

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0322.12
(601 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
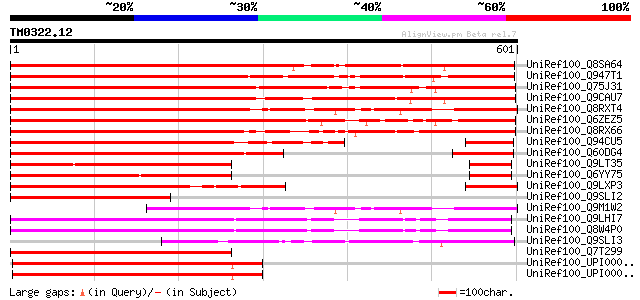
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8SA64 NIMA-related protein kinase [Populus alba x Pop... 759 0.0
UniRef100_Q947T1 LSTK-1-like kinase [Lycopersicon esculentum] 669 0.0
UniRef100_Q75J31 Putative LSTK-1-like kinase [Oryza sativa] 648 0.0
UniRef100_Q9CAU7 Putative kinase; 86849-83844 [Arabidopsis thali... 634 e-180
UniRef100_Q8RXT4 Hypothetical protein At3g63280 [Arabidopsis tha... 626 e-178
UniRef100_Q6ZEZ5 Putative NIMA-related protein kinase [Oryza sat... 608 e-172
UniRef100_Q8RX66 AT5g28290/T8M17_60 [Arabidopsis thaliana] 607 e-172
UniRef100_Q94CU5 Putative kinase [Oryza sativa] 405 e-111
UniRef100_Q60DG4 Hypothetical protein B1110B01.5 [Oryza sativa] 404 e-111
UniRef100_Q9LT35 Kinase-like protein [Arabidopsis thaliana] 384 e-105
UniRef100_Q6YY75 Serine/threonine-protein kinase Nek4-like [Oryz... 369 e-100
UniRef100_Q9LXP3 Protein kinase-like protein [Arabidopsis thaliana] 366 e-100
UniRef100_Q9SLI2 F20D21.33 protein [Arabidopsis thaliana] 346 1e-93
UniRef100_Q9M1W2 Hypothetical protein F16M2_130 [Arabidopsis tha... 339 1e-91
UniRef100_Q9LHI7 Similarity to protein kinase [Arabidopsis thali... 324 5e-87
UniRef100_Q8W4P0 Protein kinase, putative [Arabidopsis thaliana] 322 2e-86
UniRef100_Q9SLI3 F20D21.32 protein [Arabidopsis thaliana] 268 3e-70
UniRef100_Q7T299 Similar to NIMA (Never in mitosis gene a)-relat... 253 9e-66
UniRef100_UPI00003AA783 UPI00003AA783 UniRef100 entry 251 6e-65
UniRef100_UPI00003AA782 UPI00003AA782 UniRef100 entry 251 6e-65
>UniRef100_Q8SA64 NIMA-related protein kinase [Populus alba x Populus tremula]
Length = 621
Score = 759 bits (1959), Expect = 0.0
Identities = 409/626 (65%), Positives = 475/626 (75%), Gaps = 41/626 (6%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYEVLEQIGKGSFGSALLV+HKHEKK YVLKKIRLARQ+DR+RRSAHQE ELIS++RN
Sbjct: 1 MEQYEVLEQIGKGSFGSALLVKHKHEKKKYVLKKIRLARQTDRSRRSAHQEKELISRIRN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEKGC+VCII+GYCE GD+A+AIKKANGV+FPEEKLCKWLVQLLMALDYL
Sbjct: 61 PFIVEYKDSWVEKGCYVCIIIGYCEGGDMAEAIKKANGVHFPEEKLCKWLVQLLMALDYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H+NHILHRDVKCSNIFLTK QDIRLGDFGLAK+LTSDDLASS+VGTPSYMCPELLADIPY
Sbjct: 121 HMNHILHRDVKCSNIFLTKGQDIRLGDFGLAKILTSDDLASSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCCIYEM +LKPAFKAFD+QALINKINKSIVAPLPT YS AFRGLVKSMLR
Sbjct: 181 GSKSDIWSLGCCIYEMTSLKPAFKAFDMQALINKINKSIVAPLPTKYSGAFRGLVKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
KNPELRP A+ELL HPHLQPY+LKIH+K+NSPR++T PFQWPE +YM++T+F+VP+
Sbjct: 241 KNPELRPSAAELLRHPHLQPYVLKIHIKMNSPRQNTLPFQWPEPHYMKKTKFLVPEDNPL 300
Query: 301 LSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAH--------GLSTSSKE-VYEVSVGG 351
+ ++K SLSNDR+LNPSIS EQDS C T+ H LS SS E +E +V
Sbjct: 301 KAHREKRYSLSNDRALNPSISAAEQDSVCSTEGIHDTPSYLNQSLSDSSIESSHEGTV-- 358
Query: 352 ACGPWNTNKTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKF 411
C + +K +T + K S RR+T P K K ++LPV+ P+ K
Sbjct: 359 IC---RSIASKPSNFAKTTKPAPTKASVTTRRRT-EPVK------KRESLPVTRTPTKKS 408
Query: 412 PPPTRRASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAEFPLASCEDLPF 470
P TRR S PLP+R ++ + TN ++ S DVSVNAP+ID+IAEFPLAS D P+
Sbjct: 409 NPTTRRTSLPLPSRTAIQNSAHGTNNSILLQIKSPDVSVNAPRIDRIAEFPLAS-YDKPY 467
Query: 471 FPVHVPSSTSVHCYSSSAGSAYRSITKEKCIHE--DKVTVPGGSC--------------- 513
P SSTS S+S RSITK+KC + D+ +
Sbjct: 468 LPFRRTSSTSAQGSSNSLHHGDRSITKDKCTVQISDRTSAKPNFTEAWQGIEHGMFQVDE 527
Query: 514 PKGSECS-KHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELG 572
GS S ++ TAG S +S+++ +RRFDTSS+QQRAEALEGLLEFSARLLQ R+DELG
Sbjct: 528 ENGSNSSNQNATAGASSRTSSDIRRRRFDTSSFQQRAEALEGLLEFSARLLQDARYDELG 587
Query: 573 VLLKPFGLEKVSPRETAIWLAKSFKE 598
VLLKPFG KVSPRETAIWL KSFKE
Sbjct: 588 VLLKPFGPGKVSPRETAIWLTKSFKE 613
>UniRef100_Q947T1 LSTK-1-like kinase [Lycopersicon esculentum]
Length = 609
Score = 669 bits (1727), Expect = 0.0
Identities = 374/627 (59%), Positives = 443/627 (70%), Gaps = 54/627 (8%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYE+LEQIGKG+FGSA+LV+HK EKK YVLKKIRLARQ+DRTRR+AHQEM LIS ++N
Sbjct: 1 MEQYEILEQIGKGAFGSAVLVKHKLEKKKYVLKKIRLARQTDRTRRNAHQEMALISSMQN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEKGC+VCI++GYCE GD+A+AIKKA GV+F EEKLCKWLVQLLMALDYL
Sbjct: 61 PFIVEYKDSWVEKGCYVCIVIGYCEGGDMAEAIKKAKGVHFLEEKLCKWLVQLLMALDYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H NHILHRDVKCSNIFLT+ QDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY
