

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
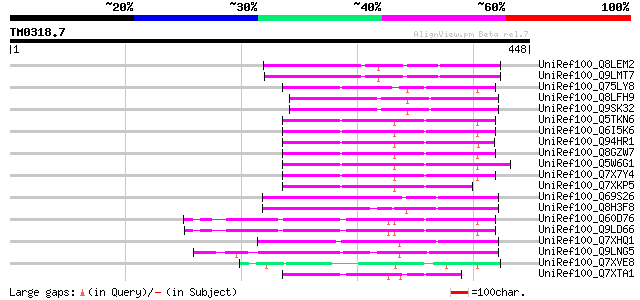
Query= TM0318.7
(448 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LEM2 Hypothetical protein [Arabidopsis thaliana] 106 1e-21
UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana] 104 4e-21
UniRef100_Q75LY8 Putative MuDR family transposase [Oryza sativa] 97 7e-19
UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana] 97 1e-18
UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana] 95 4e-18
UniRef100_Q5TKN6 Hypothetical protein OJ1058_F05.1 [Oryza sativa] 94 6e-18
UniRef100_Q6I5K6 Hypothetical protein OSJNBb0088F07.13 [Oryza sa... 94 6e-18
UniRef100_Q94HR1 Hypothetical protein OSJNBa0065C16.8 [Oryza sat... 94 6e-18
UniRef100_Q8GZW7 Putative transposable element [Oryza sativa] 94 8e-18
UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa] 94 1e-17
UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa] 93 2e-17
UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa] 90 1e-16
UniRef100_Q69S26 Serine/threonine phosphatase PP7-related-like [... 88 6e-16
UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like p... 87 1e-15
UniRef100_Q60D76 Putative polyprotein [Oryza sativa] 87 1e-15
UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein ... 86 2e-15
UniRef100_Q7XHQ1 Calcineurin-like phosphoesterase-like [Oryza sa... 83 1e-14
UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana] 83 2e-14
UniRef100_Q7XVE8 OSJNBa0083D01.11 protein [Oryza sativa] 69 4e-10
UniRef100_Q7XTA1 OSJNBa0008A08.9 protein [Oryza sativa] 67 1e-09
>UniRef100_Q8LEM2 Hypothetical protein [Arabidopsis thaliana]
Length = 478
Score = 106 bits (265), Expect = 1e-21
Identities = 70/206 (33%), Positives = 111/206 (52%), Gaps = 8/206 (3%)
Query: 220 ELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPM 279
EL+E+ G + LI ALVERW ET++FH P GEMTITLD+VS +L L +
Sbjct: 44 ELVEKAGFGWFRLIGSISLNNSLISALVERWRRETNTFHFPCGEMTITLDEVSLILGLAV 103
Query: 280 GSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNR--SQTIRFGVLQTRYEAALAEH 337
+ + + ++ + +C +L+G + + E NR ++ ++ + A + E
Sbjct: 104 DGKPVVGVKEKDEDPSQVCLKLLG---KLPKGELSGNRVTAKWLKESFAECPKGATMKEI 160
Query: 338 RYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLS 397
Y R +L+ +G+T+FA+ + Y I + ED + EYAWGA ALA LY Q+
Sbjct: 161 EYH--TRAYLIYIVGSTIFATTDPSKISVDYLI-LFEDFEKAGEYAWGAAALAFLYRQIG 217
Query: 398 QASRRGTAQMGGFTSLLLGWVYEYLS 423
AS+R + +GG +LL W Y +L+
Sbjct: 218 NASQRSQSIIGGCLTLLQCWSYFHLN 243
>UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana]
Length = 478
Score = 104 bits (260), Expect = 4e-21
Identities = 69/205 (33%), Positives = 110/205 (53%), Gaps = 8/205 (3%)
Query: 221 LIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMG 280
L+E+ G + LI ALVERW ET++FH P GEMTITLD+VS +L L +
Sbjct: 45 LVEKAGFGWFRLVGSISLNNSLISALVERWRRETNTFHFPCGEMTITLDEVSLILGLAVD 104
Query: 281 SRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNR--SQTIRFGVLQTRYEAALAEHR 338
