

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
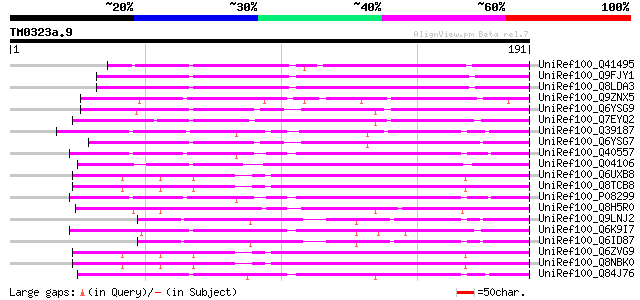
Query= TM0323a.9
(191 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q41495 STS14 protein precursor [Solanum tuberosum] 108 1e-22
UniRef100_Q9FJY1 Similarity to pathogenesis-related protein [Ara... 98 1e-19
UniRef100_Q8LDA3 Sts14 [Arabidopsis thaliana] 97 2e-19
UniRef100_Q9ZNX5 PR-1 like protein precursor [Camellia sinensis] 93 3e-18
UniRef100_Q6YSG9 Putative pathogenesis-related protein [Oryza sa... 84 1e-15
UniRef100_Q7EYQ2 Putative pathogenesis-related protein [Oryza sa... 80 4e-14
UniRef100_Q39187 Pathogenesis-related protein 1 precursor [Arabi... 80 4e-14
UniRef100_Q6YSG7 Putative pathogenesis-related protein [Oryza sa... 79 5e-14
UniRef100_Q40557 PR1a protein precursor [Nicotiana tabacum] 78 1e-13
UniRef100_Q04106 Prb-1b [Nicotiana tabacum] 78 1e-13
UniRef100_Q6UXB8 HGSC289 [Homo sapiens] 78 1e-13
UniRef100_Q8TCB8 PI16 protein [Homo sapiens] 78 1e-13
UniRef100_P08299 Pathogenesis-related protein 1A precursor [Nico... 78 1e-13
UniRef100_Q8H5R0 Putative pathogenesis-related protein [Oryza sa... 78 1e-13
UniRef100_Q9LNJ2 F6F3.11 protein [Arabidopsis thaliana] 77 2e-13
UniRef100_Q6K9I7 Putative pathogenesis related protein-1 [Oryza ... 77 2e-13
UniRef100_Q6ID87 At1g01310 [Arabidopsis thaliana] 77 2e-13
UniRef100_Q6ZVG9 Hypothetical protein FLJ42598 [Homo sapiens] 77 2e-13
UniRef100_Q8NBK0 Hypothetical protein PSEC0164 [Homo sapiens] 77 2e-13
UniRef100_Q84J76 Putative pathogenesis-related protein [Oryza sa... 76 4e-13
>UniRef100_Q41495 STS14 protein precursor [Solanum tuberosum]
Length = 214
Score = 108 bits (269), Expect = 1e-22
Identities = 66/156 (42%), Positives = 86/156 (54%), Gaps = 9/156 (5%)
Query: 37 SSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCTPGIF 96
+S ATP + A+ +L HN R E G+ L WS LAK+ SL R++RDK C+
Sbjct: 66 TSAATPPSR-AAQEFLDAHNKARSEVGVGP-LTWSPMLAKETSLLVRYQRDKQNCSFANL 123
Query: 97 DKKRYGVCCNQ-WLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIVWRK 155
+YG NQ W TV + PR + WV ++ +Y N+ + CG YTQIVW+K
Sbjct: 124 SNGKYGG--NQLWASGTV--VTPRMAVDSWVAEKKFYNYENNSCTGDDKCGVYTQIVWKK 179
Query: 156 TKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
+ LGCAQ C G L VCFY PPGNV GE+P
Sbjct: 180 SIELGCAQRTCYE--GPATLTVCFYNPPGNVIGEKP 213
>UniRef100_Q9FJY1 Similarity to pathogenesis-related protein [Arabidopsis thaliana]
Length = 185
Score = 97.8 bits (242), Expect = 1e-19
Identities = 55/159 (34%), Positives = 80/159 (49%), Gaps = 5/159 (3%)
Query: 33 LLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCT 92
++ S P + AK + HN R G+ L WS L AS AR++R++ C
Sbjct: 31 IVSTSPPPPPTISAAAKAFTDAHNKARAMVGVPP-LVWSQTLEAAASRLARYQRNQKKCE 89
Query: 93 PGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIV 152
+ +YG NQ + + P + WVK++ +Y + AN+ CG Y Q+V
Sbjct: 90 FASLNPGKYGA--NQLWAKGLVAVTPSLAVETWVKEKPFYNYKSDTCAANHTCGVYKQVV 147