Sbjct: 121 HTNHILHRDVKCSNIFLTREQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCCIYEMAA KPAFKAFD+QALINKINKSIVAPLPT YS FRGLVKSMLR
Sbjct: 181 GSKSDIWSLGCCIYEMAAFKPAFKAFDMQALINKINKSIVAPLPTKYSGPFRGLVKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
KNPELRP A+ELL +P LQPY++ HLKLN PRR++ P P+ N +++TRF + +S
Sbjct: 241 KNPELRPSAAELLRNPLLQPYVINTHLKLNGPRRNSLPACLPD-NDVKKTRFAISESTPV 299
Query: 301 LSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGA-CGPWNTN 359
+++K +S NDR+LNPS+S + +T + V E+SVG G T
Sbjct: 300 KKNREKRMSCGNDRTLNPSVSDHD-----YTFSNRRYPKTPSRVSELSVGSPDRGSTVTK 354
Query: 360 K--TKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKFPPPTRR 417
K +K+ V + P+ K +T RQ + S TL V + S TRR
Sbjct: 355 KITSKALLVNKNPQVIVPKLTTTPARQV----DLRNSDMASRTL-VKRSVST-----TRR 404
Query: 418 ASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAEFPLASCEDLPFFPVHVP 476
AS PL +A++ + ++ + S DVSVNAP+IDK+ EFPLAS ED PF P+
Sbjct: 405 ASLPLTNKAAVQELPRRPSLSFLDCIKSPDVSVNAPRIDKMLEFPLASYED-PFHPIRRT 463
Query: 477 SSTSVHCYSSSAGSAYRSITKEKC-------------------------IHEDKVTVPGG 511
SS S S S + Y S+ K+KC +H D+ +
Sbjct: 464 SSNSAQGSSGSPQAEY-SVMKDKCTIQIPDSKFDRMSSNDAWQGYEGPMVHVDREDITDS 522
Query: 512 SCPKGSECSKHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDEL 571
S ++ TAG S +S++ +RRF+ SSY+QRAEALEGLLEFSARLLQ RFDEL
Sbjct: 523 S-------DQNATAGASSRTSSDTRRRRFNMSSYKQRAEALEGLLEFSARLLQEDRFDEL 575
Query: 572 GVLLKPFGLEKVSPRETAIWLAKSFKE 598
GVLLKPFG EKVSPRETAIWL KS KE
Sbjct: 576 GVLLKPFGPEKVSPRETAIWLTKSIKE 602
>UniRef100_Q75J31 Putative LSTK-1-like kinase [Oryza sativa]
Length = 589
Score = 648 bits (1671), Expect = 0.0
Identities = 363/611 (59%), Positives = 433/611 (70%), Gaps = 36/611 (5%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYEVLEQIGKG+FGSALLVRHK EKK YVLKKIRLARQ+DRTRRSAHQEM+LI+ VRN
Sbjct: 1 MEQYEVLEQIGKGAFGSALLVRHKVEKKKYVLKKIRLARQTDRTRRSAHQEMQLIATVRN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEKGC+VCII+GYCE GD+A+AIK+A G +F EEKLCKWLVQLLMALDYL
Sbjct: 61 PFIVEYKDSWVEKGCYVCIIIGYCEGGDMAEAIKRATGDHFSEEKLCKWLVQLLMALDYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H NHILHRDVKCSNIFLT++Q IRLGDFGLAK+LTSDDLASS+VGTPSYMCPELLADIPY
Sbjct: 121 HANHILHRDVKCSNIFLTRDQSIRLGDFGLAKILTSDDLASSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G+KSDIWSLGCCIYEM AL+PAFKAFD+QALI+KI KSIV+PLPT YS AFRGL+KSMLR
Sbjct: 181 GTKSDIWSLGCCIYEMTALRPAFKAFDMQALISKITKSIVSPLPTRYSGAFRGLIKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
K+PE RP A+ELL HPHLQPY+L++HLK +SP R+ P + +++ F + S
Sbjct: 241 KSPEHRPSAAELLKHPHLQPYVLQVHLK-SSPARNIIPSHQSPIDKVKKMTF---PTESM 296
Query: 301 LSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLST--SSKEV-YEVSVGGACGPWN 357
+ + SL N+R + S E+ Q ST S K++ +VS+
Sbjct: 297 CRSKGRRNSLGNERIVTFSKPSPERKFTSSIQSIKDYSTTRSVKDLSIDVSLVEEVSSKT 356
Query: 358 TNKTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKFPPPTRR 417
T T++ ++ +TP+ K T Q P +SR+ PV+ + V RR
Sbjct: 357 TFTTRTSSIVKTPKRTPSK--TITTPQLEPPKLLSRT-------PVNRSARV-----IRR 402
Query: 418 ASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAEFPLASCEDLPFFPVH-- 474
AS PLP +S +++ +++S DVSVN+P+ID+IAEFPLAS ED PF +H
Sbjct: 403 ASLPLPLPSSETPKRGVSSISILEQLESPDVSVNSPRIDRIAEFPLASSEDPPFLKLHGR 462
Query: 475 -VPSSTSVHCYSSSAGSAYRSITKEKCIHE-----DKVTVPGGSCPKGSECSKHVTAGVS 528
P+ T HC +SITK+KC+ E D G S + A
Sbjct: 463 RSPTPTPQHCVID------QSITKDKCMVEAFHIIDVDDDDGRSDSSSGRNNAAAAASSR 516
Query: 529 IHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRET 588
SS QRRFDTSSYQQRAEALEGLLEFSA+LLQ +R+DELGVLLKPFG EKVSPRET
Sbjct: 517 AGSSESTRQRRFDTSSYQQRAEALEGLLEFSAQLLQQERYDELGVLLKPFGPEKVSPRET 576
Query: 589 AIWLAKSFKET 599
AIWL KSFKET
Sbjct: 577 AIWLTKSFKET 587
>UniRef100_Q9CAU7 Putative kinase; 86849-83844 [Arabidopsis thaliana]
Length = 606
Score = 634 bits (1634), Expect = e-180
Identities = 360/619 (58%), Positives = 423/619 (68%), Gaps = 40/619 (6%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQ+ RTRRSAHQEMELISK+ N
Sbjct: 1 MENYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQTGRTRRSAHQEMELISKIHN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEKGC+VCII+GYC+ GD+A+AIKK NGV+F EEKLCKWLVQ+L+AL+YL
Sbjct: 61 PFIVEYKDSWVEKGCYVCIIIGYCKGGDMAEAIKKTNGVHFTEEKLCKWLVQILLALEYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H NHILHRDVKCSNIFLTK+QDIRLGDFGLAK+LTSDDLASS+VGTPSYMCPELLADIPY
Sbjct: 121 HANHILHRDVKCSNIFLTKDQDIRLGDFGLAKVLTSDDLASSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCC+YEM A+KPAFKAFD+Q LIN+IN+SIV PLP YS+AFRGLVKSMLR
Sbjct: 181 GSKSDIWSLGCCMYEMTAMKPAFKAFDMQGLINRINRSIVPPLPAQYSAAFRGLVKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
