

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0331c.6
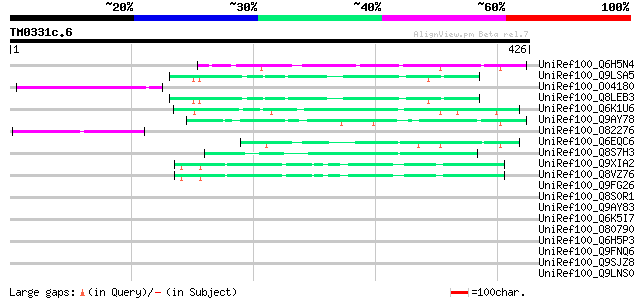
(426 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6H5N4 Hypothetical protein OJ1118_F11.28 [Oryza sativa] 67 1e-09
UniRef100_Q9LSA5 Gb|AAD43172.1 [Arabidopsis thaliana] 58 6e-07
UniRef100_O04180 Pge1 protein [Lotus japonicus] 57 1e-06
UniRef100_Q8LEB3 Hypothetical protein [Arabidopsis thaliana] 56 2e-06
UniRef100_Q6K1U6 Hypothetical protein OSJNBa0038P01.5 [Oryza sat... 55 5e-06
UniRef100_Q9AY78 Hypothetical protein OSJNBa0091J19.25 [Oryza sa... 55 5e-06
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 50 2e-04
UniRef100_Q6EQC6 Hypothetical protein P0448B03.3 [Oryza sativa] 49 3e-04
UniRef100_Q8S7H3 Hypothetical protein OSJNBa0010I09.22 [Oryza sa... 48 6e-04
UniRef100_Q9XIA2 F13F21.21 protein [Arabidopsis thaliana] 47 8e-04
UniRef100_Q8VZ76 Hypothetical protein At1g49360; F13F21.21 [Arab... 47 8e-04
UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like... 45 0.004
UniRef100_Q8S0R1 P0408G07.11 protein [Oryza sativa] 44 0.007
UniRef100_Q9AY83 Hypothetical protein OSJNBa0091J19.29 [Oryza sa... 44 0.009
UniRef100_Q6K5I7 Hypothetical protein OJ1791_B03.3 [Oryza sativa] 44 0.011
UniRef100_O80790 Putative non-LTR retroelement reverse transcrip... 43 0.015
UniRef100_Q6H5P3 Hypothetical protein OJ1118_F11.4 [Oryza sativa] 43 0.020
UniRef100_Q9FNQ6 Gb|AAF19536.1 [Arabidopsis thaliana] 43 0.020
UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcrip... 42 0.043
UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana] 40 0.17
>UniRef100_Q6H5N4 Hypothetical protein OJ1118_F11.28 [Oryza sativa]
Length = 407
Score = 67.0 bits (162), Expect = 1e-09
Identities = 66/282 (23%), Positives = 116/282 (40%), Gaps = 30/282 (10%)
Query: 155 ETYSRSIPELSGANLLTVQHGGWCVLEKGGKEKVRELLLWNPFLQEKIHLP----SLKHE 210
E S+ + E G +L + GW ++ ++ E+ LW+P +KI+LP +
Sbjct: 124 ELLSKRVDEF-GIHLYWITAQGWLLMVH---LELYEIFLWSPITNQKINLPFDEDNFLAN 179
Query: 211 NRIRNCILSCSPTETDQVCSIFLFPHEAYGYGPNILFYCEVGDKQWFAVDYRKDLVRASP 270
N + C LS P++ + + + + +YC WF +Y+ ++
Sbjct: 180 NNVVKCFLSHKPSDPNCIVLVV-------NCRDTMFWYCHPKGDVWFRHEYQSSMISTGE 232
Query: 271 PEEDGVVYNPFLYSPVYFNGRLYA-VSYSRLLVIIEKLEPPNGLQIHSADVYLPDPFPHT 329
E+ + L + GR +A ++ + ++ +E L P D PDP
Sbjct: 233 DRENVIATVKHLTA---VGGRFHAYLNQDKAILTLEFLPKPTFTTTPVKDA--PDPSYWC 287
Query: 330 FPCLRQLLVCNHELFHIQISMEY---NKIFGIAVHKLNFSHMIWERVKSIKDMVFFLSDK 386
LL ELF + +K+ I VHKLN S IW +V +I + F +
