

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0327.8
(250 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
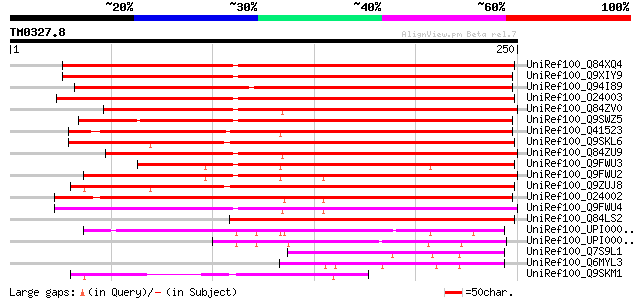
UniRef100_Q84XQ4 NtPRp27-like protein [Solanum tuberosum] 253 4e-66
UniRef100_Q9XIY9 NtPRp27 [Nicotiana tabacum] 252 7e-66
UniRef100_Q94I89 Putative NtPRp27-like protein [Atropa belladonna] 249 5e-65
UniRef100_O24003 Hypothetical protein precursor [Hordeum vulgare] 233 3e-60
UniRef100_Q84ZV0 R 14 protein [Glycine max] 223 4e-57
UniRef100_Q9SWZ5 Secretory protein [Triticum aestivum] 222 8e-57
UniRef100_Q41523 Hypothetical protein [Triticum aestivum] 217 3e-55
UniRef100_Q9SKL6 Expressed protein [Arabidopsis thaliana] 213 4e-54
UniRef100_Q84ZU9 R 13 protein [Glycine max] 202 8e-51
UniRef100_Q9FWU3 Putative secretory protein [Oryza sativa] 199 7e-50
UniRef100_Q9FWU2 Putative secretory protein [Oryza sativa] 192 5e-48
UniRef100_Q9ZUJ8 Hypothetical protein At2g15130 [Arabidopsis tha... 187 2e-46
UniRef100_O24002 Hypothetical protein precursor [Hordeum vulgare] 186 5e-46
UniRef100_Q9FWU4 Putative basic secretory protein [Oryza sativa] 184 2e-45
UniRef100_Q84LS2 Hypothetical protein [Arabidopsis thaliana] 155 7e-37
UniRef100_UPI000021A836 UPI000021A836 UniRef100 entry 102 7e-21
UniRef100_UPI000023CC63 UPI000023CC63 UniRef100 entry 96 1e-18
UniRef100_Q7S9L1 Hypothetical protein [Neurospora crassa] 92 1e-17
UniRef100_Q6MYL3 Hypothetical protein [Aspergillus fumigatus] 79 1e-13
UniRef100_Q9SKM1 Hypothetical protein At2g15170 [Arabidopsis tha... 74 3e-12
>UniRef100_Q84XQ4 NtPRp27-like protein [Solanum tuberosum]
Length = 226
Score = 253 bits (645), Expect = 4e-66
Identities = 121/223 (54%), Positives = 154/223 (68%), Gaps = 2/223 (0%)
Query: 27 IIYLMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQT 86
I ++ L ++A I AV Y VTN A TPGG RF+ +IG ++QQT+ A+ FI
Sbjct: 5 IFFISSLFVLAIFTQKIDAVDYSVTNTAANTPGGARFDRDIGAQYSQQTLVAATSFIWNI 64
Query: 87 FKQIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHE 146
F+Q KN+ VS+ V ++G A S+N I + + YI+G+SGDVR E+ G+LYHE
Sbjct: 65 FQQNSPADRKNVPKVSMFVDDMDG--VAYASNNEIHVSARYIQGYSGDVRREITGVLYHE 122
Query: 147 ATHIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCN 206
ATH+WQW GN AP GL EG+AD++RLKAG P HWV G G+ WD+GY VTA FLDYCN
Sbjct: 123 ATHVWQWNGNGGAPGGLIEGIADYVRLKAGLGPSHWVKPGQGNRWDQGYDVTAQFLDYCN 182
Query: 207 GLRDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVFG 249
LR+GFVA LN M++ YSD FFV LLGK+VDQLWSDYK+ FG
Sbjct: 183 SLRNGFVAELNKKMRNGYSDQFFVDLLGKTVDQLWSDYKAKFG 225