KNPELRP A+ELL P LQPYI KIHLK+N P + P QWPES RR F
Sbjct: 241 KNPELRPSAAELLRQPLLQPYIQKIHLKVNDPGSNVLPAQWPESESARRNSF-------- 292
Query: 301 LSDQDKCLSLSNDRSLNPS-ISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNTN 359
+ + S PS G +DS +K + + ++S T+
Sbjct: 293 --PEQRRRPAGKSHSFGPSRFRGNLEDSVSSIKKTVPAYLNRERQVDLS---------TD 341
Query: 360 KTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKFPPP-TRRA 418
+ TV R + R + P + + S LPVS + P RRA
Sbjct: 342 ASGDGTVVRRTSEASKSSRYVPVRASASPVRPRQPRSDLGQLPVSSQLKNRKPAALIRRA 401
Query: 419 SHPLPTR-ASMAKATLYTNVGTF-PRVDS-DVSVNAPQIDKIAEFPLASCEDLPFFPV-- 473
S P + A K +LY + +F +++S DVS+NAP+IDKI EFPLAS E+ PF PV
Sbjct: 402 SMPSSRKPAKEIKDSLYISKTSFLHQINSPDVSMNAPRIDKI-EFPLASYEEEPFVPVVR 460
Query: 474 -HVPSSTSVHCYSSSAGSAYR-SITKEKCIHEDKVTVPGGSC-----------PKGSECS 520
++S YS SITK+K E G +
Sbjct: 461 GKKKKASSRGSYSPPPEPPLDCSITKDKFTLEPGQNREGAIMKAVYEEDAYLEDRSESSD 520
Query: 521 KHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGL 580
++ TAG S +S+ + ++RFD SSYQQRAEALEGLLEFSARLLQ +R+DEL VLL+PFG
Sbjct: 521 QNATAGASSRASSGVRRQRFDPSSYQQRAEALEGLLEFSARLLQDERYDELNVLLRPFGP 580
Query: 581 EKVSPRETAIWLAKSFKET 599
KVSPRETAIWL+KSFKET
Sbjct: 581 GKVSPRETAIWLSKSFKET 599
>UniRef100_Q8RXT4 Hypothetical protein At3g63280 [Arabidopsis thaliana]
Length = 555
Score = 626 bits (1615), Expect = e-178
Identities = 354/612 (57%), Positives = 416/612 (67%), Gaps = 68/612 (11%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+YEVLEQIGKGSFGSALLVRHK E+K YVLKKIRLARQSDR RRSAHQEMELIS VRN
Sbjct: 1 MERYEVLEQIGKGSFGSALLVRHKQERKKYVLKKIRLARQSDRARRSAHQEMELISTVRN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PF+VEYKDSWVEKGC+VCI++GYC+ GD+ D IK+A GV+FPEEKLC+WLVQLLMALDYL
Sbjct: 61 PFVVEYKDSWVEKGCYVCIVIGYCQGGDMTDTIKRACGVHFPEEKLCQWLVQLLMALDYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H NHILHRDVKCSNIFLTK QDIRLGDFGLAK+LTSDDL SS+VGTPSYMCPELLADIPY
Sbjct: 121 HSNHILHRDVKCSNIFLTKEQDIRLGDFGLAKILTSDDLTSSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCC+YEMAA KP FKA D+Q LI KI+K I+ P+P MYS +FRGL+KSMLR
Sbjct: 181 GSKSDIWSLGCCMYEMAAHKPPFKASDVQTLITKIHKLIMDPIPAMYSGSFRGLIKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
KNPELRP A+ELLNHPHLQPYI +++KL SPRRSTFP Q+ E + +T
Sbjct: 241 KNPELRPSANELLNHPHLQPYISMVYMKLESPRRSTFPLQFSERD-------------AT 287
Query: 301 LSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLST-SSKEVYEVSVGGACGPWNTN 359
L ++ + S SNDR LNPS+S TE S + KA + ++V EV+VG
Sbjct: 288 LKERRRS-SFSNDRRLNPSVSDTEAGSVSSSGKASPTPMFNGRKVSEVTVG--------- 337
Query: 360 KTKSPTVERTPRSRADKDSTAARRQ----TMGPSKISRSGSKHDTLPVSHAPSVKFPPPT 415
+ + A K S AAR T + R+ KH+ + VS+ +
Sbjct: 338 -VVREEIVPQRQEEAKKQSGAARTPRVAGTSAKASTQRTVFKHELMKVSNPTERR----- 391
Query: 416 RRASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAEFP-----LASCEDLP 469
RR S PL T +++ ++S DVSVN P+ DKIAEFP + E
Sbjct: 392 RRVSLPLVVENPY---TYESDITALCSLNSPDVSVNTPRFDKIAEFPEDIFQNQNRETAS 448
Query: 470 FFPVHVPSSTSVHCYSSSAGSAYRSITKEKCIHEDKVTVPGGSCPKGSECSKHVTAGVSI 529
V S +S C ++ SITK+KC V
Sbjct: 449 RREVARHSFSSPPCPPHGEDNSNGSITKDKCT-------------------------VQK 483
Query: 530 HSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETA 589
S +E+ QRRFDTSSYQQRAEALEGLLEFSA+LLQ +R+DELGVLLKPFG E+VS RETA
Sbjct: 484 RSVSEVKQRRFDTSSYQQRAEALEGLLEFSAKLLQQERYDELGVLLKPFGAERVSSRETA 543
Query: 590 IWLAKSFKETVI 601
IWL KSFKE +
Sbjct: 544 IWLTKSFKEASV 555
>UniRef100_Q6ZEZ5 Putative NIMA-related protein kinase [Oryza sativa]
Length = 585
Score = 608 bits (1568), Expect = e-172
Identities = 351/618 (56%), Positives = 425/618 (67%), Gaps = 54/618 (8%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYEVLEQIGKGSFGSALLVRHK EKK YVLKKIRLARQ+DR RRSAHQEMELI+KVRN
Sbjct: 1 MEQYEVLEQIGKGSFGSALLVRHKVEKKRYVLKKIRLARQTDRCRRSAHQEMELIAKVRN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVEYKDSWVEKGC+VCI++GYCE GD+++AIKKAN F EE+LC WLVQLLMALDYL
Sbjct: 61 PYIVEYKDSWVEKGCYVCIVIGYCEGGDMSEAIKKANSNYFSEERLCMWLVQLLMALDYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
HVNHILHRDVKCSNIFLTK+Q+IRLGDFGLAK+LTSDDL SS+VGTPSYMCPELLADIPY
Sbjct: 121 HVNHILHRDVKCSNIFLTKDQNIRLGDFGLAKVLTSDDLTSSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCC+YEM ALKPAFKAFD+Q LINKI+KS++APLPT+YS AFRGL+KSMLR
Sbjct: 181 GSKSDIWSLGCCLYEMTALKPAFKAFDMQTLINKISKSVLAPLPTIYSGAFRGLIKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
K+P+ RP A+ELL HPHLQP++L++ LK +SP R+ FP S + S S
Sbjct: 241 KSPDHRPSAAELLKHPHLQPFVLELQLK-SSPARNLFPDTNKASCSDDENNWKAKYSKSH 299
Query: 301 LSDQDKCLSLSNDRSLN--PSISGTEQDSQCFTQKAHG--LSTSSKEVYEVSVGGACGPW 356
D+ + + + N PS +GT +D Q ++ L +++V + + G
Sbjct: 300 SFKVDRIVKVDKVAANNGHPSSTGTAKDYQELLKQPMDELLGQLTEKVVDEVIHG--NHS 357