Sbjct: 288 TFSTCFLLESGGELFTLSFKHPIECVDKVMQIEVHKLNLSQRIWMKVSTIDNKAFLVDCT 347
Query: 387 DLPFARRVNPGGGGL----IYFTSMNNKFVHIFNLEDNSLRI 424
F +N GL IYF +K ++++N+E + I
Sbjct: 348 G--FGASLNAEDVGLKRNCIYFVRPKDKGLYVYNMERGTTTI 387
>UniRef100_Q9LSA5 Gb|AAD43172.1 [Arabidopsis thaliana]
Length = 380
Score = 57.8 bits (138), Expect = 6e-07
Identities = 67/265 (25%), Positives = 106/265 (39%), Gaps = 36/265 (13%)
Query: 132 QSYPWLLYVHNNLAIIRS----ILDPT--ETYSRSIPELSG-ANLLTVQHGGWCVLEKGG 184
QS PWL Y + + + +P+ +T+ PEL+G N L GW ++ K
Sbjct: 90 QSRPWLFYPQSQRGGPKEGDYVLFNPSRSQTHHLKFPELTGYRNKLACAKDGWLLVVKDN 149
Query: 185 KEKVRELLLWNPFLQEKIHLPSLKHENRIRNCI-LSCSPTETDQVCSIFLFPHEAYGYGP 243
+ V L NPF E+I LP + +N R+C+ S +PT T C + F +++ Y
Sbjct: 150 PDVVFFL---NPFTGERICLPQVP-QNSTRDCLTFSAAPTSTS--CCVISFTPQSFLYAV 203
Query: 244 NILFYCEVGDKQWFAVDYRKDLVRASPPEEDGVVYNPFLYSPVYFNGRLYAVSYS-RLLV 302
+ G+ W + D Y + ++ NG Y +S S RL V
Sbjct: 204 VKVDTWRPGESVWTTHHF------------DQKRYGEVINRCIFSNGMFYCLSTSGRLSV 251
Query: 303 IIEKLEPPNGLQIHSADVYLPDPFPHTFPCLRQLLVCNHE--LFHIQISMEYNKIFGIAV 360
E N L + F +RQ+ + HE +F + N+ +
Sbjct: 252 FDPSRETWNVLPVKPCRA-----FRRKIMLVRQVFMTEHEGDIFVVTTRRVNNR--KLLA 304
Query: 361 HKLNFSHMIWERVKSIKDMVFFLSD 385
KLN +WE +K + F SD
Sbjct: 305 FKLNLQGNVWEEMKVPNGLTVFSSD 329
>UniRef100_O04180 Pge1 protein [Lotus japonicus]
Length = 210
Score = 56.6 bits (135), Expect = 1e-06
Identities = 29/120 (24%), Positives = 59/120 (49%), Gaps = 2/120 (1%)
Query: 6 IRGDGGRWVSGFAMAQGAGDAFKSKLLALQKGLLHVWELGFRPVNCFVDCSEVQKVVITN 65
+R G W+S + G AF +++LA++ G+ H ++G V C DCS+ + + +
Sbjct: 7 VRDSRGAWLSATLVCYNYGSAFLTEILAVKLGIRHALDIGHIYVFCLSDCSQALQALHAD 66
Query: 66 SDVQNYWHGAVILLIREVIARNLSVSVTPVPKEQKMVVDALAKLAVREGWDWKGQNKTEC 125
+++ +YW I + ++IA V++ + + + D L + A R ++ G C
Sbjct: 67 TNITSYWTREKICRVHDIIATLQDVNLQHIDRVKNNTADKLVREAGR--LEYMGSGSFRC 124
>UniRef100_Q8LEB3 Hypothetical protein [Arabidopsis thaliana]
Length = 380
Score = 55.8 bits (133), Expect = 2e-06
Identities = 66/265 (24%), Positives = 105/265 (38%), Gaps = 36/265 (13%)
Query: 132 QSYPWLLYVHNNLAIIRS----ILDPT--ETYSRSIPELSG-ANLLTVQHGGWCVLEKGG 184
QS PWL Y + + + +P+ +T+ PEL+G N L GW ++ K
Sbjct: 90 QSRPWLFYPQSQRGGPKEGDYVLFNPSRSQTHHLKFPELTGYRNKLACAKDGWLLVVKDN 149
Query: 185 KEKVRELLLWNPFLQEKIHLPSLKHENRIRNCI-LSCSPTETDQVCSIFLFPHEAYGYGP 243
+ V L NPF E+I LP + +N R+C+ S +PT T C + F +++ Y
Sbjct: 150 PDVVFFL---NPFTGERICLPQVP-QNSTRDCLTFSAAPTSTS--CCVISFTPQSFLYAV 203
Query: 244 NILFYCEVGDKQWFAVDYRKDLVRASPPEEDGVVYNPFLYSPVYFNGRLYAVSYS-RLLV 302
+ G+ W + D Y + ++ NG Y +S S RL