>UniRef100_Q9XIY9 NtPRp27 [Nicotiana tabacum]
Length = 242
Score = 252 bits (643), Expect = 7e-66
Identities = 120/222 (54%), Positives = 154/222 (69%), Gaps = 2/222 (0%)
Query: 27 IIYLMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQT 86
I + L ++A IHAV Y VTN A T GG RF +IG ++QQT+ A+ FI T
Sbjct: 21 IFFFYSLFVLAIFTQKIHAVDYSVTNTAANTAGGARFNRDIGAQYSQQTLAAATSFIWNT 80
Query: 87 FKQIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHE 146
F+Q KN++ VS+ V ++G A S+N I + ++YI+G+SGDVR E+ G+LYHE
Sbjct: 81 FQQNFPADRKNVQKVSMFVDDMDG--VAYASNNEIHVSASYIQGYSGDVRREITGVLYHE 138
Query: 147 ATHIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCN 206
+TH+WQW GN AP GL EG+AD++RLKAGFAP HWV G G WD+GY VTA FLDYCN
Sbjct: 139 STHVWQWNGNGGAPGGLIEGIADYVRLKAGFAPSHWVKPGQGDRWDQGYDVTARFLDYCN 198
Query: 207 GLRDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVF 248
LR+GFVA LN M+ YS+ FF+ LLGK+VDQLWSDYK+ F
Sbjct: 199 SLRNGFVAQLNKKMRTGYSNQFFIDLLGKTVDQLWSDYKAKF 240
>UniRef100_Q94I89 Putative NtPRp27-like protein [Atropa belladonna]
Length = 223
Score = 249 bits (636), Expect = 5e-65
Identities = 114/216 (52%), Positives = 156/216 (71%), Gaps = 2/216 (0%)
Query: 33 LVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQIDQ 92
L+I+A IHAV+Y VTNRA TPGG RF+ +IG+P+++ T+ ++ FI + F+Q
Sbjct: 3 LLILAIFTEKIHAVEYTVTNRAANTPGGARFDRDIGVPYSRLTLAFSTSFIWRIFEQNSP 62
Query: 93 FGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEATHIWQ 152
KN++ +S+ V ++G + A+ + I + + YI+ +SGDVR E+ G+LYHE TH+WQ
Sbjct: 63 ADRKNVQKISMFVDDMDGVAYAVNDE--IHVSARYIQSYSGDVRREIAGVLYHECTHVWQ 120
Query: 153 WFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGLRDGF 212
W GN AP GL EG+AD++RLKAG AP HWV G G WD+GY VTA FLDYCN L++GF
Sbjct: 121 WNGNGRAPGGLIEGIADYVRLKAGLAPSHWVKPGQGDRWDQGYDVTARFLDYCNSLKNGF 180
Query: 213 VAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVF 248
VA LN M++ YS+ FFV+LLGK+VDQLWSDYK+ F
Sbjct: 181 VAQLNKKMRNGYSNQFFVELLGKTVDQLWSDYKAKF 216
>UniRef100_O24003 Hypothetical protein precursor [Hordeum vulgare]
Length = 225
Score = 233 bits (594), Expect = 3e-60
Identities = 109/226 (48%), Positives = 149/226 (65%), Gaps = 2/226 (0%)
Query: 24 MSPIIYLMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFI 83
M P + + ++ + + AV + +N A+GT GG RF+N +G+ +++Q + AS FI
Sbjct: 1 MKPQVATVAFFLLVTMAATARAVTFDASNTASGTAGGQRFDNAVGLAYSKQVLSDASTFI 60
Query: 84 LQTFKQIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGIL 143
TF Q K ++ V+++V + G A S N I L + Y+ G+SGDV+ EV G+L