+ + + ++ + +C +L+G + + E NR ++ ++ + A + E
Sbjct: 105 GKPVVGVKEKDEDPSQVCLRLLG---KLPKGELSGNRVTAKWLKESFAECPKGATMKEIE 161
Query: 339 YEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQ 398
Y R +L+ +G+T+FA+ + Y I + ED + EYAWGA ALA LY Q+
Sbjct: 162 YH--TRAYLIYIVGSTIFATTDPSKISVDYLI-LFEDFEKAGEYAWGAAALAFLYRQIGN 218
Query: 399 ASRRGTAQMGGFTSLLLGWVYEYLS 423
AS+R + +GG +LL W Y +L+
Sbjct: 219 ASQRSQSIIGGCLTLLQCWSYFHLN 243
>UniRef100_Q75LY8 Putative MuDR family transposase [Oryza sativa]
Length = 1403
Score = 97.4 bits (241), Expect = 7e-19
Identities = 65/194 (33%), Positives = 101/194 (51%), Gaps = 17/194 (8%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
PV +A ++ALV+RW ET SFH+P GEMTITL DV+ +L LP+ T GR E
Sbjct: 865 PVFNAPALIALVDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVT-GRTETPGWR 923
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYEDA--------ARIWL 347
A QL G ++ I + + + I L + H ++DA AR ++
Sbjct: 924 AQVEQLFGITLNIEQGQGGKKKQNGIPLSWLSQNF-----SHLHDDAEPWRVECYARAYI 978
Query: 348 VNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRR--GTA 405
++ LG LF + +GG + WI ++ +LG + ++WG+ LA Y QL +A R ++
Sbjct: 979 LHLLGGVLFPN-AGGDIASAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSS 1037
Query: 406 QMGGFTSLLLGWVY 419
M G L+ W++
Sbjct: 1038 NMSGCVLLIQLWMW 1051
>UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana]
Length = 509
Score = 96.7 bits (239), Expect = 1e-18
Identities = 59/184 (32%), Positives = 97/184 (52%), Gaps = 7/184 (3%)
Query: 242 LILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECAALCAQL 301
LI ALVERW ET++FH+P GEMTITLD+V+ +L L + + + + +C +L
Sbjct: 75 LISALVERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSKVDDEVAMDMCGRL 134
Query: 302 MGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYEDA---ARIWLVNQLGATLFAS 358
+G E + +R ++ L+ + + ++ R +L+ +G+T+FA+
Sbjct: 135 LGKLPSAANKEVNCSR---VKLNWLKRTFSECPEDASFDVVKCHTRAYLLYLIGSTIFAT 191
Query: 359 KSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRRGTAQMGGFTSLLLGWV 418
G +V ++ + ED + YAWGA ALA LY L AS + + + G +LL W
Sbjct: 192 TDGD-KVSVKYLPLFEDFDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCWS 250
Query: 419 YEYL 422
Y +L
Sbjct: 251 YFHL 254
>UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana]
Length = 509
Score = 95.1 bits (235), Expect = 4e-18
Identities = 59/184 (32%), Positives = 96/184 (52%), Gaps = 7/184 (3%)
Query: 242 LILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECAALCAQL 301
LI ALVERW ET++FH+P GEMTITLD+V+ +L L + + + +C +L
Sbjct: 75 LISALVERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSKVGDEVAMDMCGRL 134
Query: 302 MGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYEDA---ARIWLVNQLGATLFAS 358
+G E + +R ++ L+ + + ++ R +L+ +G+T+FA+
Sbjct: 135 LGKLPSAANKEVNCSR---VKLNWLKRTFSECPEDASFDVVKCHTRAYLLYLIGSTIFAT 191
Query: 359 KSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRRGTAQMGGFTSLLLGWV 418
G +V ++ + ED + YAWGA ALA LY L AS + + + G +LL W