Sbjct: 204 VKVDTWRPGESVWTTHHF------------DQKRYGEVINRCIFSNGMFYCLSTSGRLSF 251
Query: 303 IIEKLEPPNGLQIHSADVYLPDPFPHTFPCLRQLLVCNHE--LFHIQISMEYNKIFGIAV 360
E N L + F +RQ+ + HE +F + N+ +
Sbjct: 252 FDPSRETWNVLPVKPCRA-----FRRKIMLVRQVFMTEHEGDIFVVTTRRVNNR--KLLA 304
Query: 361 HKLNFSHMIWERVKSIKDMVFFLSD 385
KLN +WE +K + F SD
Sbjct: 305 FKLNLQGNVWEEMKVPNGLTVFSSD 329
>UniRef100_Q6K1U6 Hypothetical protein OSJNBa0038P01.5 [Oryza sativa]
Length = 338
Score = 54.7 bits (130), Expect = 5e-06
Identities = 65/308 (21%), Positives = 111/308 (35%), Gaps = 37/308 (12%)
Query: 135 PWLLYVHNNLAIIRSI----LDPTETYSRSIPELSGANLLTVQHGGWCVLEKGGKEKVRE 190
P LL+ H + + S+ L P + ++ + GW ++ + G
Sbjct: 18 PLLLFGHGDATSLYSVPARALLPRRVGDGGVDDMMRGHRWWTTAQGWLLMARRGSPCT-- 75
Query: 191 LLLWNPFLQEKIHLPSLKHENRI---------RNCILSCSPTETDQVCSIFLFPHEAYGY 241
LW+PF ++ LP H+ + R C+LSC C + + H
Sbjct: 76 -FLWDPFTGRRVGLPP-DHDGTVLAAEGSSHRRRCLLSCCGPMDPASCVVLVIDH----- 128
Query: 242 GPNILFYCEVGDKQWFAVDYRKDLVRASPPEEDGVVYNPFLYSPVYFNGRLYAVSYSRLL 301
+L+YC GD W + L P E + L +G Y +
Sbjct: 129 ADTVLWYCRPGDNHWVKRHQHQYLQPGPPHHEHRGIVILALRQLTAMDGEFYTDLIDHVA 188
Query: 302 VIIEKLEPPNGLQIHSADVYLPDPFPHTFPCLRQLLV-CNHELFHIQISMEY---NKIFG 357
V+ LEP + D D P + + V N EL I S + G
Sbjct: 189 VLEFSLEPAFTVTAVDDD----DRRPAVYMKRTSIFVESNGELHSILFSHPIGCDRIVAG 244
Query: 358 IAVHKLNFS-----HMIWERVKSIKDMVFFLSDKDLPFARRVNPGG--GGLIYFTSMNNK 410
+ V++L+ + W +V S+ VFF+ + G G IY++ N K
Sbjct: 245 VGVYRLSMATTQEQRSAWVKVDSLGGRVFFVQIGCFGASLDARTTGLRGNCIYYSGFNGK 304
Query: 411 FVHIFNLE 418
+ ++++E
Sbjct: 305 ALCVYDME 312
>UniRef100_Q9AY78 Hypothetical protein OSJNBa0091J19.25 [Oryza sativa]
Length = 348
Score = 54.7 bits (130), Expect = 5e-06
Identities = 71/298 (23%), Positives = 117/298 (38%), Gaps = 47/298 (15%)
Query: 146 IIRSILDPTETYSRSIPELSGANLLTVQHGGWCVLEKGGKEKVRELLLWNPF-LQEKIHL 204
++ SI D IP L+ AN G W +L L NP Q+KIHL
Sbjct: 28 VLFSISDKKAIIDGDIPGLTNANAWFTPQG-WILLRLS-----TATFLQNPQDSQDKIHL 81
Query: 205 PSLKHENRIR-NCILSCSPTETDQVCSIFLFPHEAYGYGPNILFYCEVGDKQWFAVDYRK 263
P L R +C LS P+ C + + A +++YC +GD +W +Y
Sbjct: 82 PHLPEGLSTRCSCQLSGKPSLPG--CIVLVVEPIA-----TVIWYCRIGDDEWTRHEYDI 134
Query: 264 DLVRASPP------EEDGVVYNPFLYSPVYFNGRLYAVSY-----SRLLVIIEKLEP-PN 311
+ PP ++ + + YFNG + + + IE ++P P
Sbjct: 135 GTLPFDPPIDGKDHDDVAICQIAACHGKFYFNGFFDTIGVLEFTPTPVFSSIEIVDPTPG 194
Query: 312 GLQIHSADVYLPDPFPHTFPCLRQLLVCNHELFHIQISMEYN-KIFGIAVHKLNFSHMIW 370
GL + A H + L+ EL+ + + ++Y I+ + VHK++F W
Sbjct: 195 GLGVTGA--------AHVY-----LVESEDELYMVCLRIDYEFTIYDMTVHKMDFLSRQW 241