Sbjct: 61 WNTFNQRAAADRKPVDAVTLVVEDIGG--VAFASGNGIHLSAKYVGGYSGDVKKEVTGVL 118
Query: 144 YHEATHIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLD 203
YHEATH+WQW G A GL EG+AD++RLKAG AP HW P+G+G WD+GY +TA FLD
Sbjct: 119 YHEATHVWQWNGRGTANGGLIEGIADYVRLKAGLAPGHWRPQGSGDRWDQGYDITARFLD 178
Query: 204 YCNGLRDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVFG 249
YC+ L GFVA LNA MK YSDDFF ++LGK+V QLW DYK+ FG
Sbjct: 179 YCDSLMPGFVAQLNAKMKSGYSDDFFAQILGKNVQQLWKDYKAKFG 224
>UniRef100_Q84ZV0 R 14 protein [Glycine max]
Length = 641
Score = 223 bits (568), Expect = 4e-57
Identities = 101/205 (49%), Positives = 144/205 (69%), Gaps = 3/205 (1%)
Query: 47 QYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQIDQFGGKNIEHVSVIVT 106
++ V N A TPGG RF ++IG + +T++LA+QF+ + F+Q + KN++ VS++V
Sbjct: 437 KFTVINNALTTPGGVRFRDKIGAEYANRTLELATQFVWRIFQQKNPSDRKNVQKVSLVVD 496
Query: 107 ALNGNSAAITSDNNIFLDSNYIEGFSG-DVRNEVIGILYHEATHIWQWFGNKEAPRGLTE 165
+ G A T +N I + + Y+ G+SG DV+ E+ G+L+H+ ++WQW+GN EAP GL
Sbjct: 497 DMEG--IAYTMNNEIHVSARYVNGYSGGDVKREITGVLFHQVCYVWQWYGNGEAPGGLIG 554
Query: 166 GVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGLRDGFVAMLNAMMKDHYS 225
G+ADF+RLKA +A HW G G WDEGY +TA+FLDYC+ L+ GFVA LN +M+ YS
Sbjct: 555 GIADFVRLKANYAASHWRKPGQGQRWDEGYDITAHFLDYCDSLKSGFVAQLNQLMRTGYS 614
Query: 226 DDFFVKLLGKSVDQLWSDYKSVFGN 250
D FFV+LLGK VDQLW DYK+ +GN
Sbjct: 615 DQFFVQLLGKPVDQLWRDYKAQYGN 639
>UniRef100_Q9SWZ5 Secretory protein [Triticum aestivum]
Length = 224
Score = 222 bits (565), Expect = 8e-57
Identities = 106/214 (49%), Positives = 147/214 (68%), Gaps = 3/214 (1%)
Query: 35 IIASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQIDQFG 94
++ + + AV + +N+A+GT GG RF N+ +P++++ + AS FI +TF Q
Sbjct: 12 VLVALAAAAQAVTFDASNKASGTSGGRRF-NQAVVPYSKKVLSDASAFIWKTFNQRAVGD 70
Query: 95 GKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEATHIWQWF 154
K ++ V+++V ++G A TS N I L + Y+ G SGDV+ EV G+LYHEATH+WQW
Sbjct: 71 RKTVDAVTLVVEDISG--VAFTSANGIHLSAQYVGGISGDVKKEVTGVLYHEATHVWQWN 128
Query: 155 GNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGLRDGFVA 214
G +A GL EG+AD++RLKAGFAP HWV G G WD+GY VTA FLDYC+ L+ GFVA
Sbjct: 129 GPGQANGGLIEGIADYVRLKAGFAPGHWVKPGQGDRWDQGYDVTARFLDYCDSLKPGFVA 188
Query: 215 MLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVF 248
+NA MK Y+DDFF ++LGK+V QLW DYK+ F
Sbjct: 189 HVNAKMKSGYTDDFFAQILGKNVQQLWKDYKAKF 222
>UniRef100_Q41523 Hypothetical protein [Triticum aestivum]
Length = 231
Score = 217 bits (552), Expect = 3e-55