Query: 357 NTNKTKSPTVER---TPRSRADKDST-AARRQTMGPSKISRSGSKHDTLPVSHAPSVKFP 412
K+ +PT R TPR R + T AR PSK S +
Sbjct: 358 RVTKSPAPTPRRASSTPRIRLEPSKTFHARAAETPPSKCSLERASQ-------------- 403
Query: 413 PPTRRASHP---LPTRASMAKATLYTNVGTFPRVDSDVSVNAPQIDKIAEFPLASCEDLP 469
PTRRAS P L T A + T + + DVSVN+P+ID+IAEFP+ S +D
Sbjct: 404 -PTRRASTPVNMLQTPEKRQGADILTRLKS-----PDVSVNSPRIDRIAEFPIPSFDDEQ 457
Query: 470 FFPVHVPSSTSVHCYSSSAGSAYRSITKEKCIHE------DKVTVPGGSCPK--GSECSK 521
P +T + Y S +SITK+KC + K S P G++ +
Sbjct: 458 LHP-----TTKLKLYPPSITD--QSITKDKCTFQVLRSDSSKNHTGDSSDPSILGTDSNP 510
Query: 522 HVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLE 581
+T+ SS + QRRFDT+SY+QRAEALEGLLEFSA+LLQ +RF+ELG+LLKPFG
Sbjct: 511 LITS-----SSDWMKQRRFDTTSYRQRAEALEGLLEFSAQLLQQERFEELGILLKPFGPG 565
Query: 582 KVSPRETAIWLAKSFKET 599
K SPRETAIWL+KSFK T
Sbjct: 566 KASPRETAIWLSKSFKGT 583
>UniRef100_Q8RX66 AT5g28290/T8M17_60 [Arabidopsis thaliana]
Length = 568
Score = 607 bits (1566), Expect = e-172
Identities = 354/608 (58%), Positives = 420/608 (68%), Gaps = 57/608 (9%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQ+ RTRRSAHQEMELISK+RN
Sbjct: 1 MEHYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQTGRTRRSAHQEMELISKIRN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEKGC+VCI++GYC+ GD+A+AIKKANGV F EEKLCKWLVQLLMAL+YL
Sbjct: 61 PFIVEYKDSWVEKGCYVCIVIGYCKGGDMAEAIKKANGVEFSEEKLCKWLVQLLMALEYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H +HILHRDVKCSNIFLTK+QDIRLGDFGLAK+LTSDDLASS+VGTPSYMCPELLADIPY
Sbjct: 121 HASHILHRDVKCSNIFLTKDQDIRLGDFGLAKILTSDDLASSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCC+YEM ALKPAFKAFD+Q LIN+IN+SIVAPLP YS+AFR LVKSMLR
Sbjct: 181 GSKSDIWSLGCCMYEMTALKPAFKAFDMQGLINRINRSIVAPLPAQYSTAFRSLVKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
KNPELRP AS+LL P LQPY+ K+ LKL+ T P S RR+ +
Sbjct: 241 KNPELRPSASDLLRQPLLQPYVQKVLLKLSFREHDTLP-----SESERRSSY-------- 287
Query: 301 LSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNTNK 360
Q + + S PS G +Q+ + K P +T
Sbjct: 288 --PQQRKRTSGKSVSFGPSRFGVDQEDSVSSVK---------------------PVHTYL 324
Query: 361 TKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPV-SHAPS----VKFPPPT 415
+ V+ S D RR + S + + SK+ +PV S+ P +K T
Sbjct: 325 HRHRPVD---LSANDTSRVVVRRPAV--SSVVSNSSKY--VPVRSNQPKSGGLLKPAVVT 377
Query: 416 RRASHPLPTR-ASMAKATLY-TNVGTFPRVDS-DVSVNAPQIDKIAEFPLASCEDLPFFP 472
RRAS P+ + A K +LY N+G +++S DVSVN+P+ID+I +FPLAS E++PF P
Sbjct: 378 RRASLPISQKPAKGTKDSLYHPNIGILHQLNSPDVSVNSPRIDRI-KFPLASYEEMPFIP 436
Query: 473 VHVPSSTSVHCYSSSAGSAYRSITKEKCIHEDKVTVPGGSCPKGSECSKHVTAGVSIHSS 532
V S S + S + I +DK T+ K ++ TAG S +S
Sbjct: 437 VVRKKKGS----SRGSYSPPPEPPLDCSITKDKFTLEPERETKSDLSDQNATAGASSRAS 492
Query: 533 AELHQR-RFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIW 591
+ +R RFD SSY+QRAEALEGLLEFSARLL +R+DEL VLLKPFG KVSPRETAIW
Sbjct: 493 SGASRRQRFDPSSYRQRAEALEGLLEFSARLLIDERYDELNVLLKPFGPGKVSPRETAIW 552
Query: 592 LAKSFKET 599
L+KSFKE+
Sbjct: 553 LSKSFKES 560
>UniRef100_Q94CU5 Putative kinase [Oryza sativa]
Length = 943
Score = 405 bits (1041), Expect = e-111
Identities = 212/398 (53%), Positives = 272/398 (68%), Gaps = 33/398 (8%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+QYEV+EQIG+G+FG+A+LV HK EKK YVLKKIRLARQ++R R+SAHQEM LI+++++
Sbjct: 5 MDQYEVMEQIGRGAFGAAILVNHKTEKKKYVLKKIRLARQTERCRKSAHQEMALIARLQH 64
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVE+K++WVEKGC+VCI+ GYCE GD+A+ +KKANG FPEEKL KW QL +A+DYL
Sbjct: 65 PYIVEFKEAWVEKGCYVCIVTGYCEGGDMAELMKKANGTYFPEEKLLKWFAQLALAVDYL 124
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N +LHRD+KCSNIFLTK+QDIRLGDFGLAK L +DDL SS+VGTP+YMCPELLADIPY
Sbjct: 125 HSNFVLHRDLKCSNIFLTKDQDIRLGDFGLAKTLKADDLTSSVVGTPNYMCPELLADIPY 184
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCC+YEMAA +PAFKAFD+ LI+KIN+S + PLP YS + + L+KSMLR
Sbjct: 185 GFKSDIWSLGCCMYEMAAHRPAFKAFDMAGLISKINRSSIGPLPPCYSPSMKSLIKSMLR 244
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
K+PE RP ASE+L P+LQPY+ + P ++ + S+
Sbjct: 245 KSPEHRPTASEILKSPYLQPYVNQYR---------------PFADISHPIHSLEKPITSS 289
Query: 301 LSDQDKCLSLSNDRSLNPSISGTEQDS-QCFTQKAHGLSTSSKEVYEVSVGGACGPWNTN 359
S Q S S SISG++ DS Q + G STSS +
Sbjct: 290 RSSQK-----SMSGSQCSSISGSDIDSIQSSERNTSGPSTSSNNTIDTE--------GAE 336
Query: 360 KTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSK 397
T +V+ RS D + ++T+GP + SK
Sbjct: 337 ATDHVSVKNCSRS----DDVKSNKETVGPELERQDSSK 370
Score = 90.5 bits (223), Expect = 1e-16
Identities = 44/57 (77%), Positives = 49/57 (85%)
Query: 541 DTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKSFK 597
