

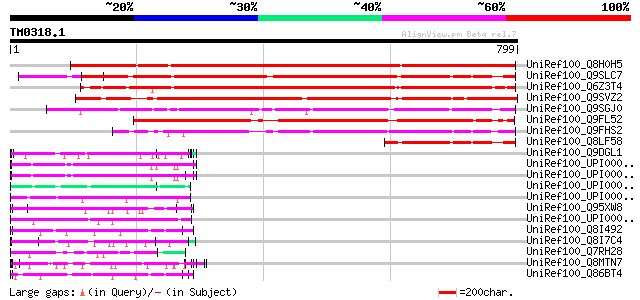
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.1
(799 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8H0H5 Hypothetical protein A20 [Nicotiana tabacum] 870 0.0
UniRef100_Q9SLC7 Hypothetical protein At2g42320 [Arabidopsis tha... 702 0.0
UniRef100_Q6Z3T4 Hypothetical protein OSJNBa0025J22.9 [Oryza sat... 697 0.0
UniRef100_Q9SVZ2 Hypothetical protein F15B8.30 [Arabidopsis thal... 667 0.0
UniRef100_Q9SGJ0 F28J7.14 protein [Arabidopsis thaliana] 512 e-143
UniRef100_Q9FL52 Gb|AAD23715.1 [Arabidopsis thaliana] 482 e-134
UniRef100_Q9FHS2 Gb|AAF03435.1 [Arabidopsis thaliana] 436 e-120
UniRef100_Q8LF58 Hypothetical protein [Arabidopsis thaliana] 214 8e-54
UniRef100_Q9DGL1 Retinitis pigmentosa GTPase regulator-like prot... 75 8e-12
UniRef100_UPI000024C743 UPI000024C743 UniRef100 entry 75 1e-11
UniRef100_UPI0000247AAF UPI0000247AAF UniRef100 entry 75 1e-11
UniRef100_UPI00001A1808 UPI00001A1808 UniRef100 entry 70 3e-10
UniRef100_UPI00003AEAEA UPI00003AEAEA UniRef100 entry 68 1e-09
UniRef100_Q95XW8 Hypothetical protein Y55B1BR.3 [Caenorhabditis ... 68 1e-09
UniRef100_UPI000045323E UPI000045323E UniRef100 entry 67 2e-09
UniRef100_Q8I492 Phoshoprotein 300 [Plasmodium falciparum] 67 2e-09
UniRef100_Q8I7C4 NAD(+) ADP-ribosyltransferase-3 [Dictyostelium ... 66 5e-09
UniRef100_Q7RH28 Mature-parasite-infected erythrocyte surface an... 66 5e-09
UniRef100_Q8MTN7 Glutamic acid-rich protein cNBL1700 [Trichinell... 66 5e-09
UniRef100_Q86BT4 Phoshoprotein 300 [Plasmodium falciparum] 65 6e-09
>UniRef100_Q8H0H5 Hypothetical protein A20 [Nicotiana tabacum]
Length = 717
Score = 870 bits (2248), Expect = 0.0
Identities = 460/710 (64%), Positives = 545/710 (75%), Gaps = 14/710 (1%)
Query: 97 VANQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKAS--KDAKSKVKVSP 154
+ + +SD E E KEG E SD DT+ DSVSSQGD EDE VE+AS K +K K
Sbjct: 10 INDHSSDMESEPKEGLEEESDIDTINDSVSSQGDPQIAEDENVERASTVKVSKKLAKSVA 69
Query: 155 SESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKAS 214
+ SS + +SD+K N Q K + S K K PS VT+KN + +NSK +K K+
Sbjct: 70 NNSSPAQRAKSDQKENNSQIKATKSTANKAKTPKKDPSTVTSKNVN-DNSKNMKVHPKSI 128
Query: 215 SESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIE 274
S+SSE DE V EV+ ++ILD S AQS+GS+DE TVN EE+ E EDK ALE KI
Sbjct: 129 SDSSEEGDEKLVKEVEPVDILDETSVSAQSVGSDDE---TVNTEESCEQEDKTALEQKIG 185
Query: 275 EMELRVEKLEEELREVAALEVSLYSVVPEHGSSAHKVHTPARRLSRLYLHASKHWTQNRR 334
+ME R+EKLEEELREVAALE++LYSVV EHGSSAHK+HTPARRLSRLYLHA K+W+Q++R
Sbjct: 186 QMESRLEKLEEELREVAALEIALYSVVSEHGSSAHKLHTPARRLSRLYLHACKYWSQDKR 245
Query: 335 ATIAKNTVSGLILVAKSCGNDVSRLTFWLSNTIVLREIISHAFGNSCQVSPLMRLAESNG 394
AT+AKNTVSGL+LVAKSCGNDV+RLTFWLSN +VLR IIS AFG+SC S L+++ ESNG
Sbjct: 246 ATVAKNTVSGLVLVAKSCGNDVARLTFWLSNAVVLRVIISQAFGSSCSPSSLVKITESNG 305
Query: 395 AGN---GKSASLKWKGFPNGKAGS--GFTQFVEDWQETGTFTSALERVESWIFSRLVESV 449
G K +S KWK P K S QF +DWQET TFT+ALERVESWIFSR+VES+
Sbjct: 306 GGKKTESKISSFKWKTHPGNKQSSKHDLLQFFDDWQETRTFTAALERVESWIFSRIVESI 365
Query: 450 WWQALTPYMQSPAGDFSSNKSFGRVLGPALGDHNQGNFSINLWRYAFEDAFQRLCPLRAG 509
WWQ LTP MQSPA D + KS GR+LGPALGD QGNFSINLW++AF++A +RLCP+RAG