Identities = 108/220 (49%), Positives = 147/220 (66%), Gaps = 6/220 (2%)
Query: 30 LMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQ 89
L+FL + A +G AV + TN + GG RF+ ++G+ + +Q + AS FI TF Q
Sbjct: 12 LLFLALAAMAG----AVTFDATNTVPDSAGGQRFDQDVGVDYAKQVLSEASSFIWTTFNQ 67
Query: 90 IDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFS-GDVRNEVIGILYHEAT 148
+ ++ + V++ V N A TS N I L + Y+ GF GDV+ EV G+LYHEAT
Sbjct: 68 PNPEDRRDYDSVTLAVVD-NIEPVAQTSGNAIQLRAQYVAGFDEGDVKKEVKGVLYHEAT 126
Query: 149 HIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGL 208
H+WQW G A GL EG+AD++RLKA AP HW P+G+G WDEGY VTA FLDYC+ L
Sbjct: 127 HVWQWNGQGRANGGLIEGIADYVRLKADLAPTHWRPQGSGDRWDEGYDVTAKFLDYCDSL 186
Query: 209 RDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVF 248
+ GFVA +N+ +KD YSDD+FV++LGKSVDQLW+DYK+ +
Sbjct: 187 KAGFVAEMNSKLKDGYSDDYFVQILGKSVDQLWNDYKAKY 226
>UniRef100_Q9SKL6 Expressed protein [Arabidopsis thaliana]
Length = 225
Score = 213 bits (542), Expect = 4e-54
Identities = 103/221 (46%), Positives = 144/221 (64%), Gaps = 3/221 (1%)
Query: 30 LMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIG-IPFTQQTIQLASQFILQTFK 88
+ F++ + S ++AV Y V + + + GG RF EIG I + QT++ A+ F+ + F+
Sbjct: 6 IFFVISLMLVVSLVNAVDYSVVDNSGDSTGGRRFRGEIGGISYGTQTLRSATDFVWRLFQ 65
Query: 89 QIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEAT 148
Q + K++ +++ + NG+ A S N I + Y+ G SGDV+ E G++YHE
Sbjct: 66 QTNPSDRKSVTKITLFME--NGDGVAYNSANEIHFNVGYLAGVSGDVKREFTGVVYHEVV 123
Query: 149 HIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGL 208
H WQW G AP GL EG+AD++RLKAG+AP HWV G G WD+GY VTA FLDYCNGL
Sbjct: 124 HSWQWNGAGRAPGGLIEGIADYVRLKAGYAPSHWVGPGRGDRWDQGYDVTARFLDYCNGL 183
Query: 209 RDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVFG 249
R+GFVA LN M++ YSD FFV LLGK V+QLW +YK+ +G
Sbjct: 184 RNGFVAELNKKMRNGYSDGFFVDLLGKDVNQLWREYKAKYG 224
>UniRef100_Q84ZU9 R 13 protein [Glycine max]
Length = 641
Score = 202 bits (513), Expect = 8e-51
Identities = 94/204 (46%), Positives = 135/204 (66%), Gaps = 3/204 (1%)
Query: 48 YQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQIDQFGGKNIEHVSVIVTA 107
+ V N A T GG RF ++IG + QT+ A+QFI + F+Q + K+++ VS+ V
Sbjct: 438 FTVINNALTTLGGVRFRDKIGAEYANQTLDSATQFIWEIFQQNNPSDRKSVQKVSLFVDD 497
Query: 108 LNGNSAAITSDNNIFLDSNYIEGFSG-DVRNEVIGILYHEATHIWQWFGNKEAPRGLTEG 166
++G A T N I + + Y+ G+SG DV+ E+ G+L+H+ ++WQW GN +AP GLT G
Sbjct: 498 MDG--IAYTRKNEIHVSARYVNGYSGGDVKREITGVLFHQVCYVWQWKGNGKAPGGLTGG 555
Query: 167 VADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGLRDGFVAMLNAMMKDHYSD 226