Query: 371 ERVKSIKDMVFFLSDKDLPFARRVNPGGGGL----IYFTSMNNKFVHIFNLEDNSLRI 424
R I FFL+ L F + GL +Y + +K + +ED+ +
Sbjct: 242 RRADEIGGRAFFLA--PLYFGASCSADEYGLEKDSVYASYAVDKCFEVSKVEDDETEV 297
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 49.7 bits (117), Expect = 2e-04
Identities = 36/109 (33%), Positives = 53/109 (48%), Gaps = 3/109 (2%)
Query: 3 GGGIRGDGGRWVSGFAMAQGAGDAFKSKLLALQKGLLHVWELGFRPVNCFVDCSEVQKVV 62
GG IR G W+ GFA+ G+ A ++L GLL W+ GFR V +DC V V
Sbjct: 1089 GGAIRNGQGEWLGGFALNIGSCAAPLAELWGAYYGLLIAWDKGFRRVELDLDCKLV--VG 1146
Query: 63 ITNSDVQN-YWHGAVILLIREVIARNLSVSVTPVPKEQKMVVDALAKLA 110
++ V N + ++ L + R+ V V+ V +E + D LA A
Sbjct: 1147 FLSTGVSNAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGLANYA 1195
>UniRef100_Q6EQC6 Hypothetical protein P0448B03.3 [Oryza sativa]
Length = 384
Score = 48.9 bits (115), Expect = 3e-04
Identities = 57/251 (22%), Positives = 95/251 (37%), Gaps = 52/251 (20%)
Query: 190 ELLLWNPFLQEKIHLPSLKH---ENRIRNCILSCSPTETDQVCSIFLFPHEAYGYGPNIL 246
E LWNPF ++I LP + C+LSC PT+ + V + ++
Sbjct: 138 ETFLWNPFTSQRISLPFDQDRFLRTNYTRCLLSCKPTDINCVVLVL-------SLNDTVI 190
Query: 247 FYCEVGDKQWFAVDYRKDLVRASPPEEDGVVYNPFLYSPVYFNGRLYAVSYSRLLVII-E 305
+YC G QWF +Y+ S + R + Y LL ++
Sbjct: 191 WYCYPGGTQWFKHEYQ---------------------SRRFHRHRGSVIGYMALLTVVGG 229
Query: 306 KLEPPNGLQIHSADVYLPDPFPHTFPCLR-----------QLLVCNHELFHIQISMEY-- 352
K G + + D + P+P P LL + ELF +
Sbjct: 230 KFYTGLGDSVITLD-FSPNPKFDIIPIKAVQNPMYNFSRLYLLESSGELFSLFFYPPMTC 288
Query: 353 -NKIFGIAVHKLNFSHMIWERVKSIKDMVFFLSDKDLPFARRVNPGGGGL----IYFTSM 407
+I I V+KL+ W +V ++ D FF++ F V+ L IYF+
Sbjct: 289 PKRIAEIEVYKLDIPRRAWVKVYTLGDRAFFVNSTKC-FGASVSAKEACLEENCIYFSRT 347
Query: 408 NNKFVHIFNLE 418
+K +++ ++E
Sbjct: 348 GDKGLYVHSME 358
>UniRef100_Q8S7H3 Hypothetical protein OSJNBa0010I09.22 [Oryza sativa]
Length = 670
Score = 47.8 bits (112), Expect = 6e-04
Identities = 47/225 (20%), Positives = 84/225 (36%), Gaps = 32/225 (14%)
Query: 161 IPELSGANLLTVQHGGWCVLEKGGKEKVRELLLWNPFLQEKIHLPSLKHENRIRNCILSC 220
+P S LL ++H C++ + V +L+ PSL+ R C+L
Sbjct: 337 LPTSSEQGLLMIKHKSKCIMTYIIRRLVDHMLM-----------PSLQQAISPRLCVLLI 385
Query: 221 SPTETDQVCSIFLFPHEAYGYGPNILFYCEVGDKQWFAVDYRKDLVRASPPEEDGVVYNP 280
P T +++YC +GD +W +Y + PP E
Sbjct: 386 EPIAT-------------------VIWYCRIGDDEWMRYEYDIGIQPLDPPFEGKDHEKV 426
Query: 281 FLYSPVYFNGRLYAVSYSRLLVIIEKLEPPNGLQIHSADVYLPDPFPHTFPCLRQLLVCN 340
+ G+ Y S + ++E P I AD + L+
Sbjct: 427 PICQIAACRGKFYFNSNFEKIEVLELTPTPTFSSIEIAD-SIAGGLGVIGGAYVYLVESE 485