Sbjct: 366 WWQTLTPNMQSPADDPMARKSVGRLLGPALGDQQQGNFSINLWKHAFQEALKRLCPVRAG 425
Query: 510 GHECGCLPVLARMVMEQCIARLDVAMFNAILRESALEIPTDPISDPILDSKVLPIPAGDL 569
HECGCLPVLAR V+EQC+ARLDVAMFNAILRESA EIPTDPISDPI+DSKVLPIPAGDL
Sbjct: 426 SHECGCLPVLARRVIEQCVARLDVAMFNAILRESAHEIPTDPISDPIVDSKVLPIPAGDL 485
Query: 570 SFGSGAQLKNSVGNWSRWLTDMFGMDVEDCVQEYQDSGENDERQGGDGEPKPFVLLNDLS 629
SFGSGAQLKNSVGNWSR LTD+FGMD ED Q + S +D R+GG+ +P+ F LLN LS
Sbjct: 486 SFGSGAQLKNSVGNWSRCLTDLFGMDAEDSGQNDEGSFGDDHRKGGN-QPEHFHLLNALS 544
Query: 630 DLLMLPKDMLIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPDPVPGTVLEALNAETIAE 689
DLLMLPKDML+DR +R EVCPSI+L L+ R+LCNF+PDEFCPDPVPGTVLEALNAE I
Sbjct: 545 DLLMLPKDMLMDRTIRMEVCPSISLPLVKRILCNFSPDEFCPDPVPGTVLEALNAECIIA 604
Query: 690 RRLS-AESVRSFPYAAAAVVYVPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYTSDEEL 748
RRLS +S SFPY AA VVY PP +++VAEKVAE GKS L+R+ SA+QR+GYTSDEEL
Sbjct: 605 RRLSGGDSSGSFPYPAAPVVYKPPVAADVAEKVAEVDGKSLLSRSASAIQRKGYTSDEEL 664
Query: 749 EELDSPLTSIIDKLPLSPTVSANGQDNQKE-HKSYTTTTNARYQLLREVW 797
EE+DSPL IIDK+P SP + NG K+ ++ + +N RY LLREVW
Sbjct: 665 EEIDSPLACIIDKMPSSPASAQNGSRKAKQKEETGSIGSNKRYDLLREVW 714
>UniRef100_Q9SLC7 Hypothetical protein At2g42320 [Arabidopsis thaliana]
Length = 669
Score = 702 bits (1812), Expect = 0.0
Identities = 375/688 (54%), Positives = 489/688 (70%), Gaps = 47/688 (6%)
Query: 114 EVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQ 173
++ T++ K + + ++E ++ + S ++ PSES + R N +
Sbjct: 24 KLQKTNSQKKTEQEKHKDLDTKEESSNISTVASDSTIQSDPSESYETVDVRYLDDDNGVI 83
Query: 174 SKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKE-- 231
+ +D++Q+ +S V +K +N ++ + E D N + +E
Sbjct: 84 AS-ADAHQESDSSS------VVDKTEKEHNLSGSICDLEKDVDEQECKDANIDKDAREGI 136
Query: 232 -IEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELREV 290
++ + ASNGA S GSE+E + E NG + + + E KIE +E R+EKLEEELREV
Sbjct: 137 SADVWEDASNGALSAGSENEAADVT--ENNGGNFEDGSSEEKIERLETRIEKLEEELREV 194
Query: 291 AALEVSLYSVVPEHGSSAHKVHTPARRLSRLYLHASKHWTQNRRATIAKNTVSGLILVAK 350
AALE+SLYSVVP+H SSAHK+HTPARR+SR+Y+HA KH+TQ +RATIA+N+VSGL+LVAK
Sbjct: 195 AALEISLYSVVPDHCSSAHKLHTPARRISRIYIHACKHFTQGKRATIARNSVSGLVLVAK 254
Query: 351 SCGNDVSRLTFWLSNTIVLREIISHAFGNSCQVSPLMRLAESNGAGNGKSASLKWKGFPN 410
SCGNDVSRLTFWLSN I LR+IIS AFG S +++ + ES + +GK +L+WK
Sbjct: 255 SCGNDVSRLTFWLSNIIALRQIISQAFGRS-RITQISEPNESGNSDSGKKTNLRWK---- 309
Query: 411 GKAGSGFTQFVEDWQETGTFTSALERVESWIFSRLVESVWWQALTPYMQSPAGDFSSNKS 470
+GF Q +EDWQET TFT+ALE++E W+FSR+VESVWWQ TP+MQSP D S++KS
Sbjct: 310 ----NGFQQLLEDWQETETFTTALEKIEFWVFSRIVESVWWQVFTPHMQSPEDDSSASKS 365
Query: 471 FGRVLGPALGDHNQGNFSINLWRYAFEDAFQRLCPLRAGGHECGCLPVLARMVMEQCIAR 530
G+++GP+LGD NQG FSI+LW+ AF DA QR+CP+R GHECGCLPVLARMVM++CI R
Sbjct: 366 NGKLMGPSLGDQNQGTFSISLWKNAFRDALQRICPMRGAGHECGCLPVLARMVMDKCIGR 425
Query: 531 LDVAMFNAILRESALEIPTDPISDPILDSKVLPIPAGDLSFGSGAQLKNSVGNWSRWLTD 590
DVAMFNAILRES +IPTDP+SDPILDSKVLPIPAGDLSFGSGAQLKN++GNWSR LT+
Sbjct: 426 FDVAMFNAILRESEHQIPTDPVSDPILDSKVLPIPAGDLSFGSGAQLKNAIGNWSRCLTE 485
Query: 591 MFGMDVEDCVQEYQDSGENDERQGGDGEPKPFVLLNDLSDLLMLPKDMLIDRHVREEVCP 650
MFGM+ +D + + + E+D E K FVLLN+LSDLLMLPKDML++ +REE+CP
