

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0327.6
(442 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
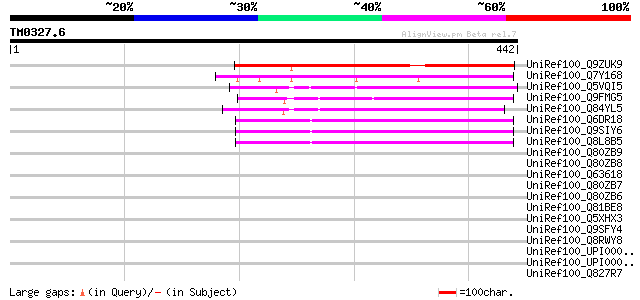
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZUK9 Hypothetical protein At2g15020 [Arabidopsis tha... 275 2e-72
UniRef100_Q7Y168 Hypothetical protein OSJNBa0090L05.5 [Oryza sat... 241 4e-62
UniRef100_Q5VQI5 Hypothetical protein P0701D05.8 [Oryza sativa] 143 1e-32
UniRef100_Q9FMG5 Gb|AAD25673.1 [Arabidopsis thaliana] 138 3e-31
UniRef100_Q84YL5 Hypothetical protein OSJNBa0086N05.123 [Oryza s... 136 1e-30
UniRef100_Q6DR18 Hypothetical protein [Arabidopsis thaliana] 133 1e-29
UniRef100_Q9SIY6 Hypothetical protein At2g40390 [Arabidopsis tha... 133 1e-29
UniRef100_Q8L8B5 Hypothetical protein At2g40390/T3G21.16 [Arabid... 133 1e-29
UniRef100_Q80ZB9 Purkinje cell espin isoform 1 [Rattus norvegicus] 39 0.39
UniRef100_Q80ZB8 Purkinje cell espin isoform 1+ [Rattus norvegicus] 39 0.39
UniRef100_Q63618 Espin [Rattus norvegicus] 39 0.39
UniRef100_Q80ZB7 Purkinje cell espin isoform 2 [Rattus norvegicus] 39 0.39
UniRef100_Q80ZB6 Purkinje cell espin isoform 2+ [Rattus norvegicus] 39 0.39
UniRef100_Q81BE8 Putative phosphohydrolases, Icc family [Bacillu... 37 1.1
UniRef100_Q5XHX3 Hypothetical protein [Rattus norvegicus] 36 1.9
UniRef100_Q9SFY4 T22C5.20 [Arabidopsis thaliana] 36 1.9
UniRef100_Q8RWY8 Hypothetical protein At1g27750 [Arabidopsis tha... 36 1.9
UniRef100_UPI000021D872 UPI000021D872 UniRef100 entry 36 2.5
UniRef100_UPI0000361403 UPI0000361403 UniRef100 entry 35 3.3
UniRef100_Q827R7 Hypothetical protein [Streptomyces avermitilis] 35 3.3
>UniRef100_Q9ZUK9 Hypothetical protein At2g15020 [Arabidopsis thaliana]
Length = 526
Score = 275 bits (703), Expect = 2e-72
Identities = 127/250 (50%), Positives = 180/250 (71%), Gaps = 18/250 (7%)
Query: 197 AHHQLEAHVQLEYTVGFHDGFIQVRARVDNIRLHVARLGFNKDGDDVD------FIDEKH 250
+H Q E VQ EY+V F++ +I+V ARVDNIR+HV++LGF+K G V+ + +E++
Sbjct: 286 SHQQAEILVQFEYSVKFYENYIKVNARVDNIRIHVSKLGFHKGGVGVENQIADCYSEERY 345
Query: 251 FPSRVRVWVGPEIGAAYVGGLSLGRSTENNEREVEIQKIVKGNFEKSEHISKVKAAARSS 310
FPSRVRVW+GPE+G+++V GLSLGRST+N ER++E+ +++KGNF K + +VKA AR +
Sbjct: 346 FPSRVRVWLGPELGSSHVSGLSLGRSTKNEERDIEVTRVLKGNFGKGKVAPRVKARARMA 405
Query: 311 RRTRTKSWRMDQDAEGNAAIFDVVLHDNMTGLEVGAWKPTGQTGDDPVHGMRGRYSGASR 370
+ + K WR++Q++EGNAA+FD VL+D +G EV KP + G
Sbjct: 406 TKRKVKDWRIEQESEGNAAVFDAVLYDRESGQEVTTVKP------------KPNQEGLKN 453