D S++QRAEALEGLLE SA LLQH R +EL V+LKPFG +KVSPRETAIWLAKSFK
Sbjct: 875 DVKSFRQRAEALEGLLELSADLLQHNRLEELAVVLKPFGKDKVSPRETAIWLAKSFK 931
>UniRef100_Q60DG4 Hypothetical protein B1110B01.5 [Oryza sativa]
Length = 943
Score = 404 bits (1038), Expect = e-111
Identities = 192/324 (59%), Positives = 251/324 (77%), Gaps = 1/324 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+QYE++EQ+G+G+FG+A+LV HK E+K YVLKKIRLARQ++R R+SAHQEM LI+++++
Sbjct: 5 MDQYEIMEQVGRGAFGAAILVNHKIERKKYVLKKIRLARQTERCRKSAHQEMALIARLQH 64
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVE+K++WVEKGC+VCI+ GYCE GD+ + +KK NG FPEEKL KW QL++A+DYL
Sbjct: 65 PYIVEFKEAWVEKGCYVCIVTGYCEGGDMDELMKKLNGTYFPEEKLLKWFAQLVLAVDYL 124
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N++LHRD+KCSNIFLTK+QDIRLGDFGLAK L DDL SS+VGTP+YMCPELL DIPY
Sbjct: 125 HSNYVLHRDLKCSNIFLTKDQDIRLGDFGLAKTLKEDDLTSSVVGTPNYMCPELLTDIPY 184
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCC+YEMAA +PAFKAFD+ LI+KIN+S + PLP YSS+ + L+KSMLR
Sbjct: 185 GFKSDIWSLGCCMYEMAAHRPAFKAFDMAGLISKINRSSIGPLPACYSSSMKTLIKSMLR 244
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
K+PE RP ASE+L +P+LQPY+ + ++P P + P S R S+
Sbjct: 245 KSPEHRPTASEILKNPYLQPYVNQCRPLSDAPTPIRMP-EKPLSTSRSNQRCTSESQSSS 303
Query: 301 LSDQDKCLSLSNDRSLNPSISGTE 324
+S D + S+DRS + T+
Sbjct: 304 ISCSDIDSTQSSDRSTSGGAPSTD 327
Score = 84.0 bits (206), Expect = 1e-14
Identities = 41/73 (56%), Positives = 53/73 (72%)
Query: 525 AGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVS 584
A + A + D +S++QRAEALEGLLE SA LL++ R +EL ++L+PFG KVS
Sbjct: 859 ASNGVKEEASPAKEALDVTSFRQRAEALEGLLELSADLLENNRLEELAIVLQPFGKNKVS 918
Query: 585 PRETAIWLAKSFK 597
PRETAIWLA+SFK
Sbjct: 919 PRETAIWLARSFK 931
>UniRef100_Q9LT35 Kinase-like protein [Arabidopsis thaliana]
Length = 416
Score = 384 bits (987), Expect = e-105
Identities = 181/262 (69%), Positives = 224/262 (85%), Gaps = 1/262 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+ YEV+EQIG+G+FGSA LV HK E++ YV+KKIRLA+Q++R + +A QEM LISK+++
Sbjct: 1 MDDYEVVEQIGRGAFGSAFLVIHKSERRKYVVKKIRLAKQTERCKLAAIQEMSLISKLKS 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVEYKDSWVEK C VCI+ YCE GD+ IKK+ GV EEKLC+W+VQLL+A+DYL
Sbjct: 61 PYIVEYKDSWVEKDC-VCIVTSYCEGGDMTQMIKKSRGVFASEEKLCRWMVQLLLAIDYL 119
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N +LHRD+KCSNIFLTK ++RLGDFGLAK+L DDLASS+VGTP+YMCPELLADIPY
Sbjct: 120 HNNRVLHRDLKCSNIFLTKENEVRLGDFGLAKLLGKDDLASSMVGTPNYMCPELLADIPY 179
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCC++E+AA +PAFKA D+ ALINKIN+S ++PLP MYSS+ + L+KSMLR
Sbjct: 180 GYKSDIWSLGCCMFEVAAHQPAFKAPDMAALINKINRSSLSPLPVMYSSSLKRLIKSMLR 239
Query: 241 KNPELRPPASELLNHPHLQPYI 262
KNPE RP A+ELL HPHLQPY+
Sbjct: 240 KNPEHRPTAAELLRHPHLQPYL 261
Score = 65.1 bits (157), Expect = 6e-09
Identities = 33/50 (66%), Positives = 39/50 (78%)
Query: 546 QQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS 595
++RAEALE LLE A LL+ ++FDEL +LKPFG E VS RETAIWL KS
Sbjct: 352 EERAEALESLLELCAGLLRQEKFDELEGVLKPFGDETVSSRETAIWLTKS 401
>UniRef100_Q6YY75 Serine/threonine-protein kinase Nek4-like [Oryza sativa]
Length = 416
Score = 369 bits (948), Expect = e-100
Identities = 170/262 (64%), Positives = 221/262 (83%), Gaps = 1/262 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYEV+EQIG+G++GSA LV HK E+K YV+KKIRL++Q+D+ +R+A+QEM L++ + N
Sbjct: 1 MEQYEVVEQIGRGAYGSAYLVVHKGERKRYVMKKIRLSKQNDKFQRTAYQEMSLMASLSN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVEYKD WV++G CI+ YCE GD+A+ IKKA GV F EE++C+W QLL+ALDYL
Sbjct: 61 PYIVEYKDGWVDEGTSACIVTSYCEGGDMAERIKKARGVLFSEERVCRWFTQLLLALDYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N +LHRD+KCSNI LTK+ +IRL DFGLAK+L +DLAS+IVGTP+YMCPE+LADIPY
Sbjct: 121 HCNRVLHRDLKCSNILLTKDNNIRLADFGLAKLL-MEDLASTIVGTPNYMCPEILADIPY 179
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCC++E+ A +PAFKA D+ +LINKIN+S ++P+P +YSS+ + +VKSMLR
Sbjct: 180 GYKSDIWSLGCCMFEILAHRPAFKAADMASLINKINRSSISPMPPIYSSSLKQIVKSMLR 239
Query: 241 KNPELRPPASELLNHPHLQPYI 262
KNPE RP A ELL HP+LQPY+
Sbjct: 240 KNPEHRPTAGELLRHPYLQPYL 261
Score = 62.4 bits (150), Expect = 4e-08
Identities = 31/50 (62%), Positives = 38/50 (76%)
Query: 546 QQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS 595
QQRA+ALE LLE A+LL+ +R +EL +L+PFG VS RETAIWL KS
Sbjct: 353 QQRADALESLLELCAKLLKQERLEELAGVLRPFGEGAVSSRETAIWLTKS 402
>UniRef100_Q9LXP3 Protein kinase-like protein [Arabidopsis thaliana]
Length = 941
Score = 366 bits (940), Expect = e-100
Identities = 189/329 (57%), Positives = 243/329 (73%), Gaps = 19/329 (5%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+QYE++EQIG+G+FG+A+LV HK E+K YVLKKIRLARQ++R RRSAHQEM LI++V++