Sbjct: 486 MFGMNSDDSSAKEKRNSEDDH-----VESKAFVLLNELSDLLMLPKDMLMEISIREEICP 540
Query: 651 SITLSLIIRVLCNFTPDEFCPDPVPGTVLEALN-AETIAERRLSAESVRSFPYAAAAVVY 709
SI+L LI R+LCNFTPDEFCPD VPG VLE LN AE+I +R+LS SFPYAA++V Y
Sbjct: 541 SISLPLIKRILCNFTPDEFCPDQVPGAVLEELNAAESIGDRKLSE---ASFPYAASSVSY 597
Query: 710 VPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYTSDEELEELDSPLTSIIDKLPLSPTVS 769
+PPS+ ++AEKVAEA K L+RNVS +QR+GYTSDEELEELDSPLTSI+DK
Sbjct: 598 MPPSTMDIAEKVAEASAK--LSRNVSMIQRKGYTSDEELEELDSPLTSIVDK-------- 647
Query: 770 ANGQDNQKEHKSYTTTTNARYQLLREVW 797
+ + T+NARY+LLR+VW
Sbjct: 648 -------ASDFTGSATSNARYKLLRQVW 668
Score = 51.6 bits (122), Expect = 9e-05
Identities = 42/139 (30%), Positives = 69/139 (49%), Gaps = 16/139 (11%)
Query: 14 QTKGSRRTERREDKLHQDNSSK-TLNEKGTESKTPQDRSAANNLVGDSNTASENSETYEN 72
Q+K S ++ R++ KL + NS K T EK + T ++ S + + DS S+ SE+YE
Sbjct: 11 QSKRSTKSVRKDQKLQKTNSQKKTEQEKHKDLDTKEESSNISTVASDSTIQSDPSESYET 70
Query: 73 VVIDYVDDVN---RSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQG 129
V + Y+DD N S +A E ++ +S +K +KE N S D KD
Sbjct: 71 VDVRYLDDDNGVIASADAHQE-------SDSSSVVDKTEKEHNLSGSICDLEKDV----- 118
Query: 130 DSFTNEDEKVEKASKDAKS 148
D +D ++K +++ S
Sbjct: 119 DEQECKDANIDKDAREGIS 137
>UniRef100_Q6Z3T4 Hypothetical protein OSJNBa0025J22.9 [Oryza sativa]
Length = 687
Score = 697 bits (1800), Expect = 0.0
Identities = 384/705 (54%), Positives = 483/705 (68%), Gaps = 46/705 (6%)
Query: 112 NGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKT-N 170
NGEV D +G E +V SK K K + S R K R+ N
Sbjct: 6 NGEVRD---------EKGVGSDYEPARVSGVSK--KLMRKDTRENSPRMAKSSGSRQVQN 54
Query: 171 KLQSKVSDSNQKKPMNSTKGPSRVTN--KNTSSNNSKPVKAPVKASSESSEGVDE----- 223
KLQ K S++ Q + + P +V N K+ S V+ P +A SE SE D+
Sbjct: 55 KLQHKASNNIQSR----SPKPRKVVNAAKSAEVRRSDTVRVPSRAPSELSEETDDIVSEA 110
Query: 224 ------NHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEME 277
E KEI++LD A + QS G++DE E EE ++K + + EE++
Sbjct: 111 GTVDDSKGNEEAKEIDVLDEAPHCDQSTGTDDEIPEI---EEKIVDDEKPVVYQRNEELQ 167
Query: 278 LRVEKLEEELREVAALEVSLYSVVPEHGSSAHKVHTPARRLSRLYLHASKHWTQNRRATI 337
+++KLE+ELREVAALEVSLYSV+PEHGSSAHK+HTPARRLSR+Y+HASK W+ ++ A++
Sbjct: 168 SKIDKLEQELREVAALEVSLYSVLPEHGSSAHKLHTPARRLSRMYIHASKFWSSDKIASV 227
Query: 338 AKNTVSGLILVAKSCGNDVSRLTFWLSNTIVLREIISHAFGNSCQVSPLMRLAESNGAG- 396
AK+TVSGL+LVAKSC ND SRLTFWLSNT+VLREII+ G SCQ S + NG+
Sbjct: 228 AKSTVSGLVLVAKSCSNDASRLTFWLSNTVVLREIIAQTIGISCQSSSTITAINMNGSAK 287
Query: 397 --NGKSASLKWKGFPNGKAGSGFT--QFVEDWQETGTFTSALERVESWIFSRLVESVWWQ 452
+G+S + W +GK + FT Q +DW ET T +ALE++ESWIFSR+VE+VWWQ
Sbjct: 288 SLDGRSMPMLWTNSSSGKQ-TKFTGMQVPDDWHETSTLLAALEKIESWIFSRIVETVWWQ 346
Query: 453 ALTPYMQSPAGDFSSNKSFGRVLGPALGDHNQGNFSINLWRYAFEDAFQRLCPLRAGGHE 512
ALTP+MQ+P S+ K+ GRVLGP+LGD QG FS+NLW+ AF DAF R+CPLRAGGHE
Sbjct: 347 ALTPHMQTPVEGSSTPKT-GRVLGPSLGDQQQGTFSVNLWKAAFHDAFNRICPLRAGGHE 405
Query: 513 CGCLPVLARMVMEQCIARLDVAMFNAILRESALEIPTDPISDPILDSKVLPIPAGDLSFG 572
CGCLPVLA++VMEQC+ RLDVAMFNAILRESA EIPTDPISDPI+D KVLPIPAGDLSFG
Sbjct: 406 CGCLPVLAKLVMEQCVGRLDVAMFNAILRESASEIPTDPISDPIVDPKVLPIPAGDLSFG 465
Query: 573 SGAQLKNSVGNWSRWLTDMFGMDVEDCVQEYQDSGENDERQGGDGEPKPFVLLNDLSDLL 632