Query: 153 WRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
WR +K LGCAQA C + L +CFY PPGNV G++P
Sbjct: 148 WRNSKELGCAQATCTKE--STVLTICFYNPPGNVIGQKP 184
>UniRef100_Q8LDA3 Sts14 [Arabidopsis thaliana]
Length = 185
Score = 97.4 bits (241), Expect = 2e-19
Identities = 54/159 (33%), Positives = 80/159 (49%), Gaps = 5/159 (3%)
Query: 33 LLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCT 92
++ S P + AK + HN R G+ L WS L AS AR++R++ C
Sbjct: 31 IVSTSPPPPPTISAAAKAFTDAHNKARAMVGVPP-LVWSQTLEAAASRLARYQRNQKKCE 89
Query: 93 PGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIV 152
+ +YG NQ + + P + WVK++ +Y + AN+ CG Y Q+V
Sbjct: 90 FASLNPGKYGA--NQLWAKGLVAVTPSLAVETWVKEKPFYNYKSDTCAANHTCGVYKQVV 147
Query: 153 WRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
WR +K LGCAQA C + L +CFY PPGN+ G++P
Sbjct: 148 WRNSKELGCAQATCTKE--STVLTICFYNPPGNIIGQKP 184
>UniRef100_Q9ZNX5 PR-1 like protein precursor [Camellia sinensis]
Length = 191
Score = 93.2 bits (230), Expect = 3e-18
Identities = 64/181 (35%), Positives = 96/181 (52%), Gaps = 24/181 (13%)
Query: 27 VLVALALLLVSSQATPATTD-----------GAKVYLFEHNFVRREKGISEHLEWSDKLA 75
V++ + LL++ +T D A+ ++ HN R E G+ + L+WS LA
Sbjct: 18 VILPVLLLVICHSSTHLLADHPIAARWVPPGAARQFVDAHNSARAEVGV-DPLKWSYSLA 76
Query: 76 KDASLYARFRRDKYLCT-PGIFDKKRYGVCCNQ-WLEDTVKIMDPREVMRQWVKD--RFY 131
AS R++++ C + + +YG NQ W + + K PREV+ WV + Y
Sbjct: 77 NAASRLVRYQKNYMHCEFADMTGQLQYG--SNQMWSDYSAK--PPREVVEYWVNSGKKHY 132
Query: 132 YEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYP-PGNVPGER 190
+N CV N CG Y Q+VW KT+ +GCAQ CG + G L++CFYYP PGN+ G+R
Sbjct: 133 RYTHNYCV-RNQNCGPYKQVVWEKTEMVGCAQGVCGNNNGS--LSICFYYPHPGNLGGQR 189
Query: 191 P 191
P
Sbjct: 190 P 190
>UniRef100_Q6YSG9 Putative pathogenesis-related protein [Oryza sativa]
Length = 170
Score = 84.3 bits (207), Expect = 1e-15
Identities = 53/171 (30%), Positives = 85/171 (48%), Gaps = 17/171 (9%)
Query: 27 VLVALALLLVSSQATPATT-----DGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLY 81
++VA+A + + AT T + V++ HN R G+ + W+D LA A +
Sbjct: 10 MVVAMAAFAMIAMATTTTAQEFSANEKAVFVQLHNNARAAVGVGP-VAWNDALAAQALQH 68
Query: 82 ARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVP 140
AR+ + +++ PG + + + W P + M WV ++ YY+ +NSC
Sbjct: 69 ARYCQTQHI--PGPYGENLW------WSYGAGTTGTPADAMSYWVGEKPYYDYSSNSC-- 118
Query: 141 ANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
C YTQ+VWR+T Y+GCA+ C + G + C YYP GN+ ERP
Sbjct: 119 GGRECRHYTQVVWRRTAYVGCARVACNTNNGIGTIIACNYYPGGNIYNERP 169
>UniRef100_Q7EYQ2 Putative pathogenesis-related protein [Oryza sativa]
Length = 169
Score = 79.7 bits (195), Expect = 4e-14
Identities = 55/169 (32%), Positives = 77/169 (45%), Gaps = 8/169 (4%)
Query: 24 FFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYAR 83
F V+ A+A++L ++ A T + HN R G+ L W D LA A YA
Sbjct: 7 FAVVMAAMAVVLATTSTAQAQTTATDIVNI-HNAARSAVGVPA-LSWDDNLAAYAQGYAN 64
Query: 84 FRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPAN 142
