

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0330.6
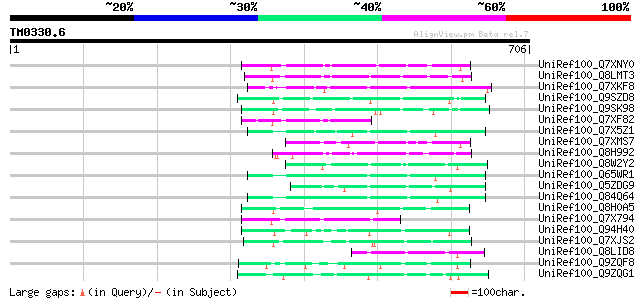
(706 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa] 104 8e-21
UniRef100_Q8LMT3 Putative retroelement [Oryza sativa] 89 4e-16
UniRef100_Q7XKF8 OSJNBb0065J09.11 protein [Oryza sativa] 86 4e-15
UniRef100_Q9SZD8 Hypothetical protein F19B15.120 [Arabidopsis th... 83 2e-14
UniRef100_Q9SK98 Very similar to retrotransposon reverse transcr... 82 5e-14
UniRef100_Q7XF82 Putative retroelement [Oryza sativa] 75 5e-12
UniRef100_Q7X5Z1 OSJNBa0006A01.14 protein [Oryza sativa] 74 1e-11
UniRef100_Q7XMS7 OSJNBa0029L02.10 protein [Oryza sativa] 74 2e-11
UniRef100_Q8H992 Oryza sativa (japonica cultivar-group) retrotra... 73 3e-11
UniRef100_Q8W2Y2 Putative reverse transcriptase [Oryza sativa] 73 3e-11
UniRef100_Q65WR1 Putative polyprotein [Oryza sativa] 72 4e-11
UniRef100_Q5ZDG9 Hypothetical protein P0480E02.9 [Oryza sativa] 72 4e-11
UniRef100_Q84Q64 Hypothetical protein OSJNBa0071M09.16 [Oryza sa... 72 7e-11
UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa] 72 7e-11
UniRef100_Q7X794 OSJNBa0035I04.8 protein [Oryza sativa] 72 7e-11
UniRef100_Q94H40 Putative reverse transcriptase [Oryza sativa] 70 2e-10
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 70 2e-10
UniRef100_Q8LID8 Cyst nematode resistance protein-like protein [... 70 2e-10
UniRef100_Q9ZQF8 Putative non-LTR retroelement reverse transcrip... 69 5e-10
UniRef100_Q9ZQG1 Putative non-LTR retroelement reverse transcrip... 69 6e-10
>UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa]
Length = 1026
Score = 104 bits (260), Expect = 8e-21
Identities = 88/324 (27%), Positives = 139/324 (42%), Gaps = 35/324 (10%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNS----------CGESSWWKEIQ 365
+ + D N A+LGKW WR E + W ++ A S G S +W+ +
Sbjct: 659 LGVLDLEAMNKAMLGKWIWRLENE-EGWWQEIIYAKYCSDKPLSGLRLKAGSSHFWQGVM 717
Query: 366 SITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMG 425
+ D F K V G T FW ++W G L ++F LY + KR ++++
Sbjct: 718 EVKDD-----FFSFCTKIVGNGEKTLFWEDSWLGGKPLAIQFPSLYGIVITKRITIADLN 772
Query: 426 KWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSS 485
+ G+ +F RR+L G ++ + +++ +G+ LVEN KD W L +G F+ S
Sbjct: 773 R--KGIDCMKF--RRDLHGDKLRDWRKIVNSWEGLNLVENCKDKLWWTLSKDGKFTVRSF 828
Query: 486 FVFLQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAIC 545
+ L+ Q P+ IW VP I+ F+W +I T+DNLL + C
Sbjct: 829 YRALKLQQTSFPN---KKIWKFRVPLKIRIFIWFFTKNKILTKDNLLKRGWRKG--DNKC 883
Query: 546 PLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWSKAQKLG 605
C E+ HL F C + LI W ++ A P + K
Sbjct: 884 QFCDKV-ETVQHLFFDCPLARLI------WNIIACALNVKPVLSRQDLFGSWIQSMDKFT 936
Query: 606 ESAIWM---SVLWSIWCLRNMVVF 626
++ + + +VLWSIW RN F
Sbjct: 937 KNLVIVGIAAVLWSIWKCRNKACF 960
>UniRef100_Q8LMT3 Putative retroelement [Oryza sativa]
Length = 764
Score = 89.0 bits (219), Expect = 4e-16
Identities = 84/323 (26%), Positives = 136/323 (42%), Gaps = 37/323 (11%)
Query: 320 D*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCG----------ESSWWKEIQSITV 369
D R N+ALL KW +R + + C +++ G S +WK + +
Sbjct: 394 DTRKMNIALLCKWIYRLESGKEDPCCVLLRNKYMKDGGGFFQSKAEESSQFWKGLHEVK- 452
Query: 370 DNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVN 429
W V G AT FW++ W G L+ ++ +Y + K + VS M +
Sbjct: 453 ----KWMDLGSSYKVGNGKATNFWSDVWIGETPLKTQYPNIYRMCADKEKTVSQM--CLE 506