Sbjct: 192 TDGD-KVSVKYLPLFEDFDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCWS 250
Query: 419 YEYL 422
Y +L
Sbjct: 251 YFHL 254
>UniRef100_Q5TKN6 Hypothetical protein OJ1058_F05.1 [Oryza sativa]
Length = 725
Score = 94.4 bits (233), Expect = 6e-18
Identities = 64/189 (33%), Positives = 96/189 (49%), Gaps = 7/189 (3%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
PV +A + ALV+RW ET SFH+P GEMTITL DV+ +L LP+ T GR E
Sbjct: 117 PVFNAPALTALVDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVT-GRTETPGWH 175
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRY---EAALAEHRYEDAARIWLVNQLG 352
A QL G + I + + + I L + + R E AR ++++ LG
Sbjct: 176 AQVQQLFGIPLNIEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLG 235
Query: 353 ATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRR--GTAQMGGF 410
LF +GG + WI ++ +LG + ++WG+ LA Y QL +A R ++ M G
Sbjct: 236 GVLFPD-AGGDIASAIWIPLVTNLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGC 294
Query: 411 TSLLLGWVY 419
L+ W++
Sbjct: 295 VLLIQMWMW 303
>UniRef100_Q6I5K6 Hypothetical protein OSJNBb0088F07.13 [Oryza sativa]
Length = 1564
Score = 94.4 bits (233), Expect = 6e-18
Identities = 64/189 (33%), Positives = 96/189 (49%), Gaps = 7/189 (3%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
PV +A + ALV+RW ET SFH+P GEMTITL DV+ +L LP+ T GR E
Sbjct: 956 PVFNAPALTALVDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVT-GRTETPGWH 1014
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRY---EAALAEHRYEDAARIWLVNQLG 352
A QL G + I + + + I L + + R E AR ++++ LG
Sbjct: 1015 AQVQQLFGIPLNIEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLG 1074
Query: 353 ATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRR--GTAQMGGF 410
LF +GG + WI ++ +LG + ++WG+ LA Y QL +A R ++ M G
Sbjct: 1075 GVLFPD-AGGDIASAIWIPLVTNLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGC 1133
Query: 411 TSLLLGWVY 419
L+ W++
Sbjct: 1134 VLLIQMWMW 1142
>UniRef100_Q94HR1 Hypothetical protein OSJNBa0065C16.8 [Oryza sativa]
Length = 681
Score = 94.4 bits (233), Expect = 6e-18
Identities = 64/188 (34%), Positives = 96/188 (51%), Gaps = 7/188 (3%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
PV +A + ALV+RW ET SFH+P GEMTITL DV+ +L LP+ T GR E
Sbjct: 165 PVFNAPALTALVDRWRPETHSFHLPSGEMTITLQDVAMILALPLQGHVVT-GRTETPGWR 223
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRY---EAALAEHRYEDAARIWLVNQLG 352
A QL G + I + + + I L + + R E AR ++++ LG
Sbjct: 224 AQVEQLFGIPLNIEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLG 283
Query: 353 ATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRRG--TAQMGGF 410
LF + +GG + WI ++ +LG + ++WG+ LA Y QL +A R ++ M G
Sbjct: 284 GVLFPN-AGGDIASAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQALSSNMSGC 342
Query: 411 TSLLLGWV 418
L+ W+
Sbjct: 343 VLLIQLWM 350
>UniRef100_Q8GZW7 Putative transposable element [Oryza sativa]
Length = 663
Score = 94.0 bits (232), Expect = 8e-18