Query: 341 HELFHIQISMEYN-KIFGIAVHKLNFSHMIWERVKSIKDMVFFLS 384
EL+ + + ++++ I+ + VH+++F W RV I FFL+
Sbjct: 486 DELYMVCLRLDHDFTIYDMTVHRMDFFSHQWRRVYEIGGRAFFLA 530
>UniRef100_Q9XIA2 F13F21.21 protein [Arabidopsis thaliana]
Length = 481
Score = 47.4 bits (111), Expect = 8e-04
Identities = 62/277 (22%), Positives = 105/277 (37%), Gaps = 33/277 (11%)
Query: 136 WLLY--VHNNLAIIRSILDPTE---TYSRSIPELSGANLLTVQHGGWCVLEKGGKEKVRE 190
WLLY + + DP E T ++PELS ++ + GW +L + +
Sbjct: 153 WLLYHDAFQDKGVSYGFFDPVEKKKTKEMNLPELSKSSGILYSKDGW-LLMNDSLSLIAD 211
Query: 191 LLLWNPFLQEKIHLPSLK-HENRIRNCILSCSPTETDQVCSIFLFPHEAYGYGPNILFYC 249
+ +NPF +E+I LP + E+ N SC+PT+ + C +F + + I +
Sbjct: 212 MYFFNPFTRERIDLPRNRIMESVHTNFAFSCAPTK--KSCLVFGINNISSSVAIKISTW- 268
Query: 250 EVGDKQWFAVDYRKDLVRASPPEEDGVVYNPFLYSPVYFNGRLYAVSYSRLLVIIEKLEP 309
G W +D P Y L + +Y +G Y S + L V
Sbjct: 269 RPGATTWL----HEDFPNLFPS------YFRRLGNILYSDGLFYTASETALGVFDPTART 318
Query: 310 PNGLQIHSADVYLPDPFPHTFPCLRQLLVCNHELFHIQISMEYNKIFGIAVHKLNFSHMI 369
N L + P P +R + +F + S V++LN +
Sbjct: 319 WNVLPV--------QPIPMAPRSIRWMTEYEGHIFLVDASS-----LEPMVYRLNRLESV 365
Query: 370 WERVKSIKDMVFFLSDKDLPFARRVNPGGGGLIYFTS 406
WE+ +++ FLSD + ++YF S
Sbjct: 366 WEKKETLDGSSIFLSDGSCVMTYGLTGSMSNILYFWS 402
>UniRef100_Q8VZ76 Hypothetical protein At1g49360; F13F21.21 [Arabidopsis thaliana]
Length = 378
Score = 47.4 bits (111), Expect = 8e-04
Identities = 62/277 (22%), Positives = 105/277 (37%), Gaps = 33/277 (11%)
Query: 136 WLLY--VHNNLAIIRSILDPTE---TYSRSIPELSGANLLTVQHGGWCVLEKGGKEKVRE 190
WLLY + + DP E T ++PELS ++ + GW +L + +
Sbjct: 50 WLLYHDAFQDKGVSYGFFDPVEKKKTKEMNLPELSKSSGILYSKDGW-LLMNDSLSLIAD 108
Query: 191 LLLWNPFLQEKIHLPSLK-HENRIRNCILSCSPTETDQVCSIFLFPHEAYGYGPNILFYC 249
+ +NPF +E+I LP + E+ N SC+PT+ + C +F + + I +
Sbjct: 109 MYFFNPFTRERIDLPRNRIMESVHTNFAFSCAPTK--KSCLVFGINNISSSVAIKISTW- 165
Query: 250 EVGDKQWFAVDYRKDLVRASPPEEDGVVYNPFLYSPVYFNGRLYAVSYSRLLVIIEKLEP 309
G W +D P Y L + +Y +G Y S + L V
Sbjct: 166 RPGATTWL----HEDFPNLFPS------YFRRLGNILYSDGLFYTASETALGVFDPTART 215
Query: 310 PNGLQIHSADVYLPDPFPHTFPCLRQLLVCNHELFHIQISMEYNKIFGIAVHKLNFSHMI 369
N L + P P +R + +F + S V++LN +
Sbjct: 216 WNVLPV--------QPIPMAPRSIRWMTEYEGHIFLVDASS-----LEPMVYRLNRLESV 262
Query: 370 WERVKSIKDMVFFLSDKDLPFARRVNPGGGGLIYFTS 406
WE+ +++ FLSD + ++YF S
Sbjct: 263 WEKKETLDGSSIFLSDGSCVMTYGLTGSMSNILYFWS 299
>UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 676
Score = 45.1 bits (105), Expect = 0.004