Query: 430 GVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFL 489
G W E RR+L R++ + +L + I L E ++D WKL G + + + L
Sbjct: 507 GDWYIEL--RRSLGERDLNEWNDLHNTPREIHLKE-ERDCIIWKLTKNGFYKAKTLYQAL 563
Query: 490 QDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCC 549
+ D V +W +P +K W +L GRIQ L + + + C LC
Sbjct: 564 --SFGGVKDKVMQDLWRSSIPLKVKILFWLMLKGRIQAAGQL---KKMKWSGSPNCKLCG 618
Query: 550 LAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWSKAQKLGESAI 609
E+ HL+F C S + C + + LL+ +I QK G+ ++
Sbjct: 619 QTEDV-DHLMFRCHISQFM--WCCYRDAFGWDNIPSSREDLLEKLNI-----QKDGQKSV 670
Query: 610 WMSVL----WSIWCLRNMVVFNG 628
++ L W+IW +RN VFNG
Sbjct: 671 LLACLAAGTWAIWLMRNDWVFNG 693
>UniRef100_Q7XKF8 OSJNBb0065J09.11 protein [Oryza sativa]
Length = 436
Score = 85.9 bits (211), Expect = 4e-15
Identities = 88/344 (25%), Positives = 140/344 (40%), Gaps = 39/344 (11%)
Query: 324 FNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITVDNGWPWFQESVQKT 383
F AL +W W +P+ W + C E K++ F S + T
Sbjct: 115 FTRALRLRWLWHEWKDPNKPWVGLD----TPCDEID--KDL-----------FAASTKIT 157
Query: 384 VFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGK----WVNGVWQWEFKWR 439
V GN T+FW W + +Y +S + R++ + GK WV+ + F
Sbjct: 158 VGDGNMTRFWDSTWIDGRRPKDLMPLVYAIS-KNRKKSLHQGKEDDTWVDDLI---FDAG 213
Query: 440 RNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPDP 499
+ V L ++ ++LV + D WK G G ++ +S++ Q LE P
Sbjct: 214 STITVNLVEQLVRLWEAVRNVQLVSEEPDQIVWKFTGNGHYTASSAY---HAQCLEAPST 270
Query: 500 VFN-WIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHL 558
N IW P K +VW ++ R+ T D L +N CPLC E+G HL
Sbjct: 271 NLNSLIWKAWAPGKCKFYVWLIIQNRVWTSDRLAIRGWQNNGH---CPLCRCEAETGLHL 327
Query: 559 LFSCSNSMLI*YECHAWLGVSTAQVS--DPKVHLLQF--SSIGWSKAQKLGESAIWMSVL 614
+ +C + I + W+G + S + + Q+ S K G ++ + V+
Sbjct: 328 VATCRYTKRIWHHVAGWVGYHQLEPSQWEEAQSVCQWWESLANTPNIPKKGLRSLILLVV 387
Query: 615 WSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWL---RSRLDDF 655
W IW RN +F+ E+ LL +I+ A W RL DF
Sbjct: 388 WEIWKERNRRIFDNKEMAVGLLLAKIKEEASVWALAGAKRLRDF 431
>UniRef100_Q9SZD8 Hypothetical protein F19B15.120 [Arabidopsis thaliana]
Length = 575
Score = 83.2 bits (204), Expect = 2e-14
Identities = 87/366 (23%), Positives = 149/366 (39%), Gaps = 48/366 (13%)
Query: 311 EADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGE----------SSW 360
+A+ I KD FN+ALLGK WR L+ P+SL +V K+ + S
Sbjct: 51 KAEGGIGFKDIEAFNLALLGKQMWRMLSRPESLMAKVFKSRYFHKSDPLNAPLGSRPSFV 110
Query: 361 WKEIQSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGS--GSLELRFRRLYNLSCQKR 418
WK I + ++ + V G W W S S LR +R + Q+
Sbjct: 111 WKSIHA-----SQEILRQGARAVVGNGEDIIIWRHKWLDSKPASAALRMQR---VPPQEY 162
Query: 419 EQVSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELMV--ILQGIRLVENKKDTWRWKLEG 476
VS++ K + + + +WR+++ E ++ + G R + D++ W
Sbjct: 163 ASVSSILKVSDLIDESGREWRKDVIEMLFPEVERKLIGELRPGGRRI---LDSYTWDYTS 219
Query: 477 EGGFSFNSSFVFL--------QDQYLEEP--DPVFNWIWMVPVPSNIKAFVWRVLLGRIQ 526
G ++ S + L Q + EP +P++ IW I+ F+W+ L +
Sbjct: 220 SGDYTVKSGYWVLTQIINKRSSPQEVSEPSLNPIYQKIWKSQTSPKIQHFLWKCLSNSLP 279
Query: 527 TRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDP 586
L H + E+ C C +E+ +HLLF C+ + L + +
Sbjct: 280 VAGAL---AYRHLSKESACIRCPSCKETVNHLLFKCTFARLTWAISSIPIPLGGEWADSI 336
Query: 587 KVHLLQFSSIG-----WSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQ 641
V+L ++G W KA +L W+ LW +W RN +VF G E + ++L + +
