

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0322.4
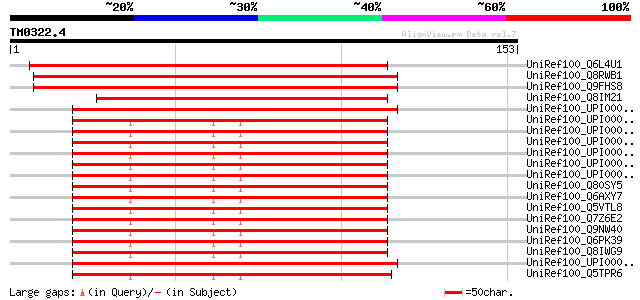
(153 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6L4U1 Hypothetical protein P0478F09.2 [Oryza sativa] 204 4e-52
UniRef100_Q8RWB1 Hypothetical protein At5g37370 [Arabidopsis tha... 187 4e-47
UniRef100_Q9FHS8 Similarity to unknown protein [Arabidopsis thal... 187 4e-47
UniRef100_Q8IM21 Hypothetical protein [Plasmodium falciparum] 110 8e-24
UniRef100_UPI0000465867 UPI0000465867 UniRef100 entry 108 2e-23
UniRef100_UPI00001CEF5E UPI00001CEF5E UniRef100 entry 105 2e-22
UniRef100_UPI00003606AF UPI00003606AF UniRef100 entry 105 2e-22
UniRef100_UPI000036821A UPI000036821A UniRef100 entry 105 2e-22
UniRef100_UPI00003AE886 UPI00003AE886 UniRef100 entry 105 2e-22
UniRef100_UPI00003AE885 UPI00003AE885 UniRef100 entry 105 2e-22
UniRef100_UPI000029C19D UPI000029C19D UniRef100 entry 105 2e-22
UniRef100_Q80SY5 RIKEN cDNA 1110021E09 [Mus musculus] 105 2e-22
UniRef100_Q6AXY7 Hypothetical protein [Rattus norvegicus] 105 2e-22
UniRef100_Q5VTL8 Novel protein [Homo sapiens] 105 2e-22
UniRef100_Q7Z6E2 Hypothetical protein [Homo sapiens] 105 2e-22
UniRef100_Q9NW40 Hypothetical protein FLJ10330 [Homo sapiens] 105 2e-22
UniRef100_Q6PK39 Hypothetical protein [Homo sapiens] 105 2e-22
UniRef100_Q8IWG9 Hypothetical protein [Homo sapiens] 105 2e-22
UniRef100_UPI000046BC38 UPI000046BC38 UniRef100 entry 103 6e-22
UniRef100_Q5TPR6 ENSANGP00000028866 [Anopheles gambiae str. PEST] 103 8e-22
>UniRef100_Q6L4U1 Hypothetical protein P0478F09.2 [Oryza sativa]
Length = 434
Score = 204 bits (518), Expect = 4e-52
Identities = 95/108 (87%), Positives = 100/108 (91%)
Query: 7 MEIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCR 66
MEIQTSG+PI+ L+EKVLCMNI+SSDYFKELYRLKTY EVIDEIYNQVDHVEPWM GNCR
Sbjct: 1 MEIQTSGKPIDLLMEKVLCMNIMSSDYFKELYRLKTYHEVIDEIYNQVDHVEPWMTGNCR 60
Query: 67 GPSTFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
GPST FCLLY FFTM LTVKQMHGLLKHPDSPYIR +GFLYLRYV DP
Sbjct: 61 GPSTAFCLLYKFFTMKLTVKQMHGLLKHPDSPYIRAIGFLYLRYVADP 108
>UniRef100_Q8RWB1 Hypothetical protein At5g37370 [Arabidopsis thaliana]
Length = 393
Score = 187 bits (475), Expect = 4e-47
Identities = 91/110 (82%), Positives = 96/110 (86%)
Query: 8 EIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRG 67
EIQ++GR ESLLEKVL MNILSSDYFKELY LKTY EVIDEIYNQV+HVEPWM GNCRG
Sbjct: 3 EIQSNGRAYESLLEKVLSMNILSSDYFKELYGLKTYHEVIDEIYNQVNHVEPWMGGNCRG 62
Query: 68 PSTFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDPNTV 117
PST +CLLY FFTM LTVKQMHGLLKH DSPYIR VGFLYLRYV D T+
Sbjct: 63 PSTAYCLLYKFFTMKLTVKQMHGLLKHTDSPYIRAVGFLYLRYVADAKTL 112
>UniRef100_Q9FHS8 Similarity to unknown protein [Arabidopsis thaliana]
Length = 385
Score = 187 bits (475), Expect = 4e-47
Identities = 91/110 (82%), Positives = 96/110 (86%)
Query: 8 EIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRG 67
EIQ++GR ESLLEKVL MNILSSDYFKELY LKTY EVIDEIYNQV+HVEPWM GNCRG
Sbjct: 3 EIQSNGRAYESLLEKVLSMNILSSDYFKELYGLKTYHEVIDEIYNQVNHVEPWMGGNCRG 62
Query: 68 PSTFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDPNTV 117
PST +CLLY FFTM LTVKQMHGLLKH DSPYIR VGFLYLRYV D T+
Sbjct: 63 PSTAYCLLYKFFTMKLTVKQMHGLLKHTDSPYIRAVGFLYLRYVADAKTL 112
>UniRef100_Q8IM21 Hypothetical protein [Plasmodium falciparum]
Length = 690
Score = 110 bits (274), Expect = 8e-24
Identities = 52/88 (59%), Positives = 65/88 (73%)
Query: 27 NILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRGPSTFFCLLY*FFTMMLTVK 86