Identities = 34/109 (31%), Positives = 54/109 (49%), Gaps = 3/109 (2%)
Query: 3 GGGIRGDGGRWVSGFAMAQGAGDAFKSKLLALQKGLLHVWELGFRPVNCFVDCSEVQKVV 62
GG IR + G W+ GFA+ G A ++L + GL+ WE G+R V VD + V V
Sbjct: 534 GGVIRDEHGSWLVGFALNIGVCSAPLAELWGVYYGLVVAWERGWRRVRLEVDSALV--VG 591
Query: 63 ITNSDV-QNYWHGAVILLIREVIARNLSVSVTPVPKEQKMVVDALAKLA 110
S + ++ ++ L I+++ V +T V +E + D LA A
Sbjct: 592 FLQSGIGDSHPLAFLVRLCHGFISKDWIVRITHVYREANRLADGLANYA 640
>UniRef100_Q8S0R1 P0408G07.11 protein [Oryza sativa]
Length = 400
Score = 44.3 bits (103), Expect = 0.007
Identities = 52/202 (25%), Positives = 69/202 (33%), Gaps = 25/202 (12%)
Query: 155 ETYSRSIPELSGANLLTVQHGGWCVLEKGGKEKVRELLLWNPFLQEKIHLPSLKHENRIR 214
ET +PEL+ + HG + E R LWNP E + LP ++
Sbjct: 104 ETTHVVVPELTSNDYHVTPHGWVFLSEPAAAGTTRRTRLWNPTTGESVELPRMEQLLPAN 163
Query: 215 -NCILSCSPTETDQVCSIFLFPHEAYGYGPNILFYCEVGDKQ---WFAVDYRKDLVRASP 270
C LS PT V + + L YC VG W A DY D+
Sbjct: 164 WKCYLSDDPTAASCVVLVLAMSEPS-------LLYCHVGATAGGGWVAHDY--DIGDVGL 214
Query: 271 PEEDGVVYNPFLYSPVYFNGRLYAVSYSRLLVIIEKLEPPNGLQIHSADVYLPDP---FP 327
P F+ NGR Y +L V+ P YL P FP
Sbjct: 215 PPSYAPPRRQFISQIAAVNGRFYFADTGKLGVLEFTPSPEFS--------YLDYPHIEFP 266
Query: 328 HTFPCLRQLLVCNH-ELFHIQI 348
+ LV +H ELF + +
Sbjct: 267 EGSNFAKSFLVASHGELFDVYV 288
>UniRef100_Q9AY83 Hypothetical protein OSJNBa0091J19.29 [Oryza sativa]
Length = 219
Score = 43.9 bits (102), Expect = 0.009
Identities = 39/182 (21%), Positives = 69/182 (37%), Gaps = 21/182 (11%)
Query: 204 LPSLKHENRIRNCILSCSPTETDQVCSIFLFPHEAYGYGPNILFYCEVGDKQWFAVDYRK 263
+PSL+ R C+L P T +++YC +GD +W +Y
Sbjct: 1 MPSLQQAISPRLCVLLIEPIAT-------------------VIWYCRIGDDEWMRYEYDI 41
Query: 264 DLVRASPPEEDGVVYNPFLYSPVYFNGRLYAVSYSRLLVIIEKLEPPNGLQIHSADVYLP 323
+ PP E + G+ Y S + ++E P I AD +
Sbjct: 42 GIQPLDPPFEGKDHEKVPICQIAACRGKFYFNSNFEKIEVLELTPTPTFSSIEIAD-SIA 100
Query: 324 DPFPHTFPCLRQLLVCNHELFHIQISMEYN-KIFGIAVHKLNFSHMIWERVKSIKDMVFF 382
L+ EL+ + + ++++ I+ + VH+++F W RV I FF
Sbjct: 101 GGLGVIGGAYVYLVESEDELYMVCLRLDHDFTIYDMTVHRMDFFSHQWRRVYEIGGRAFF 160
Query: 383 LS 384
L+
Sbjct: 161 LA 162
>UniRef100_Q6K5I7 Hypothetical protein OJ1791_B03.3 [Oryza sativa]
Length = 1254
Score = 43.5 bits (101), Expect = 0.011
Identities = 64/316 (20%), Positives = 116/316 (36%), Gaps = 49/316 (15%)
Query: 135 PWLLYVHNNLAIIRSILD--------PTETYSRSIPELSGANLLTVQHGGWCVLEKGGKE 186
P LL+ H + + S+ PT + ++ + GW ++ + G
Sbjct: 938 PLLLFGHGDATFLYSVPKRALLPMPTPTRVGDGGVDDMMRGHRWWTTAQGWLLMARRGSP 997
Query: 187 KVRELLLWNPFLQEKIHLPSLKHENRI--------RNCILSCSPTETDQVCSIFLFPHEA 238