+ADF+RLKA +A HW G G W+EGY +TA+FLDYC+ L+ GFV LN M+ YSD
Sbjct: 556 IADFVRLKANYAASHWRKPGQGQKWNEGYEITAHFLDYCDSLKSGFVGQLNQWMRTDYSD 615
Query: 227 DFFVKLLGKSVDQLWSDYKSVFGN 250
+FF LL K V+ LW DYK+++GN
Sbjct: 616 EFFFLLLAKPVNHLWRDYKAMYGN 639
>UniRef100_Q9FWU3 Putative secretory protein [Oryza sativa]
Length = 260
Score = 199 bits (505), Expect = 7e-50
Identities = 99/191 (51%), Positives = 131/191 (67%), Gaps = 7/191 (3%)
Query: 64 ENEIGIPFTQQTIQLASQFILQTFKQIDQFGG-KNIEHVSVIVTALNGNSAAITSDNNIF 122
+ E+G+ + +Q + AS FI TF+Q G K ++ V++ V ++G A TS + I
Sbjct: 71 DREVGVDYAKQMLADASSFIWDTFEQPGDGGDRKPVDAVTLTVEDIDG--VAFTSGDGIH 128
Query: 123 LDSNYIEGFS--GDVRNEVIGILYHEATHIWQWFGNKEAPRGLTEGVADFMRLKAGFAPP 180
L + Y+ G+S GDVR EV G+LYHEATH+WQW G A GL EG+ADF+RL+AG+APP
Sbjct: 129 LSARYVGGYSAAGDVRAEVTGVLYHEATHVWQWDGRGGADGGLIEGIADFVRLRAGYAPP 188
Query: 181 HWVPRGTGSGWDEGYVVTAYFLDYCN--GLRDGFVAMLNAMMKDHYSDDFFVKLLGKSVD 238
HWV G G WD+GY VTA FLDYC+ + GFVA LN MKD YSDDFFV++ GK++D
Sbjct: 189 HWVQPGQGDRWDQGYDVTARFLDYCDSPAVVQGFVAQLNGKMKDGYSDDFFVQISGKTID 248
Query: 239 QLWSDYKSVFG 249
QLW DYK+ +G
Sbjct: 249 QLWQDYKAKYG 259
>UniRef100_Q9FWU2 Putative secretory protein [Oryza sativa]
Length = 233
Score = 192 bits (489), Expect = 5e-48
Identities = 96/219 (43%), Positives = 141/219 (63%), Gaps = 7/219 (3%)
Query: 37 ASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQIDQFGG- 95
A++ + AV + TN A+ T GG RF+ E+G+ + +Q + AS FI F+Q G
Sbjct: 17 AAAATAAGAVTFDATNTASSTAGGQRFDREVGVDYAKQVLADASSFIWDAFEQPGDGGDR 76
Query: 96 KNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFS---GDVRNEVIGILYHEATHIWQ 152
K ++ V++ V ++G A TS + I L + Y+ G+S GDVR EV G+LYHEATH+WQ
Sbjct: 77 KPVDAVTLTVEDIDG--VAFTSGDGIHLSARYVGGYSSSSGDVRTEVTGVLYHEATHVWQ 134
Query: 153 W-FGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGLRDG 211
W + A + EG+ADF+RL+AG+ WV G G+ W++ Y VTA F DYC+ ++ G
Sbjct: 135 WGLQDYAAHSWVYEGIADFVRLRAGYVAAGWVQPGQGNSWEDSYSVTARFFDYCDSVKPG 194
Query: 212 FVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVFGN 250
FVA +NA +KD Y+ D+FV++ GK+V QLW DYK+ +GN
Sbjct: 195 FVADINAKLKDGYNVDYFVQITGKTVQQLWQDYKAKYGN 233
>UniRef100_Q9ZUJ8 Hypothetical protein At2g15130 [Arabidopsis thaliana]
Length = 225
Score = 187 bits (475), Expect = 2e-46
Identities = 95/221 (42%), Positives = 138/221 (61%), Gaps = 4/221 (1%)
Query: 31 MFLVI-IASSGSPIHAVQYQVTNRATGTPGGTRFENEIG-IPFTQQTIQLASQFILQTFK 88
+FLVI + + S + AV + V + +PGG RF NEIG + + +Q+++ A+ F + F+
Sbjct: 6 IFLVISLMLAVSLVSAVDFSVVDNTGDSPGGRRFRNEIGGVSYGEQSLRDATDFTWRLFQ 65