Query: 371 PFNKSGSVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVKGSYFETRYVE 430
F KSG +V DEYG+EVGWR+ +EMEGSVLKWR+GG+ W++YWPN++ ++ETR VE
Sbjct: 454 VFTKSGGMVFGRDEYGDEVGWRVGREMEGSVLKWRLGGKIWLTYWPNKLNTLFYETRCVE 513
Query: 431 WCDEVDLPLI 440
WCDEVDLPL+
Sbjct: 514 WCDEVDLPLL 523
>UniRef100_Q7Y168 Hypothetical protein OSJNBa0090L05.5 [Oryza sativa]
Length = 565
Score = 241 bits (614), Expect = 4e-62
Identities = 132/286 (46%), Positives = 168/286 (58%), Gaps = 26/286 (9%)
Query: 180 PQNVLPHRRSRHRALFHA--HHQLEAHVQLEYTVGFHDG--FIQVRARVDNIRLHVARLG 235
P LPH AL + H QLE QLEY V G F+ V RVDN+R+ VARLG
Sbjct: 272 PITALPHEAPEEPALRYGLVHQQLEVVAQLEYAVRARRGERFMTVAVRVDNVRVRVARLG 331
Query: 236 FNKDGDDVD----------FIDEKHFPSRVRVWVGPEIGAAYVGGLSLGRSTENNEREVE 285
F +D D D E+HFPSR+R+WVGP GA+Y G SLGRST N E++VE
Sbjct: 332 FRRDDADADADGGDAHDDAMDGERHFPSRLRLWVGPRFGASYATGPSLGRSTGNPEQDVE 391
Query: 286 IQKIVKGNFEKSEHIS--------KVKAAARSSRRTRTKSWRMDQDAEGNAAIFDVVLHD 337
+ VKG F + ++KA RSS R R +SWR +Q+AEG+A +F+ VL D
Sbjct: 392 TTRTVKGAFAAAGATKLANGGVPPRIKAKTRSSARARNRSWRWEQEAEGSAGVFEGVLCD 451
Query: 338 NMTGLEVGAWKPTGQTGD----DPVHGMRGRYSGASRPFNKSGSVVIAGDEYGEEVGWRL 393
TG E+ AW+ + DP +GMR RY G R F+K +V+AGDE EEV WR+
Sbjct: 452 PATGTEISAWRGDNNNNNGGAGDPRNGMRRRYGGPGRAFSKMRGLVVAGDELPEEVTWRV 511
Query: 394 SKEMEGSVLKWRIGGEFWVSYWPNQVKGSYFETRYVEWCDEVDLPL 439
+E EG L WR+G + W++Y PNQV+ +FETR VEW EVDLPL
Sbjct: 512 GREAEGRTLPWRVGLKAWLTYLPNQVRSHHFETRCVEWAHEVDLPL 557
>UniRef100_Q5VQI5 Hypothetical protein P0701D05.8 [Oryza sativa]
Length = 502
Score = 143 bits (360), Expect = 1e-32
Identities = 87/256 (33%), Positives = 134/256 (51%), Gaps = 10/256 (3%)
Query: 192 RALFH-AHHQLEAHVQLEYTVGFHDGFIQVRARVDNIRLHV----ARLGFNKDGDDVDFI 246
R LF + QLE +QL Y D +I V +VDNIR V + + + G +
Sbjct: 251 RLLFSLTYQQLEGVIQLAYKTIRRDNWIDVEIKVDNIRCDVDSLVSEILMTERGHGSE-- 308
Query: 247 DEKHFPSRVRVWVGPEIGAAYVGGLSLGRSTENNEREVEIQKIVKGNFEKSEHISKVKAA 306
EKHFPSRV + + P + + V +S+ +S++N E ++K ++G+F+ +KA+
Sbjct: 309 -EKHFPSRVMLQITP-MQQSDVLSVSVSKSSDNPTHEFGLEKGIEGSFDPPNTFG-LKAS 365
Query: 307 ARSSRRTRTKSWRMDQDAEGNAAIFDVVLHDNMTGLEVGAWKPTGQTGDDPVHGMRGRYS 366
S K W+ +Q GN + LHD + G EV + KP+ + P R RYS
Sbjct: 366 VSESLTLTMKPWKFEQSVHGNTTTLNWFLHDGVNGREVYSSKPSKLSLLQPRAWFRDRYS 425
Query: 367 GASRPFNKSGSVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVKGSYFET 426
RPF K G V+ A DEYG+ V W++ G + W I G W++YWPN+ K + ET