Sbjct: 337 YVNLYWVFNLGNGNPQWEKASQL---VPWL--LWRLWKNRNELVFRGREFNAQEVLRRAE 391
Query: 642 VTAWRW 647
W
Sbjct: 392 DDLEEW 397
>UniRef100_Q9SK98 Very similar to retrotransposon reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 82.0 bits (201), Expect = 5e-14
Identities = 89/368 (24%), Positives = 142/368 (38%), Gaps = 60/368 (16%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGE----------SSWWKEIQ 365
+ +D LFN ALL K WR L PD L+ R++K+ GE S W+ I
Sbjct: 701 LGFRDIGLFNQALLAKQAWRLLQFPDCLFARLIKSRYFPVGEFLDSDVGSRPSFGWRSIL 760
Query: 366 SITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMG 425
+G + K V G + + W + W L + + + N+
Sbjct: 761 -----HGRDLLCRGLVKRVGNGKSIRVWIDYWLDDNGLRAPW---------IKNPIINVD 806
Query: 426 KWVNGVWQWEFK-WRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNS 484
V+ + +E + WR L E +F + +V ++ R V + D W WK G +S
Sbjct: 807 LLVSDLIDYEKRDWR--LDKLEEQFFPDDVVKIRENRPVVSLDDFWIWKHNKSGDYSVKL 864
Query: 485 SFVFLQDQYLEE------PDPVFN----WIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN* 534
+ +Q L + P N +W + IK F+W+VL G I D L
Sbjct: 865 GYWLASNQNLGQVAIEAMMQPSLNDLKTQVWKLQTEPKIKVFLWKVLSGAIPVVDLL--- 921
Query: 535 QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAW---------LGVSTAQVSD 585
L++ C C ES H+LFSCS + W LG V
Sbjct: 922 SYRGMKLDSRCQTCGCEGESIQHVLFSCS------FPRQVWAMSNIHVPLLGFECGSVYA 975
Query: 586 PKVH-LLQFSSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTA 644
H L+ ++ W +L S W ++W IW RN+ F G + + +++
Sbjct: 976 NLYHFLINRDNLKW--PVELRRSFPW--IIWRIWKNRNLFFFEGKRFTVLETILKVRKDV 1031
Query: 645 WRWLRSRL 652
W +++
Sbjct: 1032 EDWFAAQV 1039
>UniRef100_Q7XF82 Putative retroelement [Oryza sativa]
Length = 586
Score = 75.5 bits (184), Expect = 5e-12
Identities = 55/187 (29%), Positives = 86/187 (45%), Gaps = 20/187 (10%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSC----------GESSWWKEIQ 365
+ I D + N+ALLGKW W L D W +++ S G+S +W+ +
Sbjct: 334 MGILDLEIMNIALLGKWLWN-LENKDGWWQEILRDKYLSKKTLLGMKKKPGDSHFWQGLM 392
Query: 366 SITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMG 425
I F + K + G T FW +NW G L L+F L+N++ K+ VS
Sbjct: 393 EIKNS-----FYKFCSKKLGDGKQTLFWEDNWIGGRPLALQFPTLFNITTTKQIIVS--- 444
Query: 426 KWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSS 485
K + G W K+RR L G + + ++ G+ L E +D WKL +G F+ S
Sbjct: 445 KVMLGGWD-AIKFRRTLHGDRLMEWNKIKQSCVGLTLQEGVRDKIWWKLTKDGSFTVKSF 503
Query: 486 FVFLQDQ 492
++ L+ Q
Sbjct: 504 YLALKMQ 510
>UniRef100_Q7X5Z1 OSJNBa0006A01.14 protein [Oryza sativa]
Length = 1189
Score = 73.9 bits (180), Expect = 1e-11
Identities = 80/332 (24%), Positives = 129/332 (38%), Gaps = 34/332 (10%)
Query: 324 FNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITVDNGWPWFQESVQKT 383
F AL +W W +P W + + C+ + F S + T
Sbjct: 868 FTRALRLRWLWNEWRDPSKPWVGL-ETPCDGVDRNL----------------FAASTKIT 910
Query: 384 VFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNGVWQWEFKWRRNLQ 443
V GN T+FW W + +Y +S + R++ GK + W + L
Sbjct: 911 VGDGNTTRFWDSAWINGRRPKDLMPLVYEIS-KNRKKSLRQGK-EDDSWVHDLTLDAGLS 968
Query: 444 GREVGWFEELMVILQGIRLVE---NKKDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPDPV 500
+ ++L+ + + +R V + D WK G ++ +S++ Q L P+
Sbjct: 969 -ITINLLDQLVRLWEAVRNVHLDSEEPDQIVWKFTSSGHYTASSAY---HAQCLGAPNTN 1024
Query: 501 FN-WIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLL 559
FN IW V P K W ++ R+ T D L +N CPLC E+ HL+