NILSS+YFK L +KT+ EV+DEI++ DHVEP+ G+ R PST FC LY FFTM L+ K
Sbjct: 190 NILSSEYFKSLIPIKTFKEVVDEIHSYADHVEPYCIGSNRAPSTLFCCLYKFFTMQLSEK 249
Query: 87 QMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
Q+ L+++ DS YIR GFLYLRYV P
Sbjct: 250 QLKSLIENKDSCYIRACGFLYLRYVHSP 277
>UniRef100_UPI0000465867 UPI0000465867 UniRef100 entry
Length = 635
Score = 108 bits (270), Expect = 2e-23
Identities = 52/98 (53%), Positives = 68/98 (69%)
Query: 20 LEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRGPSTFFCLLY*FF 79
+ +L NILSS+YF+ L LKT+ EV+DEI + DHVEP+ G+ R PST FC LY FF
Sbjct: 167 VNNLLRNNILSSEYFRSLINLKTFKEVLDEILSYADHVEPYCIGSTRAPSTLFCCLYKFF 226
Query: 80 TMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDPNTV 117
TM LT KQ+ L+ + +S Y+R GFLYLRYV P+ +
Sbjct: 227 TMQLTKKQLKSLIDNKESCYVRACGFLYLRYVHCPSNL 264
>UniRef100_UPI00001CEF5E UPI00001CEF5E UniRef100 entry
Length = 1214
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 58 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 117
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 118 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
>UniRef100_UPI00003606AF UPI00003606AF UniRef100 entry
Length = 454
Score = 105 bits (263), Expect = 2e-22
Identities = 58/114 (50%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ N+LSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 30 LNPMILTNVLSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 89
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 90 GVGTGGIVSTAFCLLYKLFTLKLTRKQLMGLITHTDSPYIRALGFMYIRYTQPP 143
>UniRef100_UPI000036821A UPI000036821A UniRef100 entry
Length = 546
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 58 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 117
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 118 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
>UniRef100_UPI00003AE886 UPI00003AE886 UniRef100 entry
Length = 546
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 61 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 120
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 121 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 174
>UniRef100_UPI00003AE885 UPI00003AE885 UniRef100 entry
Length = 488
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 61 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 120
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 121 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 174
>UniRef100_UPI000029C19D UPI000029C19D UniRef100 entry
Length = 464
Score = 105 bits (263), Expect = 2e-22
Identities = 58/114 (50%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ N+LSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 33 LNPMILTNVLSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 92
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 93 GVGTGGIVSTAFCLLYKLFTLKLTRKQLMGLITHTDSPYIRALGFMYIRYTQPP 146
>UniRef100_Q80SY5 RIKEN cDNA 1110021E09 [Mus musculus]
Length = 542
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 59 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 118
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 119 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 172
>UniRef100_Q6AXY7 Hypothetical protein [Rattus norvegicus]
Length = 542
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 58 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 117
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 118 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