R C D+ Y N +V+ + QWV ++ Y+ +NSCV
Sbjct: 65 QRAGD--CALRHSDRNNYQYGENLSWNPSVQAWTAASSVDQWVAEKGSYDYASNSCV-GG 121
Query: 143 YPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR T +GCA C + G +C Y+P GNV +RP
Sbjct: 122 AMCGHYTQVVWRDTTAVGCAAVACNANRG--VFFICTYFPAGNVQNQRP 168
>UniRef100_Q39187 Pathogenesis-related protein 1 precursor [Arabidopsis thaliana]
Length = 166
Score = 79.7 bits (195), Expect = 4e-14
Identities = 63/176 (35%), Positives = 92/176 (51%), Gaps = 14/176 (7%)
Query: 18 KMFPHPFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKD 77
KMF P VL+ALAL+L + A D + YL HN R + + H++W A+
Sbjct: 2 KMFISPQTLVLLALALVLAFAVPLKAQ-DRRQDYLDVHNHARDDVSVP-HIKWHAGAARY 59
Query: 78 ASLYA-RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRF-YYEGN 135
A YA R +RD L + RYG + L + M +R WV+++ Y+ +
Sbjct: 60 AWNYAQRRKRDCRLIHSN--SRGRYG----ENLAWSSGDMSGAAAVRLWVREKSDYFHKS 113
Query: 136 NSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
N+C A CG YTQ+VW+ ++++GCA+ +C D GG + C Y PGNV G RP
Sbjct: 114 NTC-RAGKQCGHYTQVVWKNSEWVGCAKVKC--DNGGTFV-TCNYSHPGNVRGRRP 165
>UniRef100_Q6YSG7 Putative pathogenesis-related protein [Oryza sativa]
Length = 158
Score = 79.3 bits (194), Expect = 5e-14
Identities = 46/163 (28%), Positives = 79/163 (48%), Gaps = 11/163 (6%)
Query: 30 ALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKY 89
A+ ++ ++ A + D ++ HN R G+ + W+D LA A +AR+ + ++
Sbjct: 5 AMIIMATTTTAQQFSEDEKAAFVNLHNSARAAVGVGR-VAWNDALAAQALQHARYCQTQH 63
Query: 90 LCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRF-YYEGNNSCVPANYPCGDY 148
+ PG + + + W P + M W+ ++ YY +N C C Y
Sbjct: 64 I--PGPYGENLW------WSYGAGTTGTPADAMSYWLAEKAKYYYDSNYCSAGELGCTHY 115
Query: 149 TQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
TQ+VWR+T Y+GCA+ C + G + C Y+P GN+ ERP
Sbjct: 116 TQVVWRRTAYVGCARVACNTNGIGT-IIACNYFPRGNMKNERP 157
>UniRef100_Q40557 PR1a protein precursor [Nicotiana tabacum]
Length = 168
Score = 78.2 bits (191), Expect = 1e-13
Identities = 55/170 (32%), Positives = 84/170 (49%), Gaps = 13/170 (7%)
Query: 23 PFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYA 82
P F ++ L L LV S + A + + YL HN R + G+ E L W D++A A YA
Sbjct: 10 PSFLLVSTLLLFLVISHSCRAQ-NSQQDYLDAHNTARADVGV-EPLTWDDQVAAYAQNYA 67
Query: 83 -RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPA 141
+ D L + +YG E + M + + WV ++ YY+ +++
Sbjct: 68 SQLAADCNL----VHSHGQYG---ENLAEGSGDFMTAAKAVEMWVDEKQYYDHDSNTCSQ 120
Query: 142 NYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR + +GCA+ +C + GG ++ C Y PPGN GE P
Sbjct: 121 GQVCGHYTQVVWRNSVRVGCARVQC--NNGGYVVS-CNYDPPGNYRGESP 167
>UniRef100_Q04106 Prb-1b [Nicotiana tabacum]
Length = 179
Score = 78.2 bits (191), Expect = 1e-13
Identities = 53/166 (31%), Positives = 75/166 (44%), Gaps = 15/166 (9%)
Query: 26 AVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFR 85
A + A+L SSQA + D YL HN RR+ G+ + W ++LA A YA R
Sbjct: 9 ACFITFAILFHSSQAQNSPQD----YLNPHNAARRQVGVGP-MTWDNRLAAFAQNYANQR 63