SGAQLKNS+GNWSRWLTD FG+D +D ++ D+G ER E K F LLN+LSDLL
Sbjct: 466 SGAQLKNSIGNWSRWLTDNFGIDADDSEEDGTDTG--SERSA--AESKSFQLLNELSDLL 521
Query: 633 MLPKDMLIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPDPVPGTVLEALNAETIAERRL 692
MLPKDMLI++ +R+E+CPSI L L+ R+LCNFTPDEFCPDPVP VLE LN+E++ ER
Sbjct: 522 MLPKDMLIEKSIRKEICPSIGLPLVTRILCNFTPDEFCPDPVPSIVLEELNSESLLERCT 581
Query: 693 SAESVRSFPYAAAAVVYVPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYTSDEELEELD 752
+ +FP AA VVY PPS +VAEKVA+ GG + L R S VQRRGYTSD++L++LD
Sbjct: 582 DKSATSAFPCIAAPVVYRPPSLLDVAEKVADTGGNAKLDRRASMVQRRGYTSDDDLDDLD 641
Query: 753 SPLTSIIDKLPLSPTVSANGQDNQKEHKSYTTTTNARYQLLREVW 797
SPL S+IDK +P + + G + + + NARY LREVW
Sbjct: 642 SPLASLIDK--SAPPLLSKGSAHFTAQRG-VSMENARYTFLREVW 683
>UniRef100_Q9SVZ2 Hypothetical protein F15B8.30 [Arabidopsis thaliana]
Length = 670
Score = 667 bits (1722), Expect = 0.0
Identities = 371/697 (53%), Positives = 470/697 (67%), Gaps = 62/697 (8%)
Query: 104 TEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKE 163
+E+E+ + E S+ V D ++Q ++D V +S+ K E+ GL
Sbjct: 35 SEQERLKATKEESNVSVVVDDTTTQSKLSDDDDHAVNDSSE------KTEKEETINGLAC 88
Query: 164 RSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDE 223
+ + K +SK D+ + +S P E+ EGV+
Sbjct: 89 DDEDEEEKEESKELDAIAHEKTDSVSSP------------------------ETCEGVNV 124
Query: 224 NHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKL 283
+ V +E+ D ASNG S GSE+E + EN E ED+ L+ +E +E RVEKL
Sbjct: 125 DKV-----VEVWDDASNGGLSGGSENEAGDVKEKNENFE-EDEEMLKQMVETLETRVEKL 178
Query: 284 EEELREVAALEVSLYSVVPEHGSSAHKVHTPARRLSRLYLHASKHWTQNRRATIAKNTVS 343
EEELREVAALE+SLYSVVP+H SSAHK+HTPARR+SR+Y+HA KHW+Q +RAT+A+N+VS
Sbjct: 179 EEELREVAALEISLYSVVPDHSSSAHKLHTPARRISRIYIHACKHWSQGKRATVARNSVS 238
Query: 344 GLILVAKSCGNDVSRLTFWLSNTIVLREIISHAFGNSCQVSPLMRLAESNGAGNGKSASL 403
GLIL AKSCGNDVSRLTFWLSN I LREII AFG + S + SNG+ + +
Sbjct: 239 GLILAAKSCGNDVSRLTFWLSNIISLREIILQAFGKTSVPSHFTETSASNGSEHNVLGKV 298
Query: 404 KWKGFPNGKAGSGFTQFVEDWQETGTFTSALERVESWIFSRLVESVWWQALTPYMQSPAG 463
+ K K +GF Q EDWQE+ TFT+ALE+VE WIFSR+VESVWWQ TP+MQSP
Sbjct: 299 RRKKNQWTKQSNGFKQVFEDWQESQTFTAALEKVEFWIFSRIVESVWWQVFTPHMQSP-- 356
Query: 464 DFSSNKSFGRVLGPALGDHNQGNFSINLWRYAFEDAFQRLCPLRAGGHECGCLPVLARMV 523
++ G+ LGD QG+FSI+LW+ AF+ RLCP+R HECGCLP+LA+MV
Sbjct: 357 -----ENGGKTKEHILGDIEQGSFSISLWKNAFKVTLSRLCPMRGARHECGCLPILAKMV 411
Query: 524 MEQCIARLDVAMFNAILRESALEIPTDPISDPILDSKVLPIPAGDLSFGSGAQLKNSVGN 583
ME+CIAR+DVAMFNAILRES +IPTDP+SDPILDSKVLPI +G+LSFGSGAQLKN++GN
Sbjct: 412 MEKCIARIDVAMFNAILRESEHQIPTDPVSDPILDSKVLPILSGNLSFGSGAQLKNAIGN 471
Query: 584 WSRWLTDMFGMDVEDCVQEYQDSGENDERQGGDGEPKPFVLLNDLSDLLMLPKDMLIDRH 643
WSR L +MF ++ D V+ END + K F LLN+LSDLLMLPKDML+DR
Sbjct: 472 WSRCLAEMFSINTRDSVE------ENDPIE----SEKSFSLLNELSDLLMLPKDMLMDRS 521
Query: 644 VREEVCPSITLSLIIRVLCNFTPDEFCPDPVPGTVLEALNAETIAERRLSAESVRSFPYA 703
REEVCPSI+L+LI R+LCNFTPDEFCPD VPG VLE LN E+I+E++LS SFPYA
Sbjct: 522 TREEVCPSISLALIKRILCNFTPDEFCPDDVPGAVLEELNNESISEQKLSGV---SFPYA 578
Query: 704 AAAVVYVPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYTSDEELEELDSPLTSIIDKLP 763
A+ V Y PPSS+N VAE G S ++RNVS +QR+GYTSD+ELEELDSPLTSII+ +