Identities = 64/189 (33%), Positives = 96/189 (49%), Gaps = 7/189 (3%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
PV +A + ALV+RW ET SFH+P GEMTITL DV+ +L LP+ T GR E
Sbjct: 69 PVFNAPALTALVDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVT-GRTETPGWH 127
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRY---EAALAEHRYEDAARIWLVNQLG 352
A QL G + I + + + I L + + R E AR ++++ LG
Sbjct: 128 AQVQQLFGIPLNIEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLG 187
Query: 353 ATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRR--GTAQMGGF 410
LF +GG + WI ++ +LG + ++WG+ LA Y QL +A R ++ M G
Sbjct: 188 GVLFPD-AGGDIASAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGC 246
Query: 411 TSLLLGWVY 419
L+ W++
Sbjct: 247 VLLIQIWMW 255
>UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa]
Length = 1567
Score = 93.6 bits (231), Expect = 1e-17
Identities = 66/202 (32%), Positives = 100/202 (48%), Gaps = 7/202 (3%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
PV +A + ALV RW ET SFH+P GEMTITL DV+ +L LP+ T GR E
Sbjct: 1019 PVFNAPALTALVNRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVT-GRTETPGWH 1077
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRY---EAALAEHRYEDAARIWLVNQLG 352
A QL G + I + + + I L + + R E AR ++++ LG
Sbjct: 1078 AQVQQLFGIPLNIEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRAECYARAYILHLLG 1137
Query: 353 ATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRR--GTAQMGGF 410
LF +GG + WI ++ +LG + ++WG+ LA Y QL +A R ++ M G
Sbjct: 1138 GVLFPD-AGGDIASAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGC 1196
Query: 411 TSLLLGWVYEYLSDRVIIRRVV 432
L+ W++ L + + + V
Sbjct: 1197 VLLIQMWMWLRLPNEPDMEKTV 1218
>UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa]
Length = 1575
Score = 92.8 bits (229), Expect = 2e-17
Identities = 63/189 (33%), Positives = 96/189 (50%), Gaps = 7/189 (3%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
PV +A + ALV+RW ET SFH+P GEMTITL DV+ +L LP+ T GR +
Sbjct: 961 PVFNAPALTALVDRWRPETHSFHLPSGEMTITLQDVAMILALPLRGHAVT-GRTDTPGWH 1019
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRY---EAALAEHRYEDAARIWLVNQLG 352
A QL G + I + + + I L + + R E AR ++++ LG
Sbjct: 1020 AQVQQLFGIPLNIEQGQGGKKKQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLG 1079
Query: 353 ATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRR--GTAQMGGF 410
LF +GG + WI ++ +LG + ++WG+ LA Y QL +A R ++ M G
Sbjct: 1080 EVLFPD-AGGDIASAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGC 1138
Query: 411 TSLLLGWVY 419
L+ W++
Sbjct: 1139 VLLIQMWMW 1147
>UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa]
Length = 1495
Score = 90.1 bits (222), Expect = 1e-16
Identities = 59/167 (35%), Positives = 85/167 (50%), Gaps = 5/167 (2%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
PV +A + ALV+RW ET SFH+P GEMTITL DV+ +L LP+ T GR E
Sbjct: 929 PVFNAPALTALVDRWRPETHSFHLPSGEMTITLQDVAMILALPLQGHAVT-GRTETPGWR 987
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRY---EAALAEHRYEDAARIWLVNQLG 352