LW+PF + LP H+ + R C+LSC C++ + A
Sbjct: 998 CT---FLWDPFTGRRAGLPP-DHDGTVLAAEGSHRRRCLLSCCGPMDPTSCTVLVIDL-A 1052
Query: 239 YGYGPNILFYCEVGDKQWFAVDYRKDLVRASPPEEDGVVYNPFLYSPVYFNGRLYAVSYS 298
Y L+YC GD QW + R +P D +++ L +G+ Y S
Sbjct: 1053 YPE----LWYCRPGDNQWVKLHQHPYQYR-NPAHRDAIIWG--LRQLTAIDGKFYTEELS 1105
Query: 299 RLLVIIEKLEPPNGLQIHSADVYLPDPFPHTFPCLRQLLV-CNHELFHIQISMEY---NK 354
++ ++E +I DV D P + V N EL + S
Sbjct: 1106 GIVGVLEFSPEVAFTKIAVHDV---DRRPAVYKKRTTSFVESNGELHSVVFSHPIGCDRI 1162
Query: 355 IFGIAVHKLNFS--------HMIWERVKSIKDMVFFLSDKDLPFARRVNPGG----GGLI 402
+ + V++L+ + W +V S+ FF+ F + G G +
Sbjct: 1163 VARVGVYRLSINPTTTQEQRSAAWVKVDSLGGRAFFVEIGS--FGASFDAEGTCLRGNCV 1220
Query: 403 YFTSMNNKFVHIFNLE 418
Y++ N K + ++++E
Sbjct: 1221 YYSGFNGKVLCVYDME 1236
Score = 43.1 bits (100), Expect = 0.015
Identities = 56/268 (20%), Positives = 107/268 (39%), Gaps = 43/268 (16%)
Query: 176 GWCVLEKGGKEKVRELLLWNPFLQEKIHLPSLKHENRI--------RNCILSCSPTETDQ 227
GW ++ + G LW+PF ++ LP H+ + R C+LSC
Sbjct: 67 GWLLMARRGSPCT---FLWDPFTGRRVGLPP-DHDGTVLAAEGSHRRRCLLSCCGPMDPT 122
Query: 228 VCSIFLFPHEAYGYGPNILFYCEVGDKQWFAVDYRKDLVRASPPEEDGVVYNPFLYSPVY 287
C++ + AY L+YC GD W + +++ +P D ++ FL
Sbjct: 123 SCTVLVIDL-AYPE----LWYCRPGDNHWVKL-HQQPYQYRNPAHRDAII--RFLRKFTA 174
Query: 288 FNGRLYAVSYSRLLVIIEKLEPP--NGLQIHSADVYLPDPFPHTFPCLRQLLVCNHELFH 345
+G+ Y ++ + ++E L + +H D P + C + N EL
Sbjct: 175 IDGKFYTELHTGNVGVLEFLPEVAFTKIAVHDDD-RRPAVYKKRTTC---FVESNGELHS 230
Query: 346 IQISMEY---NKIFGIAVHKLNFS--------HMIWERVKSIKDMVFFLSDKDLPFARRV 394
+ S + + V++L+ + W +V S+ FF+ K F +
Sbjct: 231 VVFSHPIGCDRIVARVGVYRLSINAAATQEQRSAAWVKVDSLGGRAFFV--KIGSFGASL 288
Query: 395 NPGGGGL----IYFTSMNNKFVHIFNLE 418
+ G GL +Y++ N K + ++++E
Sbjct: 289 DAEGTGLRGNCVYYSVFNGKVLCVYDME 316
>UniRef100_O80790 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 970
Score = 43.1 bits (100), Expect = 0.015
Identities = 32/107 (29%), Positives = 51/107 (46%), Gaps = 1/107 (0%)
Query: 4 GGIRGDGGRWVSGFAMAQGAGDAFKSKLLALQKGLLHVWELGFRPVNCFVDCSEVQKVVI 63
G I G W+ GFA+ G+ DA ++L GLL W+ GFR V +D SE+ +
Sbjct: 829 GAILNLQGEWLGGFALNIGSCDAPLAELWGAYYGLLIAWDKGFRRVELNLD-SELVVGFL 887
Query: 64 TNSDVQNYWHGAVILLIREVIARNLSVSVTPVPKEQKMVVDALAKLA 110
+ + + ++ L + R+ V V+ V +E + D LA A
Sbjct: 888 STGISKAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGLANYA 934
>UniRef100_Q6H5P3 Hypothetical protein OJ1118_F11.4 [Oryza sativa]
Length = 340
Score = 42.7 bits (99), Expect = 0.020
Identities = 23/69 (33%), Positives = 38/69 (54%), Gaps = 4/69 (5%)
