIEASDLLKGLGLLLPFG-AE 311
Query: 69 ADFTKMVKVNTPF-NELYVESIFQKVFIKVHEEGTEAAAATI-RALRGGGGPPQGLKFVA 126
AD ++M V++P LY+ SIF K F++V+E GTEAAA TI + + PP L F+
Sbjct: 312 ADLSEM--VDSPMAQNLYISSIFHKAFVEVNETGTEAAATTIAKVVLRQAPPPSVLDFIV 369
Query: 127 DHPFLFLIREDFSGTILFVGQVLNPL 152
DHPFLFLIRED SG +LF+G V+NPL
Sbjct: 370 DHPFLFLIREDTSGVVLFIGHVVNPL 395
>UniRef100_Q9ST58 Serpin [Triticum aestivum]
Length = 398
Score = 139 bits (351), Expect = 2e-32
Identities = 76/146 (52%), Positives = 106/146 (72%), Gaps = 5/146 (3%)
Query: 9 GLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGD 68
GLS L K+++EP FLE +P+ +V L FK+P+F ISF EASD+LK LG+ LPFS +
Sbjct: 253 GLSNLAEKLSAEPDFLERHIPRQRVALRQFKLPKFKISFETEASDLLKCLGLQLPFS-NE 311
Query: 69 ADFTKMVKVNTPF-NELYVESIFQKVFIKVHEEGTEAAAAT-IRALRGGGGPPQGLKFVA 126
ADF++M V++P + L V S+F + F++V+E+GTEAAA+T I+ PP + F+A
Sbjct: 312 ADFSEM--VDSPMAHGLRVSSVFHQAFVEVNEQGTEAAASTAIKMALLQARPPSVMDFIA 369
Query: 127 DHPFLFLIREDFSGTILFVGQVLNPL 152
DHPFLFL+RED SG +LF+G V+NPL
Sbjct: 370 DHPFLFLLREDISGVVLFMGHVVNPL 395
>UniRef100_Q9XET7 Barley protein Z homolog [Avena fatua]
Length = 280
Score = 137 bits (345), Expect = 1e-31
Identities = 73/145 (50%), Positives = 105/145 (72%), Gaps = 5/145 (3%)
Query: 8 DGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQG 67
DGL L ++++EP F+E +P+ KV++ FK+P+F ISF FEAS++L+ LG+ LPFS
Sbjct: 138 DGLWSLAKRLSTEPEFIENHIPKEKVEVGQFKLPKFKISFGFEASNLLQGLGLQLPFST- 196
Query: 68 DADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFVAD 127
+AD T+M V++P N L++ ++ K F++V+EEGTEA AAT + P+ + FVAD
Sbjct: 197 EADLTEM--VDSPEN-LHISAVQHKSFVEVNEEGTEAVAATATTIM-QTSMPRTIDFVAD 252
Query: 128 HPFLFLIREDFSGTILFVGQVLNPL 152
HPFLFLIRED SG +LFVG V+NP+
Sbjct: 253 HPFLFLIREDVSGVVLFVGHVVNPI 277
>UniRef100_Q8GT65 Serpin-like protein [Citrus paradisi]
Length = 389
Score = 136 bits (343), Expect = 2e-31
Identities = 78/153 (50%), Positives = 100/153 (64%), Gaps = 10/153 (6%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
+DGL L+ KM SE FL+ LP +V++ F+IPRF ISF E S VLK LG+VLPFS
Sbjct: 242 KDGLPTLLEKMGSESKFLDHHLPSQRVEVGDFRIPRFKISFGIEVSKVLKGLGLVLPFSG 301
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAAT-----IRALRGGGGPPQG 121
+MV N LYV SIFQK FI+V+EEGTEAAAA+ +R++
Sbjct: 302 EGGGLAEMVDSPVGKN-LYVSSIFQKSFIEVNEEGTEAAAASAATVVLRSIL----LLDK 356
Query: 122 LKFVADHPFLFLIREDFSGTILFVGQVLNPLGG 154
+ FVADHPF+F+IRED +G ++F+G VLNPL G
Sbjct: 357 IDFVADHPFVFMIREDMTGLVMFIGHVLNPLAG 389
>UniRef100_Q9FUV8 Phloem serpin-1 [Cucurbita maxima]
Length = 389
Score = 134 bits (337), Expect = 8e-31
Identities = 74/147 (50%), Positives = 100/147 (67%), Gaps = 5/147 (3%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
+DGL+ LI K+ SEPGF++ +P K +L F IP+F ISF E SDVLK+LG+VLPF++
Sbjct: 245 KDGLASLIEKLDSEPGFIDRHIPCKKQELGGFLIPKFKISFGIEVSDVLKKLGLVLPFTE 304
Query: 67 GDADFTKMVKVNTPF-NELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFV 125
G + V +P L V +IF K FI+V EEGT+AAA++ + P + F+
Sbjct: 305 GGL----LGMVESPVAQNLRVSNIFHKAFIEVDEEGTKAAASSAVTVGIVSLPINRIDFI 360
Query: 126 ADHPFLFLIREDFSGTILFVGQVLNPL 152
A+ PFL+LIRED SGT+LF+GQVLNPL
Sbjct: 361 ANRPFLYLIREDKSGTLLFIGQVLNPL 387
>UniRef100_Q43492 Serpin [Hordeum vulgare]
Length = 397
Score = 133 bits (335), Expect = 1e-30
Identities = 72/144 (50%), Positives = 101/144 (70%), Gaps = 5/144 (3%)
Query: 8 DGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQG 67