Sbjct: 426 TVYRPFTKKGGVIFARDEYGDSVWWKICGAALGKTMDWEIRGWIWLTYWPNKQKTFHSET 485
Query: 427 RYVEWCDEVDLPLINN 442
R +E+ + + LPL+ +
Sbjct: 486 RRLEFRECLQLPLMKS 501
>UniRef100_Q9FMG5 Gb|AAD25673.1 [Arabidopsis thaliana]
Length = 502
Score = 138 bits (348), Expect = 3e-31
Identities = 81/248 (32%), Positives = 134/248 (53%), Gaps = 14/248 (5%)
Query: 199 HQLEAHVQLEYTVGFHDGFIQVRARVDNIRLHVARLGFNK------DGDDVDFIDEKHFP 252
+QLE +Q + V D +I V ++DNIR V +L K G+ EKHFP
Sbjct: 261 NQLEGVMQFNHKVVVRDNWIDVIVKIDNIRYDVIKLVNEKLMSRRGAGEH-----EKHFP 315
Query: 253 SRVRVWVGPEIGAAYVGGLSLGRSTENNEREVEIQKIVKGNFEKSEHISKVKAAARSSRR 312
SR+ + + P + ++ +S+ +S+ N RE E+++ ++G+F+ + A +S
Sbjct: 316 SRISLQLTPTLQTDFIS-VSVSKSSNNPGREFEVERSIEGSFDPPNSLGLRVAGREASTM 374
Query: 313 TRTKSWRMDQDAEGNAAIFDVVLHDNMTG-LEVGAWKPTGQTGDDPVHGMRGRYSGASRP 371
T T W+++Q G A + +L+D+ G EV + KP+ + P + RY+ A R
Sbjct: 375 TMTP-WKLEQSVLGYTANLNWILYDSSVGGREVFSTKPSRFSIMSPRSWFKDRYARAYRS 433
Query: 372 FNKSGSVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVKGSYFETRYVEW 431
F + G V+ AGDEYGE V W++ K G ++W I G W++YWPN+ K Y ETR +E+
Sbjct: 434 FTRRGGVIFAGDEYGESVVWKIGKGALGGTMEWEIKGFIWLTYWPNKYKTFYHETRRLEF 493
Query: 432 CDEVDLPL 439
++L +
Sbjct: 494 TQLLNLTI 501
>UniRef100_Q84YL5 Hypothetical protein OSJNBa0086N05.123 [Oryza sativa]
Length = 495
Score = 136 bits (342), Expect = 1e-30
Identities = 80/251 (31%), Positives = 124/251 (48%), Gaps = 9/251 (3%)
Query: 186 HRRSRHRALFHAHHQ-LEAHVQLEYTVGFHDGFIQVRARVDNIRLHVARLGFN----KDG 240
H+ + F ++Q LEA +Q Y V F + +I V VDNIR + +L K G
Sbjct: 239 HQTKDEKLQFSLNYQHLEAVIQFIYRVTFRENWIDVTVNVDNIRCDLIQLVSETLMAKQG 298
Query: 241 DDVDFIDEKHFPSRVRVWVGPEIGAAYVGGLSLGRSTENNEREVEIQKIVKGNFEKSEHI 300
D EKHFPSR+ + + P + + L++ RST+N +EV+ + + +
Sbjct: 299 YGSD---EKHFPSRISLQLTPLVQTDILS-LTVSRSTDNPAQEVDTEMGLDATLSAAPAT 354
Query: 301 SKVKAAARSSRRTRTKSWRMDQDAEGNAAIFDVVLHDNMTGLEVGAWKPTGQTGDDPVHG 360
+ +A + + W+ + GN A + LH G EV + +P + P
Sbjct: 355 IGITMSAHETVTRTLRPWKFEHSVHGNTAALNWFLHGGAEGREVFSSEPHKRELLQPRSW 414
Query: 361 MRGRYSGASRPFNKSGSVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVK 420
R RY+ RPF + G V+ AGDEYGE V WR+ G ++W + G WV+YWPN+ +
Sbjct: 415 FRNRYTNPGRPFTRGGGVIFAGDEYGESVCWRMPAAAAGKTVEWEMKGRIWVTYWPNKKR 474
Query: 421 GSYFETRYVEW 431
+ ETR VE+
Sbjct: 475 TLHVETRRVEF 485
>UniRef100_Q6DR18 Hypothetical protein [Arabidopsis thaliana]
Length = 496
Score = 133 bits (334), Expect = 1e-29