Sbjct: 579 ASPVSYTPPSSTN----VAEVGDISRMSRNVSMIQRKGYTSDDELEELDSPLTSIIENVS 634
Query: 764 LSPTVSANGQD-NQKEHKSYTTTTNARYQLLREVWSM 799
LSP +SA G++ Q+ K T +RY+LLREVWSM
Sbjct: 635 LSP-ISAQGRNVKQEAEKIGPGVTISRYELLREVWSM 670
>UniRef100_Q9SGJ0 F28J7.14 protein [Arabidopsis thaliana]
Length = 921
Score = 512 bits (1318), Expect = e-143
Identities = 323/772 (41%), Positives = 434/772 (55%), Gaps = 69/772 (8%)
Query: 58 GDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKE------- 110
G+S +A N E Y+ I + D + S + + + + +N E++E
Sbjct: 183 GESVSALMNEEYYKEAEIASITDDDISSHSSLTVSSSTLESNGGFSVRTEEEEHERINKN 242
Query: 111 --GNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKD--AKSKVKVSPSESSRGLKERSD 166
GNG + D + V+ +S + +P+ S GL+ ++
Sbjct: 243 PRGNGHERSKSVSESRQRQIADQIPSRSSSVDLSSVFHLPEGISDSAPNTSLSGLEHCAN 302
Query: 167 ---RKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDE 223
TN+ SK++ + Q + P ++ N + +++ V + S + E + E
Sbjct: 303 VFITDTNE-SSKLASNGQHNNGEAKSVPLQIDNLSENASPRASVNSQDLTSDQEPESIVE 361
Query: 224 NHVLEVKEIEI-LD-GASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVE 281
+VK + LD SN SL SE + N H+ LE KI+ +E RV+
Sbjct: 362 KS-RKVKSVRSSLDINRSNSRLSLFSE---RKEAKVYPNSTHD--TTLESKIKNLESRVK 415
Query: 282 KLEEELREVAALEVSLYSVVPEHGSSAHKVHTPARRLSRLYLHASKHWTQNRRATIAKNT 341
KLE EL E AA+E +LYSVV EHGSS+ KVH PARRL RLYLHA + +RRA A++
Sbjct: 416 KLEGELCEAAAIEAALYSVVAEHGSSSSKVHAPARRLLRLYLHACRETHLSRRANAAESA 475
Query: 342 VSGLILVAKSCGNDVSRLTFWLSNTIVLREIISHAFGN-----SCQVSPLMRLAESNGAG 396
VSGL+LVAK+CGNDV RLTFWLSNTIVLR IIS S P + AE
Sbjct: 476 VSGLVLVAKACGNDVPRLTFWLSNTIVLRTIISDTSAEEELPVSAGPGPRKQKAERE--- 532
Query: 397 NGKSASLKWKGFPNGKAGSGFTQFVEDWQETGTFTSALERVESWIFSRLVESVWWQALTP 456
K +SLKWK P K + W + TF +ALE+VE+WIFSR+VES+WWQ LTP
Sbjct: 533 TEKRSSLKWKDSPLSKKD---IKSFGAWDDPVTFITALEKVEAWIFSRVVESIWWQTLTP 589
Query: 457 YMQSPAGDF---------SSNKSFGRVLGPALGDHNQGNFSINLWRYAFEDAFQRLCPLR 507
MQS A +S K+FGR P+ + G+FS+ LW+ AF +A +RLCPLR
Sbjct: 590 RMQSSAASTREFDKGNGSASKKTFGRT--PSSTNQELGDFSLELWKKAFREAHERLCPLR 647
Query: 508 AGGHECGCLPVLARMVMEQCIARLDVAMFNAILRESALEIPTDPISDPILDSKVLPIPAG 567
GHECGCLP+ AR++MEQC+ARLDVAMFNAILR+S PTDP+SDPI D +VLPIP+
Sbjct: 648 GSGHECGCLPIPARLIMEQCVARLDVAMFNAILRDSDDNFPTDPVSDPIADLRVLPIPSR 707
Query: 568 DLSFGSGAQLKNSVGNWSRWLTDMFGMDVEDCVQEYQDSGENDERQGGDGEPKPFVLLND 627
SFGSGAQLKNS+GNWSRWLTD+FG+D E D +DE + K F LL
Sbjct: 708 TSSFGSGAQLKNSIGNWSRWLTDLFGIDDE-------DDDSSDENSYVEKSFKTFNLLKA 760
Query: 628 LSDLLMLPKDMLIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPDPVPGTVLEALNAETI 687
LSDL+MLPKDML++ VR+EVCP LI RVL NF PDEFCPDPVP VL++L +E
Sbjct: 761 LSDLMMLPKDMLLNSSVRKEVCPMFGAPLIKRVLNNFVPDEFCPDPVPDAVLKSLESEEE 820
Query: 688 AERRLSAESVRSFPYAAAAVVYVPPSSSNVAEKVAEAGGKS--HLARNVSAVQRRGYTSD 745
AE+ + + S+P A + VY PPS ++++ + G L+R S++ R+ YTSD
Sbjct: 821 AEKSI----ITSYPCTAPSPVYCPPSRTSISTIIGNFGQPQAPQLSRIRSSITRKAYTSD 876
Query: 746 EELEELDSPLTSIIDKLPLSPTVSANGQDNQKEHKSYTTTTNARYQLLREVW 797
+EL+EL SPL ++ + Q K+ + RYQLLRE W
Sbjct: 877 DELDELSSPLAVVVLQ-----------QAGSKKINNGDADETIRYQLLRECW 917
>UniRef100_Q9FL52 Gb|AAD23715.1 [Arabidopsis thaliana]
Length = 657
Score = 482 bits (1240), Expect = e-134