Sbjct: 5 MDQYELMEQIGRGAFGAAILVHHKAERKKYVLKKIRLARQTERCRRSAHQEMSLIARVQH 64
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVE+K++WVEKGC+VCI+ GYCE GD+A+ +KK+NGV FPEEKLCKW QLL+A++YL
Sbjct: 65 PYIVEFKEAWVEKGCYVCIVTGYCEGGDMAELMKKSNGVYFPEEKLCKWFTQLLLAVEYL 124
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N++LHRD+KCSNIFLTK+QD+RLGDFGLAK L +DDL SS+VGTP+YMCPELLADIPY
Sbjct: 125 HSNYVLHRDLKCSNIFLTKDQDVRLGDFGLAKTLKADDLTSSVVGTPNYMCPELLADIPY 184
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCCIYEMAA +PAFKAFD+ LI+K S +G VK R
Sbjct: 185 GFKSDIWSLGCCIYEMAAYRPAFKAFDMAGLISK-------------KSTHQGNVKEEPR 231
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
+ + ASE+L HP+LQPY+ + L++ S P + S RR+ +S S+
Sbjct: 232 VS--AKRMASEILKHPYLQPYVEQYRPTLSA--ASITPEKPLNSREGRRSMAESQNSNSS 287
Query: 301 LSDQDKCLSLSNDRSLNPSISG--TEQDS 327
+ +S N R + PS TE DS
Sbjct: 288 SEKDNFYVSDKNIRYVVPSNGNKVTETDS 316
Score = 81.6 bits (200), Expect = 6e-14
Identities = 40/61 (65%), Positives = 48/61 (78%)
Query: 541 DTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKSFKETV 600
D S++QRAEALEGLLE SA LL+ R +EL ++L+PFG KVSPRETAIWLAKS K +
Sbjct: 865 DIKSFRQRAEALEGLLELSADLLEQSRLEELAIVLQPFGKNKVSPRETAIWLAKSLKGMM 924
Query: 601 I 601
I
Sbjct: 925 I 925
>UniRef100_Q9SLI2 F20D21.33 protein [Arabidopsis thaliana]
Length = 200
Score = 346 bits (887), Expect = 1e-93
Identities = 165/190 (86%), Positives = 178/190 (92%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYE LEQIGKGSFGSALLVRHKHEKK YVLKKIRLARQ+ RTRRSAHQE+ LISK+R+
Sbjct: 1 MEQYEFLEQIGKGSFGSALLVRHKHEKKKYVLKKIRLARQTQRTRRSAHQEVGLISKMRH 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEK C+VCI++GYCE GD+A AIKK+NGV+F EEKLCKWLVQLLM L+YL
Sbjct: 61 PFIVEYKDSWVEKACYVCIVIGYCEGGDMAQAIKKSNGVHFQEEKLCKWLVQLLMGLEYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H NHILHRDVKCSNIFLTK QDIRLGDFGLAK+LTSDDL SS+VGTPSYMCPELLADIPY
Sbjct: 121 HSNHILHRDVKCSNIFLTKEQDIRLGDFGLAKILTSDDLTSSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLG 190
GSKSDIWSLG
Sbjct: 181 GSKSDIWSLG 190
>UniRef100_Q9M1W2 Hypothetical protein F16M2_130 [Arabidopsis thaliana]
Length = 530
Score = 339 bits (870), Expect = 1e-91
Identities = 215/450 (47%), Positives = 265/450 (58%), Gaps = 68/450 (15%)
Query: 163 IVGTPSYMCPELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAP 222
+VGTPSYMCPELLADIPYGSKSDIWSLGCC+YEMAA KP FKA D+Q LI KI+K I+ P
Sbjct: 138 VVGTPSYMCPELLADIPYGSKSDIWSLGCCMYEMAAHKPPFKASDVQTLITKIHKLIMDP 197
Query: 223 LPTMYSSAFRGLVKSMLRKNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWP 282
+P MYS +FRGL+KSMLRKNPELRP A+ELLNHPHLQPYI +++KL SPRRSTFP Q+
Sbjct: 198 IPAMYSGSFRGLIKSMLRKNPELRPSANELLNHPHLQPYISMVYMKLESPRRSTFPLQFS 257
Query: 283 ESNYMRRTRFVVPDSVSTLSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLST-SS 341
E + +TL ++ + S SNDR LNPS+S TE S + KA +
Sbjct: 258 ERD-------------ATLKERRRS-SFSNDRRLNPSVSDTEAGSVSSSGKASPTPMFNG 303
Query: 342 KEVYEVSVGGACGPWNTNKTKSPTVERTPRSRADKDSTAARRQ----TMGPSKISRSGSK 397
++V EV+VG + + A K S AAR T + R+ K
Sbjct: 304 RKVSEVTVG----------VVREEIVPQRQEEAKKQSGAARTPRVAGTSAKASTQRTVFK 353
Query: 398 HDTLPVSHAPSVKFPPPTRRASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDK 456
H+ + VS+ + RR S PL T +++ ++S DVSVN P+ DK
Sbjct: 354 HELMKVSNPTERR-----RRVSLPLVVENPY---TYESDITALCSLNSPDVSVNTPRFDK 405
Query: 457 IAEFP-----LASCEDLPFFPVHVPSSTSVHCYSSSAGSAYRSITKEKCIHEDKVTVPGG 511
IAEFP + E V S +S C ++ SITK+KC
Sbjct: 406 IAEFPEDIFQNQNRETASRREVARHSFSSPPCPPHGEDNSNGSITKDKCT---------- 455
Query: 512 SCPKGSECSKHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDEL 571
V S +E+ QRRFDTSSYQQRAEALEGLLEFSA+LLQ +R+DEL
Sbjct: 456 ---------------VQKRSVSEVKQRRFDTSSYQQRAEALEGLLEFSAKLLQQERYDEL 500
Query: 572 GVLLKPFGLEKVSPRETAIWLAKSFKETVI 601
GVLLKPFG E+VS RETAIWL KSFKE +
Sbjct: 501 GVLLKPFGAERVSSRETAIWLTKSFKEASV 530
>UniRef100_Q9LHI7 Similarity to protein kinase [Arabidopsis thaliana]
Length = 571
Score = 324 bits (830), Expect = 5e-87
Identities = 209/597 (35%), Positives = 317/597 (53%), Gaps = 54/597 (9%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
++ Y V+EQ+ +G S +V H E K Y +KKI LA+ +D+ +++A QEM+L+S ++N
Sbjct: 16 LDNYHVVEQVRRGKSSSDFVVLHDIEDKKYAMKKICLAKHTDKLKQTALQEMKLLSSLKN 75
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IV Y+DSW++ CI Y E G++A+AIKKA G FPEE++ KWL QLL+A++YL
Sbjct: 76 PYIVHYEDSWIDNDNNACIFTAYYEGGNMANAIKKARGKLFPEERIFKWLAQLLLAVNYL 135
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N ++H D+ CSNIFL K+ ++LG++GLAK++ + S + G + MCPE+L D PY
Sbjct: 136 HSNRVVHMDLTCSNIFLPKDDHVQLGNYGLAKLINPEKPVSMVSGISNSMCPEVLEDQPY 195