Identities = 76/243 (31%), Positives = 128/243 (52%), Gaps = 2/243 (0%)
Query: 198 HHQLEAHVQLEYTVGFHDGFIQVRARVDNIRLHVARLGFNKDGDDVDF-IDEKHFPSRVR 256
+HQLE +QL + + + + V +DN+R + RL K + +EKHFPSR+
Sbjct: 254 YHQLEGVIQLNHRIYVREKWFNVAVNIDNVRCDIIRLVNEKLLSERGMGTEEKHFPSRIS 313
Query: 257 VWVGPEIGAAYVGGLSLGRSTENNEREVEIQKIVKGNFEKSEHISKVKAAARSSRRTRTK 316
+ + P + + +S+ +S+EN E E++K ++ + +K +A + K
Sbjct: 314 LQLTPT-NQSNILMVSVQKSSENPLTEFEVEKGIEATIDPPNTFFGLKVSANETTTKSMK 372
Query: 317 SWRMDQDAEGNAAIFDVVLHDNMTGLEVGAWKPTGQTGDDPVHGMRGRYSGASRPFNKSG 376
W+ ++ G +A LHD G EV + KP+ + +P + RYS A RPF K G
Sbjct: 373 PWKFEEWVHGYSANLTWFLHDLDDGREVSSSKPSKVSMMNPRAWFKNRYSSAFRPFTKQG 432
Query: 377 SVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVKGSYFETRYVEWCDEVD 436
VV AGD YG+ V W++ K G V+++ + G W++YWPN+ Y +TR +E+ + +
Sbjct: 433 GVVFAGDSYGQSVLWKVDKTAIGKVMEFEVKGCVWLTYWPNKHHTFYSDTRKLEFKEMLY 492
Query: 437 LPL 439
L L
Sbjct: 493 LNL 495
>UniRef100_Q9SIY6 Hypothetical protein At2g40390 [Arabidopsis thaliana]
Length = 401
Score = 133 bits (334), Expect = 1e-29
Identities = 76/243 (31%), Positives = 128/243 (52%), Gaps = 2/243 (0%)
Query: 198 HHQLEAHVQLEYTVGFHDGFIQVRARVDNIRLHVARLGFNKDGDDVDF-IDEKHFPSRVR 256
+HQLE +QL + + + + V +DN+R + RL K + +EKHFPSR+
Sbjct: 159 YHQLEGVIQLNHRIYVREKWFNVAVNIDNVRCDIIRLVNEKLLSERGMGTEEKHFPSRIS 218
Query: 257 VWVGPEIGAAYVGGLSLGRSTENNEREVEIQKIVKGNFEKSEHISKVKAAARSSRRTRTK 316
+ + P + + +S+ +S+EN E E++K ++ + +K +A + K
Sbjct: 219 LQLTPT-NQSNILMVSVQKSSENPLTEFEVEKGIEATIDPPNTFFGLKVSANETTTKSMK 277
Query: 317 SWRMDQDAEGNAAIFDVVLHDNMTGLEVGAWKPTGQTGDDPVHGMRGRYSGASRPFNKSG 376
W+ ++ G +A LHD G EV + KP+ + +P + RYS A RPF K G
Sbjct: 278 PWKFEEWVHGYSANLTWFLHDLDDGREVSSSKPSKVSMMNPRAWFKNRYSSAFRPFTKQG 337
Query: 377 SVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVKGSYFETRYVEWCDEVD 436
VV AGD YG+ V W++ K G V+++ + G W++YWPN+ Y +TR +E+ + +
Sbjct: 338 GVVFAGDSYGQSVLWKVDKTAIGKVMEFEVKGCVWLTYWPNKHHTFYSDTRKLEFKEMLY 397
Query: 437 LPL 439
L L
Sbjct: 398 LNL 400
>UniRef100_Q8L8B5 Hypothetical protein At2g40390/T3G21.16 [Arabidopsis thaliana]
Length = 496
Score = 133 bits (334), Expect = 1e-29
Identities = 76/243 (31%), Positives = 128/243 (52%), Gaps = 2/243 (0%)
Query: 198 HHQLEAHVQLEYTVGFHDGFIQVRARVDNIRLHVARLGFNKDGDDVDF-IDEKHFPSRVR 256
+HQLE +QL + + + + V +DN+R + RL K + +EKHFPSR+
Sbjct: 254 YHQLEGVIQLNHRIYVREKWFNVAVNIDNVRCDIIRLVNEKLLSERGMGTEEKHFPSRIS 313