Identities = 288/602 (47%), Positives = 367/602 (60%), Gaps = 61/602 (10%)
Query: 195 TNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHET 254
TN + S + + +P + SE V + +E + D + S+ + + +ET
Sbjct: 115 TNNGSVSWSQCELFSPEEKKSERPSMVSKIDSTPFEEGKEDDEFEDALNSVHNNESDNET 174
Query: 255 VNAEENGEHEDKAALELKIEEMELRVEKLEEELREVAALEVSLYSVVPEHGSSAHKVHTP 314
+ +E + + L KIE ME R+EKLEEELREVAALE+SLYSV PEHGSS+HK+H P
Sbjct: 175 LVYKEKKRSDVEKVLAQKIETMEARIEKLEEELREVAALEMSLYSVFPEHGSSSHKLHKP 234
Query: 315 ARRLSRLYLHASKHWTQNRRATIAKNTVSGLILVAKSCGNDVSRLTFWLSNTIVLREIIS 374
AR LSRLY A K+ ++N+ ++ KN VSGL L+ KSCG+DVSRLT+WLSNT++LREIIS
Sbjct: 235 ARNLSRLYALARKNQSENKIISVTKNIVSGLSLLLKSCGSDVSRLTYWLSNTVMLREIIS 294
Query: 375 HAFGNSCQVSPLMRLAESNGAGNGKSASLKWKGFPNGKAGSGFTQFVEDWQETGTFTSAL 434
FG+S + NG + K EDW + T +AL
Sbjct: 295 LDFGSS----------KLNGLNSLK----------------------EDWGDVRTLIAAL 322
Query: 435 ERVESWIFSRLVESVWWQALTPYMQSPAGDFSSNKSFGRVLGPALGDHNQGNFSINLWRY 494
RVES F++ VES+W Q + +M D + + G PA D Q +FS+NLW+
Sbjct: 323 RRVESCFFTQAVESIWSQVMMVHMIPQGVDSTMGEMIGNFSEPATCDRLQESFSVNLWKE 382
Query: 495 AFEDAFQRLCPLRAGGHECGCLPVLARMVMEQCIARLDVAMFNAILRESALEIPTDPISD 554
AFE+A QRLCP++A +CGCL VL RMVMEQCI RLDVAMFNAILRESA IPTD SD
Sbjct: 383 AFEEALQRLCPVQATRRQCGCLHVLTRMVMEQCIVRLDVAMFNAILRESAHHIPTDSASD 442
Query: 555 PILDSKVLPIPAGDLSFGSGAQLKNSVGNWSRWLTDMFGMDVEDCVQEYQDSGENDERQG 614
PI DS+VLPIPAG LSF SG +LKN+V WSR LTD+FG+DVE + Q
Sbjct: 443 PIADSRVLPIPAGVLSFESGVKLKNTVSYWSRLLTDIFGIDVE------------QKMQR 490
Query: 615 GDGEPKPFVLLNDLSDLLMLPKDMLIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPDPV 674
GD KPF LLN+LSDLLMLPK+M +D R+EVCPSI LSLI R++CNFTPDEFCP PV
Sbjct: 491 GDETFKPFHLLNELSDLLMLPKEMFVDSSTRDEVCPSIGLSLIKRIVCNFTPDEFCPYPV 550
Query: 675 PGTVLEALNAETIAERR-LSAESVRSFPYAAAAVVYVPPSSSNVAEKVAEAGGKSHLARN 733
PGTVLE LNA++I E R LS ++ R FP V Y PPS S++ + VAE K L
Sbjct: 551 PGTVLEELNAQSILENRSLSRDTARGFPRQVNPVSYSPPSCSHLTDIVAEFSVKLKL--- 607
Query: 734 VSAVQRRGYTSDEELEELDSPLTSIIDKLPLSPTVSANGQDNQKEHKSYTTTTNARYQLL 793
S + GY+S+E++E SP + K A +DN + TN RY+LL
Sbjct: 608 -SMTHKNGYSSNEKVETPRSPPYYNVIK-------GAVAKDNLN-----LSETNERYRLL 654
Query: 794 RE 795
E
Sbjct: 655 GE 656
>UniRef100_Q9FHS2 Gb|AAF03435.1 [Arabidopsis thaliana]
Length = 628
Score = 436 bits (1122), Expect = e-120
Identities = 276/656 (42%), Positives = 372/656 (56%), Gaps = 87/656 (13%)
Query: 162 KERSDRKTNK--LQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSE 219
+E R++N+ + S D QK+ N+ K K S S P+ P A+ + +
Sbjct: 34 EEERKRQSNQPVIVSAEIDQKQKEDTNAFK-----LKKQFSEIKSGPLSLPPDAAKKQMK 88
Query: 220 GVDENHVLEVKEIEILDGASNGAQSLGSED----ERHETVNAEENGEHEDKAALELK--- 272
L + + G ++LG E ++ +++ +G +D + +
Sbjct: 89 -------LRTNTLAL------GRKTLGMEGIPRLKQLKSIQLHFDGHKDDSSHKKASGVI 135
Query: 273 -----IEEMELRVEKLEEELREVAALEVSLYSVVPEHGSSAHKVHTPARRLSRLYLHASK 327
I + + E LE+EL+E A LE ++YSVV EH SS KVH PARRL+R YLHA K
Sbjct: 136 NKVGLITPQDSKTETLEDELKEAAVLEAAIYSVVAEHTSSMSKVHAPARRLARFYLHACK 195
Query: 328 H--WTQNRRATIAKNTVSGLILVAKSCGNDVSRLTFWLSNTIVLREIISHAFGNSCQVSP 385
++RAT A+ VSGLILV+K+CGNDV RLTFWLSN+IVLR I+S
Sbjct: 196 GNGSDHSKRATAARAAVSGLILVSKACGNDVPRLTFWLSNSIVLRAILSRGME------- 248