Query: 354 KIFGIAVHKLNFSHMIWERVKSIKDMVFFLSDKDLPFARRVNPGGGGL----IYFTSMNN 409
K+ I VHKL+ S IW +V +I +M FF+ D + +N GL IY+ +
Sbjct: 245 KVSQIEVHKLDLSRRIWVKVSTIGNMAFFVDSTDSGVSASLNAEDVGLKRNCIYYVRPKD 304
Query: 410 KFVHIFNLE 418
K ++I+++E
Sbjct: 305 KGLYIYDIE 313
>UniRef100_Q9FNQ6 Gb|AAF19536.1 [Arabidopsis thaliana]
Length = 657
Score = 42.7 bits (99), Expect = 0.020
Identities = 26/106 (24%), Positives = 50/106 (46%), Gaps = 3/106 (2%)
Query: 4 GGIRGDGGRWVSGFAMAQGAGDAFKSKLLALQKGLLHVWELGFRPVNCFVDCSEVQKVVI 63
G +R G+WV G+ + + LLA+ +GL ++W+ GFR ++ E+ +
Sbjct: 523 GIVRDQSGKWVFGYIRCHKSIPEVVAGLLAIYQGLKYLWDSGFRRIHLETTSFEIINALT 582
Query: 64 TNSDVQNYWHGAVIL-LIREVIARNLSVSVTPVPKEQKMVVDALAK 108
T S + + +L +++I + + + KEQ + LAK
Sbjct: 583 TKSSL--FCKSKTLLGACKDMILKEWECDIYHISKEQNSCAEWLAK 626
>UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 321
Score = 41.6 bits (96), Expect = 0.043
Identities = 29/108 (26%), Positives = 50/108 (45%), Gaps = 1/108 (0%)
Query: 3 GGGIRGDGGRWVSGFAMAQGAGDAFKSKLLALQKGLLHVWELGFRPVNCFVDCSEVQKVV 62
GG +R G W+ GFA+ G A ++L + GL W G R V VD S++
Sbjct: 179 GGVLRDHNGAWIGGFAVNIGVCSAPLAELWGVYYGLFIAWGRGARRVELEVD-SKMVVGF 237
Query: 63 ITNSDVQNYWHGAVILLIREVIARNLSVSVTPVPKEQKMVVDALAKLA 110
+T ++ ++ L + +++ V ++ V +E + D LA A
Sbjct: 238 LTTGIADSHPLSFLLRLCYDFLSKGWIVRISHVYREANRLADGLANYA 285
>UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana]
Length = 253
Score = 39.7 bits (91), Expect = 0.17
Identities = 28/108 (25%), Positives = 49/108 (44%), Gaps = 1/108 (0%)
Query: 3 GGGIRGDGGRWVSGFAMAQGAGDAFKSKLLALQKGLLHVWELGFRPVNCFVDCSEVQKVV 62
GG +R G W GF++ G A ++L GL WE G + +D V +
Sbjct: 111 GGVVRDGDGNWCYGFSLNIGICSAPLAELWGAYYGLNIAWERGVTQLEMEIDSEMVVGFL 170
Query: 63 ITNSDVQNYWHGAVILLIREVIARNLSVSVTPVPKEQKMVVDALAKLA 110
T D ++ ++ L +++++ SV ++ V +E + D LA A
Sbjct: 171 RTGID-DSHPLSFLVRLCHGLLSKDWSVRISHVYREANRLADGLANYA 217
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.142 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 796,129,453
Number of Sequences: 2790947
Number of extensions: 35482484
Number of successful extensions: 66260
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 66226
Number of HSP's gapped (non-prelim): 66
length of query: 426
length of database: 848,049,833
effective HSP length: 130
effective length of query: 296
effective length of database: 485,226,723
effective search space: 143627110008
effective search space used: 143627110008
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0331c.6