Query: 257 VWVGPEIGAAYVGGLSLGRSTENNEREVEIQKIVKGNFEKSEHISKVKAAARSSRRTRTK 316
+ + P + + +S+ +S+EN E E++K ++ + +K +A + K
Sbjct: 314 LQLTPT-NQSNILMVSVQKSSENPLTEFEVEKGIEATIDPPNTFFGLKVSANETTTKSMK 372
Query: 317 SWRMDQDAEGNAAIFDVVLHDNMTGLEVGAWKPTGQTGDDPVHGMRGRYSGASRPFNKSG 376
W+ ++ G +A LHD G EV + KP+ + +P + RYS A RPF K G
Sbjct: 373 PWKFEEWVHGYSANLTWFLHDLDDGREVSSSKPSKVSMMNPRAWFKNRYSSAFRPFTKQG 432
Query: 377 SVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVKGSYFETRYVEWCDEVD 436
VV AGD YG+ V W++ K G V+++ + G W++YWPN+ Y +TR +E+ + +
Sbjct: 433 GVVFAGDSYGQSVLWKVDKTAIGKVMEFEVKGCVWLTYWPNKHHTFYSDTRKLEFKEMLY 492
Query: 437 LPL 439
L L
Sbjct: 493 LNL 495
>UniRef100_Q80ZB9 Purkinje cell espin isoform 1 [Rattus norvegicus]
Length = 511
Score = 38.5 bits (88), Expect = 0.39
Identities = 27/90 (30%), Positives = 37/90 (41%), Gaps = 12/90 (13%)
Query: 35 SLNRTQSRANLRVRLRRGGDIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPT 94
S + + S+ R R D+ + L P P++ G +P S P P PS PP
Sbjct: 57 SSSHSSSKGQRSTRGARSSDLQSYMDMLNP--EPRSKQG----KPSSLPPPPPPSFPPPP 110
Query: 95 PPGNHIPLTHRAR*HLPKIPAPETEAGTHI 124
PPG +P P PAP G H+
Sbjct: 111 PPGTQLPPPP------PGYPAPNPPVGLHL 134
>UniRef100_Q80ZB8 Purkinje cell espin isoform 1+ [Rattus norvegicus]
Length = 520
Score = 38.5 bits (88), Expect = 0.39
Identities = 27/90 (30%), Positives = 37/90 (41%), Gaps = 12/90 (13%)
Query: 35 SLNRTQSRANLRVRLRRGGDIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPT 94
S + + S+ R R D+ + L P P++ G +P S P P PS PP
Sbjct: 57 SSSHSSSKGQRSTRGARSSDLQSYMDMLNP--EPRSKQG----KPSSLPPPPPPSFPPPP 110
Query: 95 PPGNHIPLTHRAR*HLPKIPAPETEAGTHI 124
PPG +P P PAP G H+
Sbjct: 111 PPGTQLPPPP------PGYPAPNPPVGLHL 134
>UniRef100_Q63618 Espin [Rattus norvegicus]
Length = 837
Score = 38.5 bits (88), Expect = 0.39
Identities = 27/90 (30%), Positives = 37/90 (41%), Gaps = 12/90 (13%)
Query: 35 SLNRTQSRANLRVRLRRGGDIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPT 94
S + + S+ R R D+ + L P P++ G +P S P P PS PP
Sbjct: 383 SSSHSSSKGQRSTRGARSSDLQSYMDMLNP--EPRSKQG----KPSSLPPPPPPSFPPPP 436
Query: 95 PPGNHIPLTHRAR*HLPKIPAPETEAGTHI 124
PPG +P P PAP G H+
Sbjct: 437 PPGTQLPPPP------PGYPAPNPPVGLHL 460
>UniRef100_Q80ZB7 Purkinje cell espin isoform 2 [Rattus norvegicus]
Length = 435
Score = 38.5 bits (88), Expect = 0.39
Identities = 27/90 (30%), Positives = 37/90 (41%), Gaps = 12/90 (13%)
Query: 35 SLNRTQSRANLRVRLRRGGDIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPT 94