Query: 386 LMRLAESNGAGNGKSASLKWKGFPNGKAGSGFTQFVEDWQETGTFTSALERVESWIFSRL 445
K K P KAGS ++W++ F +ALE+ ESWIFSR+
Sbjct: 249 ------------------KMKIVPE-KAGS------DEWEDPRAFLAALEKFESWIFSRV 283
Query: 446 VESVWWQALTPYMQSPA--GDFSSNKSFGRVLGPALGDHNQGNFSINLWRYAFEDAFQRL 503
V+SVWWQ++TP+MQSPA G + S R LG NQG ++I LW+ AF A +RL
Sbjct: 284 VKSVWWQSMTPHMQSPAVKGSIARKVSGKR----RLGHRNQGLYAIELWKNAFRAACERL 339
Query: 504 CPLRAGGHECGCLPVLARMVMEQCIARLDVAMFNAILRESALEIPTDPISDPILDSKVLP 563
CPLR ECGCLP+LA++VMEQ I+RLDVAMFNAILRESA E+PTDP+SDPI D VLP
Sbjct: 340 CPLRGSRQECGCLPMLAKLVMEQLISRLDVAMFNAILRESAGEMPTDPVSDPISDINVLP 399
Query: 564 IPAGDLSFGSGAQLKNSVGNWSRWLTDMFGMDVEDCVQEYQDSGENDERQGGDGEPKPFV 623
IPAG SFG+GAQLKN++G WSRWL D F ED +D ND+ + + F
Sbjct: 400 IPAGKASFGAGAQLKNAIGTWSRWLEDQFEQK-EDKSGRNKDEDNNDKEKPECEHFRLFH 458
Query: 624 LLNDLSDLLMLPKDMLIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPDPVPGTVLEALN 683
LLN L DL+MLP ML D+ R+EVCP++ +I RVL NF PDEF P +P + + LN
Sbjct: 459 LLNSLGDLMMLPFKMLADKSTRKEVCPTLGPPIIKRVLRNFVPDEFNPHRIPRRLFDVLN 518
Query: 684 AETIAERRLSAESVRSFPYAAAAVVYVPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYT 743
+E + E +V FP AA+ VY+ PS+ ++ + E S ++ S+V ++ YT
Sbjct: 519 SEGLTEEDNGCITV--FPSAASPTVYLMPSTDSIKRFIGELNNPS-ISETGSSVFKKQYT 575
Query: 744 SDEELEELDSPLTSIIDKLPLSPTVSANGQDNQKE--HKSYTTTTNARYQLLREVW 797
SD+EL++LD+ + SI SA G N E K Y RYQLLRE+W
Sbjct: 576 SDDELDDLDTSINSIF---------SAPGTTNSSEWMPKGYGRRKTVRYQLLREIW 622
>UniRef100_Q8LF58 Hypothetical protein [Arabidopsis thaliana]
Length = 184
Score = 214 bits (545), Expect = 8e-54
Identities = 119/208 (57%), Positives = 150/208 (71%), Gaps = 26/208 (12%)
Query: 591 MFGMDVEDCVQEYQDSGENDERQGGDGEPKPFVLLNDLSDLLMLPKDMLIDRHVREEVCP 650
MFGM+ +D + + + E+D E K FVLLN+LSDLLMLPKDML++ +REE+CP
Sbjct: 1 MFGMNSDDSSAKEKRNSEDDHV-----ESKAFVLLNELSDLLMLPKDMLMEISIREEICP 55
Query: 651 SITLSLIIRVLCNFTPDEFCPDPVPGTVLEALN-AETIAERRLSAESVRSFPYAAAAVVY 709
SI+L LI R+LCNFTPDEFCPD VPG VLE LN AE+I +R+LS SFPYAA++V Y
Sbjct: 56 SISLPLIKRILCNFTPDEFCPDQVPGAVLEELNAAESIGDRKLSE---ASFPYAASSVSY 112
Query: 710 VPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYTSDEELEELDSPLTSIIDKLPLSPTVS 769
+PPS+ ++AEKVAEA K L+RNVS +QR+GYTSDEELEELDSPLTSI+DK
Sbjct: 113 MPPSTMDIAEKVAEASAK--LSRNVSMIQRKGYTSDEELEELDSPLTSIVDK-------- 162
Query: 770 ANGQDNQKEHKSYTTTTNARYQLLREVW 797
+ + T+NARY+LLR+VW
Sbjct: 163 -------ASDFTGSATSNARYKLLRQVW 183
>UniRef100_Q9DGL1 Retinitis pigmentosa GTPase regulator-like protein [Fugu rubripes]
Length = 791
Score = 75.1 bits (183), Expect = 8e-12
Identities = 66/294 (22%), Positives = 130/294 (43%), Gaps = 18/294 (6%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTL--NEKGTESKTPQDRSAANNLVGD 59
+E E+R ++ S+++ ++ ER+E + ++ + +E+G++ ++ + +++ + G+
Sbjct: 389 EEEEERSSTAESESEDEKKGERKERDVEEEEGESEVEESEEGSDHESERKKASESEEEGE 448
Query: 60 SNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQA-SDTEKEQKEGNGEVSDT 118
A SE E D + +EE+ AE + A + S TEKE + E S T
Sbjct: 449 EEEADSLSEEEEG------DSKSDAEESDAESETEGEGAEEEQSSTEKEGESNEEEQSST 502
Query: 119 DTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSD 178