Query: 89 QIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEAT 148
Q + K++ +++ + N N A +S + I ++ + G VR G++YHE
Sbjct: 66 QTNPSDRKDVTKITLFME--NSNGIAYSSQDEIHYNAGSLVDDKGYVRRGFTGVVYHEVV 123
Query: 149 HIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGL 208
H WQW G AP GL EG+AD++RLKAG+ HWV G G WD+GY VTA FL+YCN L
Sbjct: 124 HSWQWNGAGRAPGGLIEGIADYVRLKAGYVASHWVRPGGGDRWDQGYDVTARFLEYCNDL 183
Query: 209 RDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVFG 249
R+GFVA LN M+ Y+D FFV LLGK V+QLW +YK+ +G
Sbjct: 184 RNGFVAELNKKMRSDYNDGFFVDLLGKDVNQLWREYKANYG 224
>UniRef100_O24002 Hypothetical protein precursor [Hordeum vulgare]
Length = 229
Score = 186 bits (472), Expect = 5e-46
Identities = 95/228 (41%), Positives = 139/228 (60%), Gaps = 5/228 (2%)
Query: 23 KMSPIIYLMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQF 82
K+S + L+ +A++ S AV + VTN A+ T GG RF+ E G + +Q + AS F
Sbjct: 2 KISIAAAAVLLLALAATAS---AVTFDVTNEASSTAGGQRFDREYGAAYAKQVLSDASSF 58
Query: 83 ILQTFKQIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGD-VRNEVIG 141
F Q D + + +V + + N A TS + I L + + G +GD ++ +V G
Sbjct: 59 TWGIFNQPDPSDRRPADGDTVTLAVRDTNGIASTSGSTIELSARSVGGITGDNLKEQVDG 118
Query: 142 ILYHEATHIWQW-FGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAY 200
+LYHE H+WQW + G+ EG+AD++RLKAG+ +WV G GS WDEGY VTA
Sbjct: 119 VLYHEVVHVWQWGLQDYHEHHGIFEGIADYVRLKAGYVAANWVKEGGGSRWDEGYDVTAR 178
Query: 201 FLDYCNGLRDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVF 248
FLDYC+ + GFVA +N +KD Y+DD+FV++LG S DQLW+DYK+ +
Sbjct: 179 FLDYCDSRKPGFVAEMNGKLKDGYNDDYFVQILGTSADQLWNDYKAKY 226
>UniRef100_Q9FWU4 Putative basic secretory protein [Oryza sativa]
Length = 229
Score = 184 bits (467), Expect = 2e-45
Identities = 93/230 (40%), Positives = 138/230 (59%), Gaps = 4/230 (1%)
Query: 23 KMSPIIYLMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQF 82
K+ + ++ V +SS P AV Y+V+N A T GG RF+ E G + +Q + AS F
Sbjct: 2 KLHVVAAILLAVAASSSPPPAAAVTYEVSNEAASTAGGQRFDREYGAGYAKQVLAAASSF 61
Query: 83 ILQTFKQIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSG-DVRNEVIG 141
F Q + ++ V + V ++G A TS N I L + Y+ G +G D + +V G
Sbjct: 62 TWSIFSQPSAADRRPVDAVVLAVRDVDG--IASTSGNTITLGAGYVAGVTGNDFKTQVTG 119
Query: 142 ILYHEATHIWQW-FGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAY 200
+LYHE H+WQW + A + EG+ADF+RL+AG+ WV G G+ W++ Y VTA
Sbjct: 120 VLYHEVVHVWQWGLQDYGAHSWVYEGIADFVRLRAGYPAAGWVQPGQGNSWEDSYSVTAR 179
Query: 201 FLDYCNGLRDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVFGN 250