S + + S+ R R D+ + L P P++ G +P S P P PS PP
Sbjct: 57 SSSHSSSKGQRSTRGARSSDLQSYMDMLNP--EPRSKQG----KPSSLPPPPPPSFPPPP 110
Query: 95 PPGNHIPLTHRAR*HLPKIPAPETEAGTHI 124
PPG +P P PAP G H+
Sbjct: 111 PPGTQLPPPP------PGYPAPNPPVGLHL 134
>UniRef100_Q80ZB6 Purkinje cell espin isoform 2+ [Rattus norvegicus]
Length = 444
Score = 38.5 bits (88), Expect = 0.39
Identities = 27/90 (30%), Positives = 37/90 (41%), Gaps = 12/90 (13%)
Query: 35 SLNRTQSRANLRVRLRRGGDIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPT 94
S + + S+ R R D+ + L P P++ G +P S P P PS PP
Sbjct: 57 SSSHSSSKGQRSTRGARSSDLQSYMDMLNP--EPRSKQG----KPSSLPPPPPPSFPPPP 110
Query: 95 PPGNHIPLTHRAR*HLPKIPAPETEAGTHI 124
PPG +P P PAP G H+
Sbjct: 111 PPGTQLPPPP------PGYPAPNPPVGLHL 134
>UniRef100_Q81BE8 Putative phosphohydrolases, Icc family [Bacillus cereus]
Length = 410
Score = 37.0 bits (84), Expect = 1.1
Identities = 34/131 (25%), Positives = 50/131 (37%), Gaps = 15/131 (11%)
Query: 303 VKAAARSSRRTRTKSWRMDQDAEGNAAIFDVVLHD--NMTGLEVGAWKPTGQTGDDPVHG 360
V A + R ++ + D +G+ FD VL D +T L + GD G
Sbjct: 20 VHAEGKEQARKSATTFNVISDIQGDLGDFDHVLKDMNKVTPLS----RALIMNGDITPTG 75
Query: 361 MRGRYSGASRPFNKSGSVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVK 420
+ +Y R NK+ +Y E V W E KW G+ S WPN V
Sbjct: 76 QQSQYDDVKRVLNKN--------KYPENV-WSTVGNHEFYAGKWTADGKLSQSTWPNGVN 126
Query: 421 GSYFETRYVEW 431
RY+++
Sbjct: 127 EETLFNRYLKF 137
>UniRef100_Q5XHX3 Hypothetical protein [Rattus norvegicus]
Length = 526
Score = 36.2 bits (82), Expect = 1.9
Identities = 34/108 (31%), Positives = 42/108 (38%), Gaps = 3/108 (2%)
Query: 15 LERVRLTTQVRASE*GQQLNSLNRTQSRANLRVRLRRGGDIHRVLARLPPIQHPK--ASL 72
LER+ Q R E ++L L R Q R+ G + A PP P A
Sbjct: 224 LERLERERQDRERE-RERLEQLEREQVEWERERRMSNAGHVLGPPAPPPPPPLPSGPAYA 282
Query: 73 GIREMQPFSR*PKPLPSSAPPTPPGNHIPLTHRAR*HLPKIPAPETEA 120
P P PLPS+ PP PP PL ++ P PAP A
Sbjct: 283 SALPPPPGPPPPPPLPSAGPPPPPPPPPPLPNQVPPPPPPPPAPPLPA 330
>UniRef100_Q9SFY4 T22C5.20 [Arabidopsis thaliana]
Length = 1840
Score = 36.2 bits (82), Expect = 1.9
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 3/54 (5%)
Query: 63 PPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGNHIPLTHRAR*HLPKIPAP 116
PP+ H S+ +QP S+ P+P P PP P PL H H P +P P
Sbjct: 832 PPLGHSLPSVLQPPLQPQSQPPEPPPEMMPPPPQALPPPLPHS---HPPLVPPP 882
>UniRef100_Q8RWY8 Hypothetical protein At1g27750 [Arabidopsis thaliana]
Length = 1075