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCC+YE+ A +PAFKA D+ LINKIN+S+++PLP +YSS + ++K MLR
Sbjct: 196 GYKSDIWSLGCCMYEITAHQPAFKAPDMAGLINKINRSLMSPLPIVYSSTLKQMIKLMLR 255
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSV-- 298
K PE RP A ELL +P LQPY+L+ L+ FP + S + R +P
Sbjct: 256 KKPEYRPTACELLRNPSLQPYLLQCQ-NLSPIYLPVFPIKPVNSPKDKARRNSLPGKFGK 314
Query: 299 STLSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNT 358
+S + +S S + +L P + TE S +Q A + + ++ + +C +T
Sbjct: 315 ERVSREKSEVSRSLE-NLYPFWTNTETGSSSSSQPASSTNGAEDKLETKRIDPSC---DT 370
Query: 359 NKTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKFPPPTRRA 418
K T +++ S D D +T P
Sbjct: 371 LKISEFTSQKSDESLIDPDIAVYSTET----------------------------PAEEN 402
Query: 419 SHPLPTRASMAKATLYTNVGTFPRVDSDVSVNAPQIDKIAEFPLASCEDLPFFPVHVPSS 478
+ P T ++ + +V +V+ + P+ + AE A + P + VP S
Sbjct: 403 ALPKETENIFSEESQLRDVDVGVVSAQEVACSPPRAIEEAETQEALPK--PKEQITVPIS 460
Query: 479 TSVHCYSSSAGSAYRSITKEKCIHEDKVTVPGGSCPKGSECSKHVTAGVSIHSSAELHQR 538
+ H SS A + DK +K V S SS
Sbjct: 461 SVAH---SSTEVAAAKDHLSGSLEGDK--------------AKMVKLTASEMSSVLSKLT 503
Query: 539 RFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS 595
+ ++RA+ALE LLE A L++ ++++EL LL PFG + VS R+TAIW AK+
Sbjct: 504 KLGPPQSKERADALECLLEKCAGLVKQEKYEELAGLLTPFGEDGVSARDTAIWFAKT 560
>UniRef100_Q8W4P0 Protein kinase, putative [Arabidopsis thaliana]
Length = 571
Score = 322 bits (825), Expect = 2e-86
Identities = 208/597 (34%), Positives = 317/597 (52%), Gaps = 54/597 (9%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
++ Y V+EQ+ +G S +V H E K Y +KKI LA+ +D+ +++A QEM+L+S ++N
Sbjct: 16 LDNYHVVEQVRRGKSSSDFVVLHDIEDKKYAMKKICLAKHTDKLKQTALQEMKLLSSLKN 75
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IV Y+DSW++ CI Y E G++A+AIKKA G FPEE++ KWL QLL+A++YL
Sbjct: 76 PYIVHYEDSWIDNDNNACIFTAYYEGGNMANAIKKARGKLFPEERIFKWLAQLLLAVNYL 135
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N ++H D+ CSNIFL K+ ++LG++GLAK++ + S + G + MCPE+L D P+
Sbjct: 136 HSNRVVHMDLTCSNIFLPKDDHVQLGNYGLAKLINPEKPVSMVSGISNSMCPEVLEDQPH 195
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCC+YE+ A +PAFKA D+ LINKIN+S+++PLP +YSS + ++K MLR
Sbjct: 196 GYKSDIWSLGCCMYEITAHQPAFKAPDMAGLINKINRSLMSPLPIVYSSTLKQMIKLMLR 255
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSV-- 298
K PE RP A ELL +P LQPY+L+ L+ FP + S + R +P
Sbjct: 256 KKPEYRPTACELLRNPSLQPYLLQCQ-NLSPIYLPVFPIKPVNSPKDKARRNSLPGKFGK 314
Query: 299 STLSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNT 358
+S + +S S + +L P + TE S +Q A + + ++ + +C +T
Sbjct: 315 ERVSREKSEVSRSLE-NLYPFWTNTETGSSSSSQPASSTNGAEDKLETKRIDPSC---DT 370
Query: 359 NKTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKFPPPTRRA 418
K T +++ S D D +T P
Sbjct: 371 LKISEFTSQKSDESLIDPDIAVYSTET----------------------------PAEEN 402
Query: 419 SHPLPTRASMAKATLYTNVGTFPRVDSDVSVNAPQIDKIAEFPLASCEDLPFFPVHVPSS 478
+ P T ++ + +V +V+ + P+ + AE A + P + VP S
Sbjct: 403 ALPKETENIFSEESQLRDVDVGVVSAQEVACSPPRAIEEAETQEALPK--PKEQITVPIS 460
Query: 479 TSVHCYSSSAGSAYRSITKEKCIHEDKVTVPGGSCPKGSECSKHVTAGVSIHSSAELHQR 538
+ H SS A + DK +K V S SS
Sbjct: 461 SVAH---SSTEVAAAKDHLSGSLEGDK--------------AKMVKLTASEMSSVLSKLT 503
Query: 539 RFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS 595
+ ++RA+ALE LLE A L++ ++++EL LL PFG + VS R+TAIW AK+
Sbjct: 504 KLGPPQSKERADALECLLEKCAGLVKQEKYEELAGLLTPFGEDGVSARDTAIWFAKT 560
>UniRef100_Q9SLI3 F20D21.32 protein [Arabidopsis thaliana]
Length = 425
Score = 268 bits (686), Expect = 3e-70
Identities = 189/434 (43%), Positives = 245/434 (55%), Gaps = 52/434 (11%)
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y D + GCCIYEMA LKPAFKAFD+QALINKINK+IV+PLP YS FRGLVKSML
Sbjct: 7 YSRTHDHRTAGCCIYEMAYLKPAFKAFDMQALINKINKTIVSPLPAKYSGPFRGLVKSML 66
Query: 240 RKNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESN-YMRRTRFVVPDSV 298
RKNPE+RP PY+L + L+LN+ RR T P + P S M++ F P
Sbjct: 67 RKNPEVRP-----------SPYVLDVKLRLNNLRRKTLPPELPSSKRIMKKAHFSEPAVT 115
Query: 299 STLSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNT 358
+ + SL NDR+LNP E+D+ A + S+ + ++S+
Sbjct: 116 CPAFGERQHRSLWNDRALNPE---AEEDT------ASSIKCISRRISDLSI--------E 158
Query: 359 NKTKSPTVERTPRSRADKDSTAARRQTMGPSKIS--RSGSKHDTLPVSHAPSV-KFPPPT 415
+ +K + + S A + + R T+ + G +H PVS + K P
Sbjct: 159 SSSKGTLICKQVSSSALIYALSCRTNTLDTNDHGCLLRGIQH---PVSGGGTTSKIIPSA 215
Query: 416 RRASHPLPTRASMAKATLYTN-VGTFPRVDS-DVSVNAPQIDKIAEFPLASCE-DLPFFP 472
RR S PL RA+ + Y VG V S + S+N PQ+DKIA FPLA E D+ F P
Sbjct: 216 RRTSLPLTKRATNQEVAAYNPIVGILQNVKSPEYSINEPQVDKIAIFPLAPYEQDIFFTP 275
Query: 473 VHVPSSTSVHCYSSSAGSAYRSITKEKCIHEDKVTVPG------GSCPKGSECS-KHVTA 525