F DYC+ ++ GFVA LNA +K+ Y+ D+FV++ GK+V QLW DYK+ +GN
Sbjct: 180 FFDYCDSVKPGFVADLNAKLKNGYNVDYFVQITGKTVQQLWQDYKAKYGN 229
>UniRef100_Q84LS2 Hypothetical protein [Arabidopsis thaliana]
Length = 144
Score = 155 bits (393), Expect = 7e-37
Identities = 72/141 (51%), Positives = 92/141 (65%)
Query: 109 NGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEATHIWQWFGNKEAPRGLTEGVA 168
N N A +S + I ++ + G VR G++YHE H WQW G AP GL EG+A
Sbjct: 3 NSNGIAYSSQDEIHYNAGSLVDDKGYVRRGFTGVVYHEVVHSWQWNGAGRAPGGLIEGIA 62
Query: 169 DFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGLRDGFVAMLNAMMKDHYSDDF 228
D++RLKAG+ HWV G G WD+GY VTA FL+YCN LR+GFVA LN M+ Y+D F
Sbjct: 63 DYVRLKAGYVASHWVRPGGGDRWDQGYDVTARFLEYCNDLRNGFVAELNKKMRSDYNDGF 122
Query: 229 FVKLLGKSVDQLWSDYKSVFG 249
FV LLGK V+QLW +YK+ +G
Sbjct: 123 FVDLLGKDVNQLWREYKANYG 143
>UniRef100_UPI000021A836 UPI000021A836 UniRef100 entry
Length = 313
Score = 102 bits (255), Expect = 7e-21
Identities = 72/226 (31%), Positives = 112/226 (48%), Gaps = 21/226 (9%)
Query: 37 ASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQIDQFGGK 96
+SS P+ ++ +V R G RF + + T T Q IL +
Sbjct: 71 SSSTFPLPTLRLEV--RDVMHTGAHRFLSRVDASATLPTAVANVQRILYNTPKDHTTHLP 128
Query: 97 NIEHVSVIVTALNG----NSAAITSDNN-IFLDSNYIEGFS------GD----VRNEVIG 141
V++I+ ++G + + SD+ I +YIE S GD + +E++G
Sbjct: 129 PTRSVTLILRDMDGVAYTTGSELDSDHKEIHFSLHYIESISKREKCSGDENDGIAHEIVG 188
Query: 142 ILYHEATHIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYF 201
++ HE H +QW P GL EG+ADF+RL+ G +PPHW GS WD GY TAYF
Sbjct: 189 VITHELVHCYQWDAKGTCPGGLIEGIADFVRLRCGLSPPHWEKDLEGS-WDRGYQHTAYF 247
Query: 202 LDYCN-GLRDGFVAMLNAMMKDHYSDD--FFVKLLGKSVDQLWSDY 244
LDY DG V +N ++ ++ F+ +++G+ V+QLW+DY
Sbjct: 248 LDYLECRFGDGTVRKINEGLRTRKYEEKPFWTEIVGRPVEQLWADY 293
>UniRef100_UPI000023CC63 UPI000023CC63 UniRef100 entry
Length = 263
Score = 95.5 bits (236), Expect = 1e-18
Identities = 57/155 (36%), Positives = 86/155 (54%), Gaps = 11/155 (7%)
Query: 101 VSVIVTALNG----NSAAITSDNN-IFLDSNYIEGFSGDVR--NEVIGILYHEATHIWQW 153
V+VI+ +++G + + SD+ I ++YI R +E+ G+L HE H Q+
Sbjct: 105 VTVILRSMSGVAYTTGSELDSDHKEIHFSTDYISNIHPISRRSDEINGVLTHELVHCLQY 164
Query: 154 FGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYC-NGLRDGF 212
G+ P GL EG+AD++RL +PPHW R +G WD GY TAYFLDY G
Sbjct: 165 NGHGHCPGGLIEGIADWVRLHCLLSPPHW-KRESGGKWDAGYQQTAYFLDYLEERFGKGT 223
Query: 213 VAMLNAMMK--DHYSDDFFVKLLGKSVDQLWSDYK 245
+ LN ++ + F+ +L+G+ V+QLW DYK
Sbjct: 224 IRRLNEKLRIQKYEEKSFWTELVGRPVEQLWDDYK 258