Score = 36.2 bits (82), Expect = 1.9
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 3/54 (5%)
Query: 63 PPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGNHIPLTHRAR*HLPKIPAP 116
PP+ H S+ +QP S+ P+P P PP P PL H H P +P P
Sbjct: 849 PPLGHSLPSVLQPPLQPQSQPPEPPPEMMPPPPQALPPPLPHS---HPPLVPPP 899
>UniRef100_UPI000021D872 UPI000021D872 UniRef100 entry
Length = 860
Score = 35.8 bits (81), Expect = 2.5
Identities = 24/74 (32%), Positives = 31/74 (41%), Gaps = 12/74 (16%)
Query: 51 RGGDIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGNHIPLTHRAR*HL 110
R D+ + L P P++ G +P S P P PS PP PPG +P
Sbjct: 403 RSSDLQSYMDMLNP--EPRSKQG----KPSSLPPPPPPSFPPPPPPGTQLPPPP------ 450
Query: 111 PKIPAPETEAGTHI 124
P PAP G H+
Sbjct: 451 PGYPAPNPPVGLHL 464
>UniRef100_UPI0000361403 UPI0000361403 UniRef100 entry
Length = 696
Score = 35.4 bits (80), Expect = 3.3
Identities = 38/139 (27%), Positives = 47/139 (33%), Gaps = 30/139 (21%)
Query: 84 PKPLPSSAPPTPPGNHIPLTHRAR*HLPKIPAPETEAGTHIMDHGLTHTGISLQFLQPRL 143
P P SAPP P G+ +P P PAP T T + H + H+
Sbjct: 467 PSTAPPSAPPAPAGSTVPPP------APPPPAPPTTNATTV-HHPVPHSSF--------- 510
Query: 144 HRAPLLALRL*RAQRSWVTLLPIPARSHSRDGVVEDPQ--------NVLPHRRSRHRALF 195
A L+ L A + P +HS D + NV PH H
Sbjct: 511 --ATSLSSHLPSANAPAASAEAYPNPTHSPASYERDRENLGRLQMLNVTPH----HHQHS 564
Query: 196 HAHHQLEAHVQLEYTVGFH 214
H H L H Q T G H
Sbjct: 565 HIHSHLHLHQQDTATGGVH 583
>UniRef100_Q827R7 Hypothetical protein [Streptomyces avermitilis]
Length = 296
Score = 35.4 bits (80), Expect = 3.3
Identities = 22/61 (36%), Positives = 25/61 (40%), Gaps = 4/61 (6%)
Query: 62 LPPIQHPKASLGIREMQPFSR*PKPLPSSA----PPTPPGNHIPLTHRAR*HLPKIPAPE 117
L HP A G P P P P A PP PP +P H H+P PAP+
Sbjct: 24 LSAAHHPGAHPGPAPAPPPGPAPAPPPGFAGAPVPPGPPHTQVPQHHAPPQHVPVPPAPD 83
Query: 118 T 118
T
Sbjct: 84 T 84
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.141 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 762,029,966
Number of Sequences: 2790947
Number of extensions: 34137147
Number of successful extensions: 125961
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 125827
Number of HSP's gapped (non-prelim): 97
length of query: 442
length of database: 848,049,833
effective HSP length: 130
effective length of query: 312
effective length of database: 485,226,723
effective search space: 151390737576
effective search space used: 151390737576
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0327.6