Sbjct: 1025 FNSLIWKVWAPGKCKFHAWLIIQNRVWTSDRLATRGWRNNGH---CPLCRCDTETAFHLV 1081
Query: 560 FSCSNSMLI*YECHAWLG---VSTAQVSDPK-VHLLQFSSIGWSKAQKLGESAIWMSVLW 615
C + I W G + AQ + + VH S K G ++ + V+W
Sbjct: 1082 AMCRYTKRIWRLVATWAGYQQLEPAQWEEARSVHQWWESLANTPGIPKKGLRSLILLVVW 1141
Query: 616 SIWCLRNMVVFNGGELDKDQLLEQIQVTAWRW 647
IW RN +F+ E+ LL +I+ A W
Sbjct: 1142 EIWKERNRRIFDHKEMATGVLLAKIKEEASLW 1173
>UniRef100_Q7XMS7 OSJNBa0029L02.10 protein [Oryza sativa]
Length = 389
Score = 73.6 bits (179), Expect = 2e-11
Identities = 70/257 (27%), Positives = 108/257 (41%), Gaps = 29/257 (11%)
Query: 376 FQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNGVWQWE 435
F + V G T FW +NW G L +FR LYN++ K+ V++ V V
Sbjct: 7 FYKFCTNVVGNGRNTLFWEDNWIGGKPLAEQFRDLYNITLTKKITVAD----VKQVGIDS 62
Query: 436 FKWRRNLQGREVGWFEELMVILQG---IRLVENKKDTWRWKLEGEGGFSFNSSFVFLQDQ 492
K+RR++ G +L+ I Q + L + +DT RW L +G F+ S L+ Q
Sbjct: 63 IKFRRDVLG------NKLLKIKQSWSDLVLEDGTRDTLRWNLTKDGKFTVKSFHTALKMQ 116
Query: 493 YLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAE 552
+ P IW + VP IK F+W +I T+DNL +A C C E
Sbjct: 117 QVIFP---HKKIWGLKVPLKIKIFIWMFTKNKILTKDNLFKRGWRKG--KANCQFCDQTE 171
Query: 553 ESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWSKAQKLGESAIWM- 611
LF C + L+ W V A + ++ +K K + + +
Sbjct: 172 T--LQPLFYCPLARLL------WNIVKCALNINSVLNRQDLFGSWMNKIDKFTKKLVVVG 223
Query: 612 --SVLWSIWCLRNMVVF 626
+++W+IW RN F
Sbjct: 224 LAAIIWTIWKFRNKACF 240
>UniRef100_Q8H992 Oryza sativa (japonica cultivar-group) retrotransposon Karma DNA,
complete sequence [Oryza sativa]
Length = 1197
Score = 73.2 bits (178), Expect = 3e-11
Identities = 68/288 (23%), Positives = 119/288 (40%), Gaps = 38/288 (13%)
Query: 358 SSW----WKE------IQSITVDNGWPWFQESVQKT-------VFAGNATQFWAENWTGS 400
SSW WKE ++S+ + W FQ+ + + + G+ T FW + WTG+
Sbjct: 883 SSWTNWIWKEFDGRSLLKSLPLGQHWSVFQKLLPELFKITTVHIGDGSRTSFWHDRWTGN 942
Query: 401 GSLELRFRRLYNLSCQKREQVSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGI 460
+L +F L++ S V + + G++ L +EL ILQ
Sbjct: 943 MTLAAQFESLFSHSTDDLATVEQILSF--GIYDI---LTPRLSSAAQNDLQELQTILQNF 997
Query: 461 RLVENKKDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRV 520
L N+ DT +L G + + ++ P P + +IW +P IK F W +
Sbjct: 998 SLT-NESDT---RLNNRLG-NLTTKTIYDLRSPPGFPSPNWKFIWDCRMPLKIKLFAWLL 1052
Query: 521 LLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVST 580
+ R+ T+ NLL +++ A C +C +E+ HL F+C + W +
Sbjct: 1053 VRDRLSTKLNLLKKKIVQT---ATCDICSTTDETADHLSFNCP------FAISFWQALHI 1103
Query: 581 AQVSDPKVHLLQFSSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVFNG 628
+ + +L Q + + + +M W +W R+ VVF G
Sbjct: 1104 QPIINETKYLSQLKAPATIPTRHF--QSFFMLCFWVLWNHRHDVVFRG 1149
>UniRef100_Q8W2Y2 Putative reverse transcriptase [Oryza sativa]
Length = 581
Score = 72.8 bits (177), Expect = 3e-11
Identities = 64/285 (22%), Positives = 109/285 (37%), Gaps = 24/285 (8%)
Query: 376 FQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNM---GKWVNGVW 432
F + TV G +FW W + + +L ++ S +K V + KW+ +
Sbjct: 296 FAATTTITVGDGKIARFWGSKWLDNQTPQLIAPDIFVASRRKNRTVHDALAENKWIRDI- 354
Query: 433 QWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFLQDQ 492
R + + + L +++ +R + D+ RW L G +S S++ Q
Sbjct: 355 ----DVSRIFEANHLQQYIRLWRMIRDMRPLRENHDSIRWNLTANGQYSAKSAYRL---Q 407
Query: 493 YLEEPDPVF-NWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLA 551
+L F IW P K F W V R+ T D L + N + ICPLC