Query: 86 RDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPC 145
G + G + L + ++ WV ++ +Y N++ A C
Sbjct: 64 A-------GDCRMQHSGGPYGENLAAAYPQLHAAGAVKMWVDEKQFYNYNSNTCAAGNVC 116
Query: 146 GDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
G YTQ+VWR + LGCA+ C G C Y PPGN G+RP
Sbjct: 117 GHYTQVVWRNSVRLGCARVRCN---NGWYFITCNYDPPGNWRGQRP 159
>UniRef100_Q6UXB8 HGSC289 [Homo sapiens]
Length = 463
Score = 78.2 bits (191), Expect = 1e-13
Identities = 59/178 (33%), Positives = 81/178 (45%), Gaps = 18/178 (10%)
Query: 24 FFAVLVALALLLVSSQA-TPATTDGAKVYLFE-HNFVRREKGISE----HLEWSDKLAKD 77
F +L+ L LLLV++ A TD K + E HN R + + H+ W ++LA
Sbjct: 7 FLMLLLPLLLLLVATTGPVGALTDEEKRLMVELHNLYRAQVSPTASDMLHMRWDEELAAF 66
Query: 78 ASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNS 137
A YAR C G K G T + MD M +W +R +Y + +
Sbjct: 67 AKAYAR------QCVWG--HNKERGRRGENLFAITDEGMDVPLAMEEWHHEREHYNLSAA 118
Query: 138 CVPANYPCGDYTQIVWRKTKYLGCAQAEC----GPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VW KT+ +GC C G + + L VC Y PPGNV G+RP
Sbjct: 119 TCSPGQMCGHYTQVVWAKTERIGCGSHFCEKLQGVEETNIELLVCNYEPPGNVKGKRP 176
>UniRef100_Q8TCB8 PI16 protein [Homo sapiens]
Length = 408
Score = 78.2 bits (191), Expect = 1e-13
Identities = 59/178 (33%), Positives = 81/178 (45%), Gaps = 18/178 (10%)
Query: 24 FFAVLVALALLLVSSQA-TPATTDGAKVYLFE-HNFVRREKGISE----HLEWSDKLAKD 77
F +L+ L LLLV++ A TD K + E HN R + + H+ W ++LA
Sbjct: 7 FLMLLLPLLLLLVATTGPVGALTDEEKRLMVELHNLYRAQVSPTASDMLHMRWDEELAAF 66
Query: 78 ASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNS 137
A YAR C G K G T + MD M +W +R +Y + +
Sbjct: 67 AKAYAR------QCVWG--HNKERGRRGENLFAITDEGMDVPLAMEEWHHEREHYNLSAA 118
Query: 138 CVPANYPCGDYTQIVWRKTKYLGCAQAEC----GPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VW KT+ +GC C G + + L VC Y PPGNV G+RP
Sbjct: 119 TCSPGQMCGHYTQVVWAKTERIGCGSHFCEKLQGVEETNIELLVCNYEPPGNVKGKRP 176
>UniRef100_P08299 Pathogenesis-related protein 1A precursor [Nicotiana tabacum]
Length = 168
Score = 78.2 bits (191), Expect = 1e-13
Identities = 55/170 (32%), Positives = 84/170 (49%), Gaps = 13/170 (7%)
Query: 23 PFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYA 82
P F ++ L L LV S + A + + YL HN R + G+ E L W D++A A YA
Sbjct: 10 PSFLLVSTLLLFLVISHSCRAQ-NSQQDYLDAHNTARADVGV-EPLTWDDQVAAYAQNYA 67
Query: 83 -RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPA 141
+ D L + +YG E + M + + WV ++ YY+ +++
Sbjct: 68 SQLAADCNL----VHSHGQYG---ENLAEGSGDFMTAAKAVEMWVDEKQYYDHDSNTCAQ 120
Query: 142 NYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR + +GCA+ +C + GG ++ C Y PPGN GE P
Sbjct: 121 GQVCGHYTQVVWRNSVRVGCARVQC--NNGGYVVS-CNYDPPGNYRGESP 167
>UniRef100_Q8H5R0 Putative pathogenesis-related protein [Oryza sativa]
Length = 176
Score = 77.8 bits (190), Expect = 1e-13
Identities = 53/172 (30%), Positives = 83/172 (47%), Gaps = 12/172 (6%)
Query: 25 FAVLVALALLLVSSQATPAT--TDGAKVYLFE-HNFVRREKGISEHLEWSDKLAKDASLY 81