DGLS L K+++EP FLE R+P +V++ F +P+F ISF FEA+ +LK LG+ LPFS
Sbjct: 255 DGLSRLAQKLSTEPDFLENRIPTEEVEVGQFMLPKFKISFGFEANKLLKTLGLQLPFSL- 313
Query: 68 DADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFVAD 127
+A+ ++M VN+P LY+ S+F K F++V EEGT+A AAT + P + + FVA+
Sbjct: 314 EANLSEM--VNSPMG-LYISSVFHKTFVEVDEEGTKAGAATGDVIVDRSLPIR-MDFVAN 369
Query: 128 HPFLFLIREDFSGTILFVGQVLNP 151
HPFLFLIRED +G +LF+G V NP
Sbjct: 370 HPFLFLIREDIAGVVLFIGHVANP 393
>UniRef100_Q9S7T8 F16N3.3 protein [Arabidopsis thaliana]
Length = 391
Score = 132 bits (331), Expect = 4e-30
Identities = 71/147 (48%), Positives = 100/147 (67%), Gaps = 3/147 (2%)
Query: 8 DGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQG 67
+GLS L+ K+ S PGFL+ +P+ +VK+ FKIP+F SF F+AS+VLK LG+ PFS G
Sbjct: 245 NGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFSFGFDASNVLKGLGLTSPFS-G 303
Query: 68 DADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAAT--IRALRGGGGPPQGLKFV 125
+ T+MV+ L V +IF K I+V+EEGTEAAAA+ + LRG + FV
Sbjct: 304 EEGLTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAAASAGVIKLRGLLMEEDEIDFV 363
Query: 126 ADHPFLFLIREDFSGTILFVGQVLNPL 152
ADHPFL ++ E+ +G +LF+GQV++PL
Sbjct: 364 ADHPFLLVVTENITGVVLFIGQVVDPL 390
>UniRef100_Q5PP54 At2g25240 [Arabidopsis thaliana]
Length = 324
Score = 127 bits (320), Expect = 8e-29
Identities = 67/145 (46%), Positives = 96/145 (66%), Gaps = 2/145 (1%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
++GL+PL+ K+ SEP F + +P H + + +F+IP+F SF F AS+VLK++G+ PF+
Sbjct: 179 KEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVLKDMGLTSPFNN 238
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFVA 126
G T+MV + ++LYV SI K I+V EEGTEAAA ++ + + FVA
Sbjct: 239 G-GGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEAAAVSVGVV-SCTSFRRNPDFVA 296
Query: 127 DHPFLFLIREDFSGTILFVGQVLNP 151
D PFLF +RED SG ILF+GQVL+P
Sbjct: 297 DRPFLFTVREDKSGVILFMGQVLDP 321
>UniRef100_Q9SIR9 Putative serpin [Arabidopsis thaliana]
Length = 385
Score = 127 bits (320), Expect = 8e-29
Identities = 67/145 (46%), Positives = 96/145 (66%), Gaps = 2/145 (1%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
++GL+PL+ K+ SEP F + +P H + + +F+IP+F SF F AS+VLK++G+ PF+
Sbjct: 240 KEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVLKDMGLTSPFNN 299
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFVA 126
G T+MV + ++LYV SI K I+V EEGTEAAA ++ + + FVA
Sbjct: 300 G-GGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEAAAVSVGVV-SCTSFRRNPDFVA 357
Query: 127 DHPFLFLIREDFSGTILFVGQVLNP 151
D PFLF +RED SG ILF+GQVL+P
Sbjct: 358 DRPFLFTVREDKSGVILFMGQVLDP 382
>UniRef100_O48706 Putative serpin [Arabidopsis thaliana]
Length = 389
Score = 127 bits (320), Expect = 8e-29
Identities = 68/149 (45%), Positives = 101/149 (67%), Gaps = 10/149 (6%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
+DGL+ L+ K+++EPGFL+ +P H+ + + +IP+ + SF F+AS+VLK++G+ PF+
Sbjct: 244 KDGLAALLEKISTEPGFLDSHIPLHRTPVDALRIPKLNFSFEFKASEVLKDMGLTSPFT- 302
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGL---- 122
+ T+MV + ++L+V SI K I+V EEGTEAAA ++ + PQ L
Sbjct: 303 SKGNLTEMVDSPSNGDKLHVSSIIHKACIEVDEEGTEAAAVSVAIMM-----PQCLMRNP 357
Query: 123 KFVADHPFLFLIREDFSGTILFVGQVLNP 151
FVADHPFLF +RED SG ILF+GQVL+P
Sbjct: 358 DFVADHPFLFTVREDNSGVILFIGQVLDP 386
>UniRef100_Q40076 Protein z-type serpin [Hordeum vulgare]
Length = 400
Score = 125 bits (313), Expect = 5e-28