Sbjct: 408 FLGAAKSTFYQSIWKAWAPPKCKFFSWLVTQNRVWTADRLEK-RGWQN--QKICPLCYNV 464
Query: 552 EESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWSKAQKLGES---- 607
+ES LL C + I E W G + H ++ W+ + +
Sbjct: 465 DESALLLLAKCKLTKRIWAEMQRWTGTDLQMDNSESCHSVE---QWWTLVTESPNTPCKP 521
Query: 608 --AIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWLRS 650
+ + + W IWC RN +F +L +I+ W+++
Sbjct: 522 LRTLVILISWEIWCERNARIFRNNASTPSSILAKIKEEGRAWVKA 566
>UniRef100_Q65WR1 Putative polyprotein [Oryza sativa]
Length = 1023
Score = 72.4 bits (176), Expect = 4e-11
Identities = 75/329 (22%), Positives = 123/329 (36%), Gaps = 28/329 (8%)
Query: 324 FNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITVDNGWPWFQESVQKT 383
F AL +W W +P W + + C+ F S + T
Sbjct: 702 FTRALRLRWLWNEWRDPSKPWVGL-ETPCDGVDRDL----------------FAASTKIT 744
Query: 384 VFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNGVWQWEFKWRRNLQ 443
V GN T+FW W + +Y +S +++ + + + V ++
Sbjct: 745 VGDGNTTRFWDSAWINGRRPKDLMPLVYEISKNRKKSLRQGKEDDSWVHDLTLDAGSSIT 804
Query: 444 GREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPDPVFN- 502
+ L ++ + L + D WK G ++ +S++ Q L P+ FN
Sbjct: 805 INLLDQLVRLWEAVRNVHLDSEEPDQIVWKFTSSGHYTASSAY---HAQCLGAPNTNFNS 861
Query: 503 WIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSC 562
IW V P K W ++ R+ T D L +N CPLC E+ HL+ C
Sbjct: 862 LIWKVWAPGKCKFHAWLIIQNRVWTSDRLATRGWRNNGH---CPLCRCDTETALHLVAMC 918
Query: 563 SNSMLI*YECHAWLG---VSTAQVSDPK-VHLLQFSSIGWSKAQKLGESAIWMSVLWSIW 618
+ I W G + AQ + + VH S K G ++ + V+W IW
Sbjct: 919 RYTKRIWRLVATWAGYQQLEPAQWEEARSVHQWWESLANTPGIPKKGLRSLILLVVWEIW 978
Query: 619 CLRNMVVFNGGELDKDQLLEQIQVTAWRW 647
RN +F+ E+ LL +I+ A W
Sbjct: 979 KERNRRIFDHKEMATGVLLAKIKEEASLW 1007
>UniRef100_Q5ZDG9 Hypothetical protein P0480E02.9 [Oryza sativa]
Length = 385
Score = 72.4 bits (176), Expect = 4e-11
Identities = 70/279 (25%), Positives = 106/279 (37%), Gaps = 29/279 (10%)
Query: 383 TVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNGVWQWEFKWRRNL 442
TV G FW W L+ +Y S +K + ++ W + N
Sbjct: 112 TVGDGTKVSFWDSGWLQGRRLKDVAPLVYAASKKKTSTLQQAS--LSDQWMQDLDLPEN- 168
Query: 443 QGREVGWFEELM-------VILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFLQDQYLE 495
GW EL+ ++Q + L+E+++D WKL G ++ S++ + Q L
Sbjct: 169 ----TGWSIELIDQLIEVWSVVQNLHLIEHEEDKITWKLTSHGEYTTTSAY---KAQLLG 221
Query: 496 EPDPVF-NWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEES 554
F N IW P K F W ++ R+ T + L + +ICPLC +E+
Sbjct: 222 TTATNFNNLIWKPWAPHKCKTFAWLIIQNRVWTSNRLATRGWPNG---SICPLCRHRQET 278
Query: 555 GSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGW------SKAQKLGESA 608
HLL C + I W + Q++ H S W A K
Sbjct: 279 AIHLLAECRYTRRIWGAIAEW--TACEQLNPSLWHQASTVSEWWEATANTKDAPKKALRT 336
Query: 609 IWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRW 647
+ + V W IW RN F EL LL +I+ A W
Sbjct: 337 LTLLVAWEIWNERNRRTFQQKELSMGSLLAKIKEEAKSW 375
>UniRef100_Q84Q64 Hypothetical protein OSJNBa0071M09.16 [Oryza sativa]
Length = 1143
Score = 71.6 bits (174), Expect = 7e-11
Identities = 75/329 (22%), Positives = 122/329 (36%), Gaps = 28/329 (8%)
Query: 324 FNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITVDNGWPWFQESVQKT 383
F AL +W W +P W + + C+ F S + T
Sbjct: 822 FTRALRLRWLWNEWRDPSKPWVGL-ETPCDGVDRDL----------------FAASTKIT 864
Query: 384 VFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNGVWQWEFKWRRNLQ 443
V GN T+FW W + +Y +S +++ + + + V ++