A QL G + I + + I L + + R E AR ++++ LG
Sbjct: 988 AQVEQLFGIQLNIEQGQGGKKMQNGIPLSWLSQNFSHLDDDAEPWRVECYARAYILHLLG 1047
Query: 353 ATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQA 399
LF +GG + WI ++ +LG + ++WG+ LA Y QL +A
Sbjct: 1048 GVLFPD-AGGDIASAIWIPLVANLGDLGRFSWGSAVLAWTYRQLCEA 1093
>UniRef100_Q69S26 Serine/threonine phosphatase PP7-related-like [Oryza sativa]
Length = 619
Score = 87.8 bits (216), Expect = 6e-16
Identities = 64/205 (31%), Positives = 98/205 (47%), Gaps = 4/205 (1%)
Query: 219 RELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLP 278
++L+ +GL L + D L+ A ERW+ ET++ H EM +L DVS +L +P
Sbjct: 52 QQLVNASGLGHLALTTGFTIDRSLLTAFCERWNNETNTAHFMGFEMAPSLRDVSYILGIP 111
Query: 279 MGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHR 338
+ T + +C +G S G E R + Q + E
Sbjct: 112 VTGHVVTAEPIGDEAVRRMCLHFLGESPGNGEQLCGLIRLTWLYRKFHQLPENPTINEIA 171
Query: 339 YEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQ 398
Y + R +L+ +G+TLF G+ + Y + +L D ++ EYAWG+ ALA LY LS
Sbjct: 172 Y--STRAYLLYLVGSTLFPDTMRGFVSPRY-LPLLADFRKIREYAWGSAALAHLYRGLSV 228
Query: 399 A-SRRGTAQMGGFTSLLLGWVYEYL 422
A + T Q G +LL+ W+YEYL
Sbjct: 229 AVTPNATTQFLGSATLLMAWIYEYL 253
>UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like protein [Oryza
sativa]
Length = 589
Score = 87.0 bits (214), Expect = 1e-15
Identities = 61/209 (29%), Positives = 104/209 (49%), Gaps = 12/209 (5%)
Query: 219 RELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLP 278
++L++ +GL L + + V D ++A E W +ET++ + EM +L D + +L +P
Sbjct: 39 QQLVDASGLGNLIHTAGLVIDRIALMAFFELWSKETNTAQLNGFEMAPSLRDAAYILGIP 98
Query: 279 MGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHR 338
+ R T G LC Q +G + R ++ LQ+++ + + E
Sbjct: 99 VTGRVVTTGAVLNKSVEDLCFQYLGQVPDCRDC-----RGSHVKLSWLQSKF-SRIPERP 152
Query: 339 YED----AARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYD 394
D R +L+ +G+ L + GY + Y + +L D +V EYAWGA ALA LY
Sbjct: 153 TNDQTMYGTRAYLLFLIGSALLPERDRGYVSPKY-LPLLSDFDKVQEYAWGAAALAHLYK 211
Query: 395 QLSQA-SRRGTAQMGGFTSLLLGWVYEYL 422
LS A + ++ G +LL+GW+YEY+
Sbjct: 212 ALSIAVAHSARKRLFGSAALLMGWIYEYI 240
>UniRef100_Q60D76 Putative polyprotein [Oryza sativa]
Length = 1754
Score = 86.7 bits (213), Expect = 1e-15
Identities = 77/279 (27%), Positives = 126/279 (44%), Gaps = 36/279 (12%)
Query: 151 QRLPPFPGGPVELSLLTYYAD-HKAPWAWHALLRTDERYVDRRQLKVATAGGKVWNLAYD 209
+R+ FPG L +Y + H+AP + +LK G ++ +D
Sbjct: 885 RRMEHFPG------LEDFYEEKHRAP-----------QIASGERLKTLRVRGHTAHILFD 927
Query: 210 GDSDSHRRVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLD 269
+ R +L+ + Q P P+ +A + ALV+RW ET +FH+P GE+T+TL+
Sbjct: 928 DRYVPYLRRAKLLAFVTMAQRPV---PLYNAAALTALVDRWRPETHTFHLPCGELTVTLE 984
Query: 270 DVSALLHLPMGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGV---- 325
DV+ +L LP+ + T D + + + +G+ + QT GV
Sbjct: 985 DVAMILGLPIRGQAVT-----GDTASGNWRERVEEYLGLEPPVAPDGQRQTKTSGVPLSW 1039
Query: 326 LQTRY---EAALAEHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEY 382