FA+ VA+A +++++ T A +D K + HN R G+ + WS+ +A A +
Sbjct: 11 FALAVAIATIVMATTTTMAADLSDAEKAQFVKLHNDARAAVGVKAQVSWSEAVAAKAREH 70
Query: 82 ARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVP 140
A R ++ P + +G W + P + M WV ++ YY+ +N CV
Sbjct: 71 ASTCRTDHIQGP-YGENLWWG-----WSSTAGWVGKPADAMGSWVGEKPYYDRSSNKCVG 124
Query: 141 ANYPCGDYTQIVWRKTKYLGCAQAE-CGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VW +T +GCA+ C + L C Y P GN+ GERP
Sbjct: 125 GKV-CGHYTQVVWSRTTQIGCARVTGCNINGRSSTLIACNYNPRGNINGERP 175
>UniRef100_Q9LNJ2 F6F3.11 protein [Arabidopsis thaliana]
Length = 283
Score = 77.4 bits (189), Expect = 2e-13
Identities = 49/147 (33%), Positives = 72/147 (48%), Gaps = 16/147 (10%)
Query: 48 AKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRD--KYLCTPGIFDKKRYGVCC 105
++ +L HN VR G +W +LA A +A R + + + G + + +
Sbjct: 127 SREFLIAHNLVRARVG-EPPFQWDGRLAAYARTWANQRVGDCRLVHSNGPYGENIFWAGK 185
Query: 106 NQWLEDTVKIMDPREVMRQWV-KDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQA 164
N W PR+++ W +D+FY N+C P + CG YTQIVWR + +GCA
Sbjct: 186 NNW--------SPRDIVNVWADEDKFYDVKGNTCEP-QHMCGHYTQIVWRDSTKVGCASV 236
Query: 165 ECGPDYGGVRLNVCFYYPPGNVPGERP 191
+C GGV +C Y PPGN GE P
Sbjct: 237 DC--SNGGV-YAICVYNPPGNYEGENP 260
>UniRef100_Q6K9I7 Putative pathogenesis related protein-1 [Oryza sativa]
Length = 178
Score = 77.4 bits (189), Expect = 2e-13
Identities = 59/177 (33%), Positives = 83/177 (46%), Gaps = 14/177 (7%)
Query: 23 PFFAVLVALALLLVSSQATPATTDG-----AKVYLFEHNFVRREKGISEHLEWSDKLAKD 77
PF AV + LL ++ A +++ G A +L HN RR+ G+ L W ++LA
Sbjct: 7 PFAAVTAVILLLHGAADAKSSSSSGKTKSLASGFLDAHNAARRQVGVPP-LRWDERLASY 65
Query: 78 ASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWV-KDRFYYEG-N 135
A+ YA R + YG + P +V+ WV ++R Y+ +
Sbjct: 66 AARYAAARSGAGGGCALVHSHGPYG---ENLFHGSGVGWAPADVVAAWVSRERALYDAAS 122
Query: 136 NSCVPANYP-CGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
NSC + CG YTQ+VWR+T +GCA A C G VC Y PPGN G RP
Sbjct: 123 NSCRGGDAAACGHYTQVVWRRTTAVGCALATCAGGRG--TYGVCSYNPPGNYVGVRP 177
>UniRef100_Q6ID87 At1g01310 [Arabidopsis thaliana]
Length = 241
Score = 77.4 bits (189), Expect = 2e-13
Identities = 49/147 (33%), Positives = 72/147 (48%), Gaps = 16/147 (10%)
Query: 48 AKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRD--KYLCTPGIFDKKRYGVCC 105
++ +L HN VR G +W +LA A +A R + + + G + + +
Sbjct: 85 SREFLIAHNLVRARVG-EPPFQWDGRLAAYARTWANQRVGDCRLVHSNGPYGENIFWAGK 143
Query: 106 NQWLEDTVKIMDPREVMRQWV-KDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQA 164
N W PR+++ W +D+FY N+C P + CG YTQIVWR + +GCA
Sbjct: 144 NNW--------SPRDIVNVWADEDKFYDVKGNTCEP-QHMCGHYTQIVWRDSTKVGCASV 194
Query: 165 ECGPDYGGVRLNVCFYYPPGNVPGERP 191
+C GGV +C Y PPGN GE P
Sbjct: 195 DC--SNGGV-YAICVYNPPGNYEGENP 218
>UniRef100_Q6ZVG9 Hypothetical protein FLJ42598 [Homo sapiens]
Length = 270
Score = 77.4 bits (189), Expect = 2e-13
Identities = 59/178 (33%), Positives = 80/178 (44%), Gaps = 18/178 (10%)