Sbjct: 865 VGDGNTTRFWDSAWINGQRPKDLMPLVYEISKNRKKSLRQGKEDDSWVHDLTLDAGSSIT 924
Query: 444 GREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPDPVFN- 502
+ L ++ + L + D WK G ++ +S++ Q L P+ FN
Sbjct: 925 INLLDQLVRLWEAVRNVHLDSEEPDQIVWKFTSSGHYTASSAY---HAQCLGAPNTNFNS 981
Query: 503 WIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSC 562
IW V P K W ++ R+ T D L +N CPLC E+ HL+ C
Sbjct: 982 LIWKVWAPGKCKFHAWLIIQNRVWTSDRLATRGWRNNGH---CPLCRCDTETALHLVAMC 1038
Query: 563 SNSMLI*YECHAWLGVST---AQVSDPK-VHLLQFSSIGWSKAQKLGESAIWMSVLWSIW 618
+ I W G AQ + + VH S K G ++ + V+W IW
Sbjct: 1039 RYTKRIWRLVATWAGYQQFEPAQWEEARSVHQWWESLANTPGIPKKGLRSLILLVVWEIW 1098
Query: 619 CLRNMVVFNGGELDKDQLLEQIQVTAWRW 647
RN +F+ E+ LL +I+ A W
Sbjct: 1099 KERNRRIFDHKEMATGVLLAKIKEEASLW 1127
>UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa]
Length = 1557
Score = 71.6 bits (174), Expect = 7e-11
Identities = 75/334 (22%), Positives = 135/334 (39%), Gaps = 51/334 (15%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGE----------SSWWKEIQ 365
+ KD RLFN ALL + WR + PDSL RV+KA G S W+
Sbjct: 1222 LGFKDIRLFNQALLARQAWRLIDNPDSLCARVLKAKYYPNGSIVDTSFGGNASPGWQ--- 1278
Query: 366 SITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMG 425
+++G ++ + + G + + W + W +LS + +N
Sbjct: 1279 --AIEHGLELVKKGIIWRIGNGRSVRVWQDPWLPR-----------DLSRRPITPKNNCR 1325
Query: 426 -KWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNS 484
KWV + W N + ++ ++ +IL+ +++D W + G FS +
Sbjct: 1326 IKWVADLMLDNGMWDAN-KINQIFLPVDVEIILKLRTSSRDEEDFIAWHPDKLGNFSVRT 1384
Query: 485 SFVFLQDQYLEEPD---------PVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*Q 535
++ ++ EE + +W VPS +K F WR + T DN +
Sbjct: 1385 AYRLAENWAKEEASSSSSDVNIRKAWELLWKCNVPSKVKIFTWRATSNCLPTWDNK---K 1441
Query: 536 VLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKV--HLLQF 593
+ + C +C + +E H L C + H WL + + ++ HLL
Sbjct: 1442 KRNLEISDTCVICGMEKEDTMHALCRCPQAK------HLWLAMKESNDLSLRMDDHLLGP 1495
Query: 594 SSIGWSKAQKL--GESAIWMSVLWSIWCLRNMVV 625
S + +++ L E +++ VLW IW + N ++
Sbjct: 1496 SWL-FNRLALLPDHEQPMFLMVLWRIWFVHNEII 1528
>UniRef100_Q7X794 OSJNBa0035I04.8 protein [Oryza sativa]
Length = 357
Score = 71.6 bits (174), Expect = 7e-11
Identities = 57/226 (25%), Positives = 99/226 (43%), Gaps = 24/226 (10%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNS----------CGESSWWKEIQ 365
+ I++ L N +L KW ++ E + +W +++ S G+S +W +
Sbjct: 124 LGIQNLELQNKYMLSKWLYKLWNE-EGVWQNLLRRKYLSKKTLTHVDKKSGDSHFWSGLM 182
Query: 366 SITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMG 425
+ F V G +FW + W G+ R+ LYNL K E V+
Sbjct: 183 MVK-----NIFLSCGSLKVHNGTQVRFWEDKWNGNTPFAARYPALYNLVINKNESVA--- 234
Query: 426 KWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSS 485
V + +RR + G + + E++ + I+L E + T+ W+++ G FS NS
Sbjct: 235 -VVMSKRPLKVSFRRAMVGHNLKAWLEVVSKVISIKLTE-QNGTFLWEIQKNGCFSVNSM 292
Query: 486 FVFLQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNL 531
+ + E P + IW + +P IK F+W + G I T+DNL
Sbjct: 293 YKAIM---YREVVPKKDMIWKLRIPLKIKIFLWYLKNGVILTKDNL 335
>UniRef100_Q94H40 Putative reverse transcriptase [Oryza sativa]
Length = 1185
Score = 70.5 bits (171), Expect = 2e-10
Identities = 81/340 (23%), Positives = 130/340 (37%), Gaps = 55/340 (16%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGES-----------SWWKEI 364
+ +D FN A+L K WRFL +PDSL RV++ G +W +