L+ + A E + R +++ G+ LF SGG + W+ +L D +
Sbjct: 1040 LRANFGQCPAEADEATVQRYCRAYVLYIFGSILFPD-SGGDMASWMWLPLLADWDEAGTF 1098
Query: 383 AWGAIALATLYDQLSQASRR--GTAQMGGFTSLLLGWVY 419
+WG+ ALA LY QL A RR G A + G LL W++
Sbjct: 1099 SWGSAALAWLYRQLCDACRRQGGDANLAGCVWLLQVWMW 1137
>UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein [Oryza sativa]
Length = 1230
Score = 86.3 bits (212), Expect = 2e-15
Identities = 77/278 (27%), Positives = 125/278 (44%), Gaps = 36/278 (12%)
Query: 152 RLPPFPGGPVELSLLTYYAD-HKAPWAWHALLRTDERYVDRRQLKVATAGGKVWNLAYDG 210
R+ FPG L +Y + H+AP + +LK G ++ +D
Sbjct: 504 RMEHFPG------LEDFYEEKHRAP-----------QIASGERLKTLRVRGHTAHILFDD 546
Query: 211 DSDSHRRVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDD 270
+ R +L+ + Q P P+ +A + ALV+RW ET +FH+P GE+T+TL+D
Sbjct: 547 RYVPYLRRAKLLAFVTMAQRPV---PLYNAAALTALVDRWRPETHTFHLPCGELTVTLED 603
Query: 271 VSALLHLPMGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGV----L 326
V+ +L LP+ + T D + + + +G+ + QT GV L
Sbjct: 604 VAMILGLPIRGQAVT-----GDTASGNWRERVEEYLGLEPPVAPDGQRQTKTSGVPLSWL 658
Query: 327 QTRY---EAALAEHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYA 383
+ + A E + R +++ G+ LF SGG + W+ +L D ++
Sbjct: 659 RANFGQCPAEADEATVQRYCRAYVLYIFGSILFPD-SGGDMASWMWLPLLADWDEAGTFS 717
Query: 384 WGAIALATLYDQLSQASRR--GTAQMGGFTSLLLGWVY 419
WG+ ALA LY QL A RR G A + G LL W++
Sbjct: 718 WGSAALAWLYRQLCDACRRQGGDANLAGCVWLLQVWMW 755
>UniRef100_Q7XHQ1 Calcineurin-like phosphoesterase-like [Oryza sativa]
Length = 327
Score = 83.2 bits (204), Expect = 1e-14
Identities = 67/223 (30%), Positives = 101/223 (45%), Gaps = 19/223 (8%)
Query: 215 HRRVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSAL 274
H + +E G Q+ D LI ALVERW +T++FH+P GEMTITL DVS L
Sbjct: 43 HSLCQTALENLGFFQIARMKEINNDKYLISALVERWRPKTNTFHLPVGEMTITLQDVSCL 102
Query: 275 LHLPMGSRFYTPGRGERD-ECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAA 333
LP+ R P G+ D + +L+G + + + + V +RY +
Sbjct: 103 WGLPIHGR---PITGQADGSWVDMIERLLGIPMEEQHMKQKKRKKEDDMTMVSYSRYSIS 159
Query: 334 LA--------------EHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRV 379
L+ E R +++ +G+ +F SG +Y + + +L
Sbjct: 160 LSKLRDRFRVMPKNATEREINWYTRALVLDIIGSMVFTDTSGDGVPAMY-LQFMMNLSEQ 218
Query: 380 CEYAWGAIALATLYDQLSQASRRGTAQMGGFTSLLLGWVYEYL 422
EY WGA AL+ LY QLS AS + A++ LL W + L
Sbjct: 219 TEYNWGAAALSMLYRQLSIASEKERAEISRPLLLLQLWSWSRL 261
>UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana]
Length = 1340
Score = 82.8 bits (203), Expect = 2e-14
Identities = 74/269 (27%), Positives = 122/269 (44%), Gaps = 28/269 (10%)
Query: 159 GPVELSLLTYYADHKAPWAWHALLRTDERYVDRRQL--KVATAGGKVWNLAYDGDSDSHR 216
GPV+ S+L + +H++ W E V R+L + G + W L
Sbjct: 14 GPVDQSILVWQHEHRSAAIW-------EDEVPPRELTCRHKLLGMRDWPL--------DP 58
Query: 217 RVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLH 276
V + + + GL+ + ++ D LI ALVERW ET +FH+P GE+T+TL DV+ LL
Sbjct: 59 LVCQKLIEFGLYGVYKVAFIQLDYALITALVERWRPETHTFHLPAGEITVTLQDVNILLG 118