Query: 24 FFAVLVALALLLVSSQA-TPATTDGAKVYLFE-HNFVRREKGISE----HLEWSDKLAKD 77
F +L+ L LLLV++ A TD K + E HN R + H+ W ++LA
Sbjct: 7 FLMLLLPLLLLLVATTGPVGALTDEEKRLMVELHNLYRAQVSPPASDMLHMRWDEELAAF 66
Query: 78 ASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNS 137
A YAR C G K G T + MD M +W +R +Y + +
Sbjct: 67 AKAYAR------QCVWG--HNKERGRRGENLFAITDEGMDVPLAMEEWHHEREHYNLSAA 118
Query: 138 CVPANYPCGDYTQIVWRKTKYLGCAQAEC----GPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VW KT+ +GC C G + + L VC Y PPGNV G+RP
Sbjct: 119 TCSPGQMCGHYTQVVWAKTERIGCGSHFCEKLQGVEETNIELLVCNYEPPGNVKGKRP 176
>UniRef100_Q8NBK0 Hypothetical protein PSEC0164 [Homo sapiens]
Length = 463
Score = 77.4 bits (189), Expect = 2e-13
Identities = 59/178 (33%), Positives = 80/178 (44%), Gaps = 18/178 (10%)
Query: 24 FFAVLVALALLLVSSQA-TPATTDGAKVYLFE-HNFVRREKGISE----HLEWSDKLAKD 77
F +L+ L LLLV++ A TD K + E HN R + H+ W ++LA
Sbjct: 7 FLMLLLPLLLLLVATTGPVGALTDEEKRLMVELHNLYRAQVSPPASDMLHMRWDEELAAF 66
Query: 78 ASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNS 137
A YAR C G K G T + MD M +W +R +Y + +
Sbjct: 67 AKAYAR------QCVWG--HNKERGRRGENLFAITDEGMDVPLAMEEWHHEREHYNLSAA 118
Query: 138 CVPANYPCGDYTQIVWRKTKYLGCAQAEC----GPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VW KT+ +GC C G + + L VC Y PPGNV G+RP
Sbjct: 119 TCSPGQMCGHYTQVVWAKTERIGCGSHFCEKLQGVEETNIELLVCNYEPPGNVKGKRP 176
>UniRef100_Q84J76 Putative pathogenesis-related protein [Oryza sativa]
Length = 179
Score = 76.3 bits (186), Expect = 4e-13
Identities = 52/168 (30%), Positives = 83/168 (48%), Gaps = 9/168 (5%)
Query: 26 AVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFR 85
A+ A+A++L + + + ++ HN RR +G+ E + W D +A A YA R
Sbjct: 10 ALAAAIAVVLAMATTPAVAQNSPQDFVDLHNAARRVEGVGE-VVWDDAVAAYAENYAAER 68
Query: 86 R-DKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPANY 143
D L G ++K YG + + + WV ++ Y+ +NSC+ +
Sbjct: 69 AGDCALIHSGSWEKAGYG---ENLFGGSGSEWTAADAVNSWVGEKDLYDYDSNSCLGSWD 125
Query: 144 PCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
C YTQ++W +T +GCA+ +C D GGV + C Y P GN GERP
Sbjct: 126 SCLHYTQVMWSRTTAIGCARVDC--DNGGVFI-TCNYNPAGNFQGERP 170
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.142 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 369,453,932
Number of Sequences: 2790947
Number of extensions: 15085537
Number of successful extensions: 32534
Number of sequences better than 10.0: 443
Number of HSP's better than 10.0 without gapping: 235
Number of HSP's successfully gapped in prelim test: 208
Number of HSP's that attempted gapping in prelim test: 32003
Number of HSP's gapped (non-prelim): 483
length of query: 191
length of database: 848,049,833
effective HSP length: 120
effective length of query: 71
effective length of database: 513,136,193
effective search space: 36432669703
effective search space used: 36432669703
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0323a.9