Sbjct: 671 LGFRDLYCFNRAMLAKQIWRFLCDPDSLCARVMRCKYYPDGNILRARPKKGSSYAWQSIL 730
Query: 365 QSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGS----LELRFRRLYNLSCQKREQ 420
G+ W + G+ W ++ S S L R R + K E+
Sbjct: 731 SGSECFKGYIW-------RIGDGSQVNIWEDSLIPSSSNGKVLTPRGRNIIT----KVEE 779
Query: 421 VSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGF 480
+ N V+G W + + W + ILQ I + ++D W G F
Sbjct: 780 LINP---VSGGWDEDLI-------DTLFWPIDANRILQ-IPISAGREDFVAWHHTRSGIF 828
Query: 481 SFNSSFVF-----------LQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRD 529
S S++ L D PV++ +W + +P IK F WR+L G I +
Sbjct: 829 SVRSAYHCQWNAKFGSKERLPDGASSSSRPVWDNLWKLEIPPKIKIFAWRMLHGVIPCKG 888
Query: 530 NLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVH 589
L + H A++ CP+C E H++F+C + LI + W + D
Sbjct: 889 ILAD---KHIAVQGGCPVCSSGVEDIKHMVFTCRRAKLIWKQLGIWSRIQPILGFDRSGS 945
Query: 590 LLQFSSI----GWSKAQKLGESAIWMSVLWSIWCLRNMVV 625
+L I S +G + + ++ W IW R +V
Sbjct: 946 VLVEEMIRKGGKVSHLNAIGMTELILTGAWYIWWERRQLV 985
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 70.5 bits (171), Expect = 2e-10
Identities = 78/334 (23%), Positives = 134/334 (39%), Gaps = 47/334 (14%)
Query: 319 KD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGE----------SSWWKEIQSIT 368
KD + FN ALL K WR L E SL+ RV ++ S + S W+ I
Sbjct: 834 KDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILF-- 891
Query: 369 VDNGWPWFQESVQKTVFAGNATQFWAENW--TGSGSLELRFRRLYNLSCQKREQVSNMGK 426
G + ++ + G T W + W GS L RR N+ + + + +
Sbjct: 892 ---GRELLMQGLRTVIGNGQKTFVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSR 948
Query: 427 WVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSF 486
W N+ W + +++ Q R + K+D++ W G +S + +
Sbjct: 949 ----------NWNLNMLRDLFPWKDVEIILKQ--RPLFFKEDSFCWLHSHNGLYSVKTGY 996
Query: 487 VFLQDQ-----YLE-----EPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QV 536
FL Q Y E + +F+ IW + I+ F+W+ L G I D L +
Sbjct: 997 EFLSKQVHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKALHGAIPVEDRLRTRGI 1056
Query: 537 LHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSI 596
+ C +C E+ +H+LF C + + H L + ++ S+ V+ I
Sbjct: 1057 ---RSDDGCLMCDTENETINHILFECPLARQVWAITH--LSSAGSEFSN-SVYTNMSRLI 1110
Query: 597 GWSKAQKLGESAIWMS--VLWSIWCLRNMVVFNG 628
++ L ++S +LW +W RN ++F G
Sbjct: 1111 DLTQQNDLPHHLRFVSPWILWFLWKNRNALLFEG 1144
>UniRef100_Q8LID8 Cyst nematode resistance protein-like protein [Oryza sativa]
Length = 210
Score = 70.1 bits (170), Expect = 2e-10
Identities = 55/193 (28%), Positives = 79/193 (40%), Gaps = 22/193 (11%)
Query: 465 NKKDTWRWKLEGEGGFSFNSSFVFLQDQYL--EEPDPVFNWIWMVPVPSNIKAFVWRVLL 522
N D RWK +G FS +S++ D + + P IW PS ++ F W
Sbjct: 18 NDPDKIRWKPGPDGAFSVSSAY----DMFFMARQSYPFGQHIWQTNAPSRVRFFFWLAAR 73
Query: 523 GRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQ 582
GR QT DNL H E C LC +E HL +C S + AW+ V
Sbjct: 74 GRCQTADNLAKKGWPH---EDSCALCMREQEDCHHLFVTCEFSGRVWELMRAWISVDFPI 130
Query: 583 VSDPKVHLLQFSSIGWSKAQKLGES-------AIWMSVLWSIWCLRNMVVFNGGELDKDQ 635
L+ + W +A++ + A+ M V W IW RN +F +Q
Sbjct: 131 PGQIDCSLIDW----WFEARRCFRTGYREIFDAVLMLVCWLIWKERNARIFEQRLRSPEQ 186
Query: 636 LLEQI--QVTAWR 646
L+E I ++ WR
Sbjct: 187 LVEDIKEEINVWR 199
>UniRef100_Q9ZQF8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1225
Score = 68.9 bits (167), Expect = 5e-10
Identities = 83/355 (23%), Positives = 133/355 (37%), Gaps = 78/355 (21%)