Query: 277 LPMGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALA- 335
L + T + A LC L+G G + + L+ + A
Sbjct: 119 LRVDGPAVT--GSTKYNWADLCEDLLGHRPGPKDL-----HGSHVSLAWLRENFRNLPAD 171
Query: 336 --EHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLY 393
E + R +++ + L+ KS + + ++ +L D V + +WG+ LA LY
Sbjct: 172 PDEVTLKCHTRAFVLALMSGFLYGDKS-KHDVALTFLPLLRDFDEVAKLSWGSATLALLY 230
Query: 394 DQLSQASRRGTAQMGGFTSLLLGWVYEYL 422
+L +AS+R + + G LL W +E L
Sbjct: 231 RELCRASKRTVSTICGPLVLLQLWAWERL 259
>UniRef100_Q7XVE8 OSJNBa0083D01.11 protein [Oryza sativa]
Length = 938
Score = 68.6 bits (166), Expect = 4e-10
Identities = 67/240 (27%), Positives = 98/240 (39%), Gaps = 50/240 (20%)
Query: 199 AGGKVWNLAYDGDSDSHRRVRE--------LIEQTGLHQLPYCSYPVTDAGLILALVERW 250
AGG W++ + R+RE L+E H P + + D L+ ALV+RW
Sbjct: 562 AGGSAWDVP-----GTDMRLREAGLLPMCRLVEAAAGHVDPARRWTM-DRSLLAALVDRW 615
Query: 251 HEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECAALCAQLMGGSVGIYE 310
ET +FH+P GE+ TL DVS LL LP L G +VG
Sbjct: 616 RPETHTFHLPCGEVAPTLQDVSYLLGLP----------------------LAGDAVGPVT 653
Query: 311 AEFDTNRSQTIRFGVLQTRYE---AALAEHRYEDAARIWLVNQLGATLFASKSGGYHTTV 367
D RF ++Q LA HR + WL+ G + V
Sbjct: 654 TAADWQDDLMARFALVQRAPHLPLQPLAHHRNTGPTKRWLLQFTGHAVDKG-------LV 706
Query: 368 YWIGMLED--LGRVCEYAWGAIALATLYDQLSQASRR--GTAQMGGFTSLLLGWVYEYLS 423
++ + D +G V +++WG+ LA LY L +A + +A GG L W E ++
Sbjct: 707 HYARSIADAAVGEVPQWSWGSALLAALYRGLCEACTKTDPSATFGGCPLFLSIWAAERIA 766
>UniRef100_Q7XTA1 OSJNBa0008A08.9 protein [Oryza sativa]
Length = 1560
Score = 66.6 bits (161), Expect = 1e-09
Identities = 49/165 (29%), Positives = 80/165 (47%), Gaps = 19/165 (11%)
Query: 236 PVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECA 295
P+ +A + ALV+RW ET +FH+P GE+T+TL+DV+ +L LP+ + T D +
Sbjct: 870 PLYNAAALTALVDRWRPETHTFHLPCGELTVTLEDVAMILGLPIRGQAVT-----SDTAS 924
Query: 296 ALCAQLMGGSVGIYEAEFDTNRSQTIRFGV----LQTRYEAALAE------HRYEDAARI 345
+ + +G+ + QT GV L+ + AE RY R
Sbjct: 925 GNWRERVEEYLGVEPPVAPDGQRQTKTSGVPLSWLRANFGQCPAEADKATVQRY---CRA 981
Query: 346 WLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALA 390
+++ G+ LF SGG + W+ +L D ++WG+ ALA
Sbjct: 982 YVLYIFGSILFPD-SGGDMASWMWLPLLADWDEAGTFSWGSAALA 1025
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 787,671,656
Number of Sequences: 2790947
Number of extensions: 35466397
Number of successful extensions: 200161
Number of sequences better than 10.0: 2426
Number of HSP's better than 10.0 without gapping: 763
Number of HSP's successfully gapped in prelim test: 1800
Number of HSP's that attempted gapping in prelim test: 181436
Number of HSP's gapped (non-prelim): 11299
length of query: 448
length of database: 848,049,833
effective HSP length: 131
effective length of query: 317
effective length of database: 482,435,776
effective search space: 152932140992
effective search space used: 152932140992
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0318.7