Identities = 69/151 (45%), Positives = 100/151 (65%), Gaps = 12/151 (7%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
QDGL L ++++EP F+E +P+ V++ F++P+F IS+ FEAS +L+ LG+ LPFS+
Sbjct: 254 QDGLWSLAKRLSTEPEFIENHIPKQTVEVGRFQLPKFKISYQFEASSLLRALGLQLPFSE 313
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLK--- 123
+AD ++MV + L + +F K F++V+EEGTEA AAT+ G LK
Sbjct: 314 -EADLSEMVDSS---QGLEISHVFHKSFVEVNEEGTEAGAATVAM---GVAMSMPLKVDL 366
Query: 124 --FVADHPFLFLIREDFSGTILFVGQVLNPL 152
FVA+HPFLFLIRED +G ++FVG V NPL
Sbjct: 367 VDFVANHPFLFLIREDIAGVVVFVGHVTNPL 397
>UniRef100_P06293 Protein Z [Hordeum vulgare]
Length = 399
Score = 125 bits (313), Expect = 5e-28
Identities = 69/151 (45%), Positives = 100/151 (65%), Gaps = 12/151 (7%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
QDGL L ++++EP F+E +P+ V++ F++P+F IS+ FEAS +L+ LG+ LPFS+
Sbjct: 253 QDGLWSLAKRLSTEPEFIENHIPKQTVEVGRFQLPKFKISYQFEASSLLRALGLQLPFSE 312
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLK--- 123
+AD ++MV + L + +F K F++V+EEGTEA AAT+ G LK
Sbjct: 313 -EADLSEMVDSS---QGLEISHVFHKSFVEVNEEGTEAGAATVAM---GVAMSMPLKVDL 365
Query: 124 --FVADHPFLFLIREDFSGTILFVGQVLNPL 152
FVA+HPFLFLIRED +G ++FVG V NPL
Sbjct: 366 VDFVANHPFLFLIREDIAGVVVFVGHVTNPL 396
>UniRef100_Q9M1T7 Serpin-like protein [Arabidopsis thaliana]
Length = 393
Score = 123 bits (309), Expect = 1e-27
Identities = 71/157 (45%), Positives = 102/157 (64%), Gaps = 22/157 (14%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
+DGL L+ +++S+P FL+ +P+ ++ +FKIP+F SF F+ASDVLKE+G+ LPF+
Sbjct: 244 RDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVLKEMGLTLPFTH 303
Query: 67 GDADFTKMVK-VNTPFN-----ELYVESIFQKVFIKVHEEGTEAAAATIRA------LRG 114
G T+MV+ + P N L+V ++F K I+V EEGTEAAA ++ + L G
Sbjct: 304 G--SLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAVSVASMTKDMLLMG 361
Query: 115 GGGPPQGLKFVADHPFLFLIREDFSGTILFVGQVLNP 151
FVADHPFLF +RE+ SG ILF+GQVL+P
Sbjct: 362 --------DFVADHPFLFTVREEKSGVILFMGQVLDP 390
>UniRef100_Q94DW6 Putative serpin [Oryza sativa]
Length = 411
Score = 113 bits (282), Expect = 2e-24
Identities = 58/149 (38%), Positives = 92/149 (60%), Gaps = 6/149 (4%)
Query: 9 GLSPLIGKMASEPGFL--EGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
GL ++ K+ S+P L KV + +F +PRF +S+ A++ L++LG+ LPF
Sbjct: 259 GLPDMLRKLCSDPAALIESSAALTEKVPVGAFMVPRFTLSYKTNAAETLRQLGLRLPFEY 318
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQG----L 122
AD ++MV+ + ++ V +++ + F++V+EEGTEAAAAT + G P +
Sbjct: 319 PGADLSEMVESSPEAEKIVVSAVYHESFVEVNEEGTEAAAATAVVMTLGCAAPSAPVHVV 378
Query: 123 KFVADHPFLFLIREDFSGTILFVGQVLNP 151
FVADHPF+FLI+ED +G ++F GQV NP
Sbjct: 379 DFVADHPFMFLIKEDLTGVVVFAGQVTNP 407
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.143 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 283,238,616
Number of Sequences: 2790947
Number of extensions: 11199971
Number of successful extensions: 23989
Number of sequences better than 10.0: 895
Number of HSP's better than 10.0 without gapping: 540
Number of HSP's successfully gapped in prelim test: 355
Number of HSP's that attempted gapping in prelim test: 22309
Number of HSP's gapped (non-prelim): 913
length of query: 162
length of database: 848,049,833
effective HSP length: 117
effective length of query: 45
effective length of database: 521,509,034
effective search space: 23467906530
effective search space used: 23467906530
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0333b.4