Query: 312 ADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVV--------------KANCNSCGE 357
+D + + FN+ LL K WR + P+SL RV+ KAN S G
Sbjct: 729 SDGGLGFRTLEEFNLVLLAKQLWRLIRFPNSLLSRVLRGRYFRYSDPIQIGKANRPSFG- 787
Query: 358 SSWWKEIQSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGS-------GSLELRFRRL 410
W+ I + P +++T+ +G T+ W + W S L +R L
Sbjct: 788 ---WRSIMAAK-----PLLLSGLRRTIGSGMLTRVWEDPWIPSFPPRPAKSILNIRDTHL 839
Query: 411 YNLSCQKREQVSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELM-----VILQGIRLVEN 465
Y V+++ V W+ +G +EL+ ++ GIR
Sbjct: 840 Y---------VNDLIDPVTKQWK-------------LGRLQELVDPSDIPLILGIRPSRT 877
Query: 466 -KKDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPDPVFNW---------IWMVPVPSNIKA 515
K D + W G ++ S + +D D F +W + K
Sbjct: 878 YKSDDFSWSFTKSGNYTVKSGYWAARDLSRPTCDLPFQGPSVSALQAQVWKIKTTRKFKH 937
Query: 516 FVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAW 575
F W+ L G + T L + H E +CP C EES +HLLF C S I A
Sbjct: 938 FEWQCLSGCLATNQRLFS---RHIGTEKVCPRCGAEEESINHLLFLCPPSRQI----WAL 990
Query: 576 LGVSTAQVSDPKVHLLQFSSIGWSKAQKLGES----AIWMSVLWSIWCLRNMVVF 626
+ +++ P+ L S+ ++ + I+ +LW IW RN +F
Sbjct: 991 SPIPSSEYIFPRNSLFYNFDFLLSRGKEFDIAEDIMEIFPWILWYIWKSRNRFIF 1045
>UniRef100_Q9ZQG1 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1138
Score = 68.6 bits (166), Expect = 6e-10
Identities = 80/372 (21%), Positives = 145/372 (38%), Gaps = 58/372 (15%)
Query: 310 AEADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITV 369
A+ + +D FN ALL K WR L P L+ R K S ++ + + +
Sbjct: 623 AKESGGLGFRDIESFNQALLAKQSWRILQYPSFLFARFFK--------SRYFDDEEFLEA 674
Query: 370 D-------------NGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQ 416
D G + ++K V G + W + W + L +R +++
Sbjct: 675 DLGVRPSYACCSILFGRELLAKGLRKEVGNGKSLNVWMDPWIFDIAPRLPLQRHFSV--- 731
Query: 417 KREQVSNMGKWVNGVWQWEFK-WRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLE 475
N+ VN + +E + W+R+L E ++ + ++ V N D W W
Sbjct: 732 ------NLDLKVNDLINFEDRCWKRDL--LEELFYPTDVELITKRNPVVNMDDFWVWLHA 783
Query: 476 GEGGFSFNSSF----------VFLQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRI 525
G +S S + + Q L + + +W IK F+WR+L +
Sbjct: 784 KTGEYSVKSGYWLAFQSNKLDLIQQANLLPSTNGLKEQVWSTKSSPKIKMFLWRILSAAL 843
Query: 526 QTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSD 585
D ++ + +++ C +C ES +H+LF+CS + + A GV T +
Sbjct: 844 PVADQIIRRGM---SIDPRCQICGDEGESTNHVLFTCSMAR----QVWALSGVPTPEFGF 896
Query: 586 PKVHLLQFSSIGW---SKAQKLGESAI---WMSVLWSIWCLRNMVVFNGGELDKDQLLEQ 639
+ F++I + K L + W VLW +W RN + F+G + +
Sbjct: 897 QNASI--FANIQFLFELKKMILVPDLVKRSWPWVLWRLWKNRNKLFFDGITFCPLNSIVK 954
Query: 640 IQVTAWRWLRSR 651
IQ W +++
Sbjct: 955 IQEDTLEWFQAQ 966
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.345 0.151 0.582
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,129,283,118
Number of Sequences: 2790947
Number of extensions: 44521806
Number of successful extensions: 158177
Number of sequences better than 10.0: 160
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 122
Number of HSP's that attempted gapping in prelim test: 157778
Number of HSP's gapped (non-prelim): 259
length of query: 706
length of database: 848,049,833
effective HSP length: 135
effective length of query: 571
effective length of database: 471,271,988
effective search space: 269096305148
effective search space used: 269096305148
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0330.6