>UniRef100_Q5VTL8 Novel protein [Homo sapiens]
Length = 546
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 58 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 117
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 118 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
>UniRef100_Q7Z6E2 Hypothetical protein [Homo sapiens]
Length = 190
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 58 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 117
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 118 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
>UniRef100_Q9NW40 Hypothetical protein FLJ10330 [Homo sapiens]
Length = 546
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 58 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 117
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 118 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
>UniRef100_Q6PK39 Hypothetical protein [Homo sapiens]
Length = 465
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 58 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 117
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 118 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
>UniRef100_Q8IWG9 Hypothetical protein [Homo sapiens]
Length = 303
Score = 105 bits (263), Expect = 2e-22
Identities = 59/114 (51%), Positives = 70/114 (60%), Gaps = 19/114 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NILSS YFK +LY LKTY EV+DEIY +V HVEPW M G R
Sbjct: 58 LNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCGGVR 117
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDP 114
G ST FCLLY FT+ LT KQ+ GL+ H DSPYIR +GF+Y+RY P
Sbjct: 118 GVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
>UniRef100_UPI000046BC38 UPI000046BC38 UniRef100 entry
Length = 654
Score = 103 bits (258), Expect = 6e-22
Identities = 49/98 (50%), Positives = 67/98 (68%)
Query: 20 LEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRGPSTFFCLLY*FF 79
+ +L NILSS+YF+ L LKT+ EV+DEI + DH EP+ G+ R PST FC LY F
Sbjct: 189 VNNLLRNNILSSEYFRSLITLKTFKEVLDEILSYADHAEPYCIGSTRAPSTLFCCLYKLF 248
Query: 80 TMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDPNTV 117
TM L+ KQ+ L+++ +S Y+R GFLYLRYV P+ +
Sbjct: 249 TMHLSKKQLKSLIENKESCYVRACGFLYLRYVHSPSNL 286
>UniRef100_Q5TPR6 ENSANGP00000028866 [Anopheles gambiae str. PEST]
Length = 336
Score = 103 bits (257), Expect = 8e-22
Identities = 57/115 (49%), Positives = 70/115 (60%), Gaps = 19/115 (16%)
Query: 20 LEKVLCMNILSSDYFK-ELYRLKTYLEVIDEIYNQVDHVEPW------------MAGNCR 66
L ++ NI S YFK L++LKTY EV+DEIY QV H+EPW M G R
Sbjct: 57 LNPLILANIQGSSYFKVSLFKLKTYHEVVDEIYYQVKHLEPWERGSRKTAGQTGMCGGVR 116
Query: 67 GP------STFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDPN 115
G ST FCLLY +T+ LT KQ++GLL H DSPYIR +GF+YLRY P+
Sbjct: 117 GVGAGGIVSTAFCLLYKLYTLRLTRKQVNGLLSHGDSPYIRALGFMYLRYTQPPS 171
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.352 0.162 0.599
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 239,256,601
Number of Sequences: 2790947
Number of extensions: 8695205
Number of successful extensions: 34918
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 34818
Number of HSP's gapped (non-prelim): 63
length of query: 153
length of database: 848,049,833
effective HSP length: 129
effective length of query: 24
effective length of database: 488,017,670
effective search space: 11712424080
effective search space used: 11712424080
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0322.4