+ +S S S+ + RSITK+KC + T G + GS S ++ TA
Sbjct: 276 MQRKTS------SKSSSVSDRSITKDKCTVQTHTTWQGIQLNMVDNISDGSSSSDQNATA 329
Query: 526 GVSIHSSAELHQR-RFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVS 584
G S H+++ +R RFD SSY+QRA+ALEGLLEFSARLLQ R+DEL VLLKPFG KVS
Sbjct: 330 GASSHTTSSSSRRCRFDPSSYRQRADALEGLLEFSARLLQEGRYDELNVLLKPFGPGKVS 389
Query: 585 PRETAIWLAKSFKE 598
PRETAIW+AKS KE
Sbjct: 390 PRETAIWIAKSLKE 403
>UniRef100_Q7T299 Similar to NIMA (Never in mitosis gene a)-related expressed kinase
4 [Brachydanio rerio]
Length = 849
Score = 253 bits (647), Expect = 9e-66
Identities = 120/263 (45%), Positives = 177/263 (66%), Gaps = 1/263 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+ Y + +GKGS+G LVRHK ++K YV+KK+ L S R RR+A QE +L+S++++
Sbjct: 1 MDGYLFVRVVGKGSYGEVNLVRHKSDRKQYVIKKLNLRTSSRRERRAAEQEAQLLSQLKH 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P IV Y++SW + C + I++G+CE GDL +K+ G PE ++ +W VQ+ MAL YL
Sbjct: 61 PNIVMYRESWEGEDCQLYIVMGFCEGGDLYHRLKQQKGELLPERQVVEWFVQIAMALQYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKML-TSDDLASSIVGTPSYMCPELLADIP 179
H HILHRD+K NIFLTK I++GD G+A++L +D+AS+++GTP YM PEL ++ P
Sbjct: 121 HEKHILHRDLKTQNIFLTKTNIIKVGDLGIARVLENQNDMASTLIGTPYYMSPELFSNKP 180
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y KSD+W+LGCC+YEMA LK AF A D+ +L+ +I + + +P+ Y L+K ML
Sbjct: 181 YNYKSDVWALGCCVYEMATLKHAFNAKDMNSLVYRIVEGKLPQMPSKYDPQLGELIKRML 240
Query: 240 RKNPELRPPASELLNHPHLQPYI 262
K PE RP +L P+++ I
Sbjct: 241 CKKPEDRPDVKHILRQPYIKHQI 263
>UniRef100_UPI00003AA783 UPI00003AA783 UniRef100 entry
Length = 753
Score = 251 bits (640), Expect = 6e-65
Identities = 124/308 (40%), Positives = 187/308 (60%), Gaps = 12/308 (3%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y L +GKGS+G LVRH+ + K YV+KK+ L S R R++A QE +L+S++++P I
Sbjct: 6 YCFLRAVGKGSYGEVSLVRHRQDSKQYVIKKLNLKHASSRERKAAEQEAQLLSQLKHPNI 65
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
V Y++SW + + I++G+CE GDL +K+ G PE ++ +W VQ+ MAL YLH
Sbjct: 66 VAYRESWQGEDGLLYIVMGFCEGGDLYHRLKEQKGKLLPENQVVEWFVQIAMALQYLHEK 125
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIPYGS 182
HILHRD+K N+FLT+ I++GD G+A++L + D+AS+++GTP YM PEL ++ PY
Sbjct: 126 HILHRDLKTQNVFLTRTNIIKVGDLGIARVLENQCDMASTLIGTPYYMSPELFSNKPYNY 185
Query: 183 KSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKN 242
KSD+W+LGCC+YEMA LK AF A D+ +L+ +I + + P+P YS ++++ML K
Sbjct: 186 KSDVWALGCCVYEMATLKHAFNAKDMNSLVYRIIEGKLPPMPKDYSPQLVEIIQTMLSKR 245
Query: 243 PELRPPASELLNHPHLQPYI-----------LKIHLKLNSPRRSTFPFQWPESNYMRRTR 291
PE RP +L P+++ I K H K+ + P N
Sbjct: 246 PEQRPSVKSILRQPYIKHQISLFLEATKMKAAKSHKKMAGSKLKDSSSVVPVKNESHNKN 305
Query: 292 FVVPDSVS 299
+ PDS S
Sbjct: 306 VIPPDSSS 313
>UniRef100_UPI00003AA782 UPI00003AA782 UniRef100 entry
Length = 785
Score = 251 bits (640), Expect = 6e-65
Identities = 124/308 (40%), Positives = 187/308 (60%), Gaps = 12/308 (3%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y L +GKGS+G LVRH+ + K YV+KK+ L S R R++A QE +L+S++++P I
Sbjct: 6 YCFLRAVGKGSYGEVSLVRHRQDSKQYVIKKLNLKHASSRERKAAEQEAQLLSQLKHPNI 65
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
V Y++SW + + I++G+CE GDL +K+ G PE ++ +W VQ+ MAL YLH
Sbjct: 66 VAYRESWQGEDGLLYIVMGFCEGGDLYHRLKEQKGKLLPENQVVEWFVQIAMALQYLHEK 125
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIPYGS 182
HILHRD+K N+FLT+ I++GD G+A++L + D+AS+++GTP YM PEL ++ PY
Sbjct: 126 HILHRDLKTQNVFLTRTNIIKVGDLGIARVLENQCDMASTLIGTPYYMSPELFSNKPYNY 185
Query: 183 KSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKN 242
KSD+W+LGCC+YEMA LK AF A D+ +L+ +I + + P+P YS ++++ML K
Sbjct: 186 KSDVWALGCCVYEMATLKHAFNAKDMNSLVYRIIEGKLPPMPKDYSPQLVEIIQTMLSKR 245
Query: 243 PELRPPASELLNHPHLQPYI-----------LKIHLKLNSPRRSTFPFQWPESNYMRRTR 291
PE RP +L P+++ I K H K+ + P N
Sbjct: 246 PEQRPSVKSILRQPYIKHQISLFLEATKMKAAKSHKKMAGSKLKDSSSVVPVKNESHNKN 305
Query: 292 FVVPDSVS 299
+ PDS S
Sbjct: 306 VIPPDSSS 313
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 988,461,610
Number of Sequences: 2790947
Number of extensions: 41194857
Number of successful extensions: 162662
Number of sequences better than 10.0: 18872
Number of HSP's better than 10.0 without gapping: 12663
Number of HSP's successfully gapped in prelim test: 6213
Number of HSP's that attempted gapping in prelim test: 118716
Number of HSP's gapped (non-prelim): 22989
length of query: 601
length of database: 848,049,833
effective HSP length: 133
effective length of query: 468
effective length of database: 476,853,882
effective search space: 223167616776
effective search space used: 223167616776
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0322.12