>UniRef100_Q7S9L1 Hypothetical protein [Neurospora crassa]
Length = 292
Score = 92.0 bits (227), Expect = 1e-17
Identities = 45/113 (39%), Positives = 67/113 (58%), Gaps = 6/113 (5%)
Query: 138 EVIGILYHEATHIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGT--GSGWDEGY 195
E+ G+L HE H +QW + AP GL EG+AD++RL +PPHW WD+GY
Sbjct: 167 EITGVLTHELVHCYQWDAHGSAPGGLIEGIADWVRLNCDLSPPHWKQGDVKIDDPWDKGY 226
Query: 196 VVTAYFLDYCNG-LRDGFVAMLNAMM---KDHYSDDFFVKLLGKSVDQLWSDY 244
TAYFL Y G +G V LN + +++ + +F+V +LG+ V++LW +Y
Sbjct: 227 QHTAYFLQYLEGRFGEGTVRRLNEALRSTREYEAREFWVGVLGEEVEELWGEY 279
>UniRef100_Q6MYL3 Hypothetical protein [Aspergillus fumigatus]
Length = 312
Score = 79.0 bits (193), Expect = 1e-13
Identities = 50/137 (36%), Positives = 69/137 (49%), Gaps = 26/137 (18%)
Query: 134 DVRNEVIGILYHEATHIWQWF---GNKEA-----PRGLTEGVADFMRLKAGFAPPHW--- 182
D E+ G+L HE H +Q G++ A P GL EG+ADF+RLKAG PPHW
Sbjct: 155 DPAAEITGVLTHELVHCYQHTTPNGDESASVPRPPGGLIEGIADFVRLKAGLEPPHWKRP 214
Query: 183 -VPRGTGSGWDEGYVVTAYFLDYCNGLR--DGFVAMLNAMM------------KDHYSDD 227
+ S WD+GY TAYFL + +R G V MLN + ++ +
Sbjct: 215 TSAKERPSKWDQGYQHTAYFLAWLEDVRVGKGAVGMLNDRLLRVGYVGEDSESENQDTQS 274
Query: 228 FFVKLLGKSVDQLWSDY 244
F+ L GK + +LW +Y
Sbjct: 275 FWRGLFGKGISELWEEY 291
>UniRef100_Q9SKM1 Hypothetical protein At2g15170 [Arabidopsis thaliana]
Length = 134
Score = 73.9 bits (180), Expect = 3e-12
Identities = 50/149 (33%), Positives = 72/149 (47%), Gaps = 31/149 (20%)
Query: 31 MFLVI-IASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQ 89
+FLVI + + S ++AV + V + +PGG +F +EIG
Sbjct: 6 IFLVICLMLAVSMVNAVDFFVVDNTGDSPGGRKFRDEIG--------------------- 44
Query: 90 IDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEATH 149
G + S IVTA T + ++ + G SGDVR G++YHE H
Sbjct: 45 -----GVSYGKQSRIVTAW---PTTRTWERRYIFNARSLAGNSGDVRVGFTGVIYHEVAH 96
Query: 150 IWQWFGNKE-APRGLTEGVADFMRLKAGF 177
WQWFG + P GL EG+AD++RLKAG+
Sbjct: 97 SWQWFGAADRVPGGLIEGIADYVRLKAGY 125
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 441,088,043
Number of Sequences: 2790947
Number of extensions: 19045394
Number of successful extensions: 39308
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 39242
Number of HSP's gapped (non-prelim): 41
length of query: 250
length of database: 848,049,833
effective HSP length: 124
effective length of query: 126
effective length of database: 501,972,405
effective search space: 63248523030
effective search space used: 63248523030
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0327.8