+ K+ + + S T ++E E+ + + + + E S KE S+ + K +
Sbjct: 503 E--KEESNEEEQSSTEKEESNEEEQSSTEKEGESNEEEQSSTEKEESNEEEQSSTEKEEE 560
Query: 179 SNQKKPMNSTKGPSRVTNKN---TSSNNSKPVKAPVKASSESSEGVDENHVLEVKEI--- 232
SN+++ ++ K S + + SN + + K E G ++ E E
Sbjct: 561 SNEEEQSSTEKEESNEEEEEEEESESNEEESEEEDQKDEEEEETGEEDEEEEEESEEQNQ 620
Query: 233 -EILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEE 285
E + +G + E+E E EE E E+ E EE RVE+ E+
Sbjct: 621 EEEAEEEEDGETDVQEEEEEEEETEKEEEDEEEETEKEEEDEEEETDRVEEEED 674
Score = 67.8 bits (164), Expect = 1e-09
Identities = 60/285 (21%), Positives = 122/285 (42%), Gaps = 22/285 (7%)
Query: 4 TEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTA 63
TEK S + + + E E+ + SS E E ++ ++ +N S+T
Sbjct: 488 TEKEGESNEEEQSSTEKEESNEE----EQSSTEKEESNEEEQSSTEKEGESNEEEQSSTE 543
Query: 64 SENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKD 123
E S E + ++ N E++ E + + + E+E+ E N E S+ + KD
Sbjct: 544 KEESNEEEQSSTEKEEESNEEEQSSTEKE-----ESNEEEEEEEESESNEEESEEEDQKD 598
Query: 124 SVSSQ-GDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQK 182
+ G+ E+E+ E+ +++ +++ + E+ +E + +T K + + +K
Sbjct: 599 EEEEETGEEDEEEEEESEEQNQEEEAEEE-EDGETDVQEEEEEEEETEKEEEDEEEETEK 657
Query: 183 KPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASS-ESSEGVDE----NHVLEVKEIEILDG 237
+ + + RV + + N + + + E S+G +E + E +E E
Sbjct: 658 EEEDEEEETDRVEEEEDAEENEEEEEEESNSEEDEESDGEEEEDEESDSEEDEEEESESE 717
Query: 238 ASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEK 282
G +S E+E E+ + EE GE D+ EE E++ +K
Sbjct: 718 EEEGEESESEEEEGEESESEEEEGEESDRE------EEEEIKKKK 756
Score = 66.6 bits (161), Expect = 3e-09
Identities = 68/308 (22%), Positives = 131/308 (42%), Gaps = 34/308 (11%)
Query: 3 ETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQ-DRSAANNLVGDSN 61
E +++++ + ++ + E ED+ + +S ++E+ E+ T D+++ GDS+
Sbjct: 297 EEDEKESIQGTEEEEEEEEEEEEDQKEEQKTSADVSEEEEENVTQSSDKASDEEKEGDSD 356
Query: 62 TASENSETYENVVIDYVDDVNRSE------EALAEMKVNAMVANQASDTEK--------- 106
T E+ +V D + +RSE E E + + A S+ EK
Sbjct: 357 TLHPEDESQSDVE-DEEEGESRSEGQEEEEEEEEEEEERSSTAESESEDEKKGERKERDV 415
Query: 107 EQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSD 166
E++EG EV + S +G +E +K ++ ++ + + S SE G +SD
Sbjct: 416 EEEEGESEVEE--------SEEGSDHESERKKASESEEEGEEEEADSLSEEEEG-DSKSD 466
Query: 167 RKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHV 226
+ + +S+ ++ +ST+ + SS + ++S+E E +E
Sbjct: 467 AEESDAESETEGEGAEEEQSSTEKEGESNEEEQSSTEKEESNEEEQSSTEKEESNEEEQS 526
Query: 227 LEVKEIEILDGASNGAQSLGS-EDERHETVNAEENGEHEDKAA-------LELKIEEMEL 278
KE E + + + S E+E+ T EE+ E E + E + EE E
Sbjct: 527 STEKEGESNEEEQSSTEKEESNEEEQSSTEKEEESNEEEQSSTEKEESNEEEEEEEESES 586
Query: 279 RVEKLEEE 286
E+ EEE
Sbjct: 587 NEEESEEE 594
Score = 58.2 bits (139), Expect = 1e-06
Identities = 55/292 (18%), Positives = 116/292 (38%), Gaps = 13/292 (4%)
Query: 3 ETEKRKASRNSQTKGSRRTER-----REDKLHQDNSSKTLNEKGTESKTPQDRSAANNLV 57
+++ ++ S+T+G E +E + +++ S T E+ E + +N
Sbjct: 464 KSDAEESDAESETEGEGAEEEQSSTEKEGESNEEEQSSTEKEESNEEEQSSTEKEESNEE 523
Query: 58 GDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSD 117
S+T E E ++ N E++ E + + Q+S TEKE+ E +
Sbjct: 524 EQSSTEKEGESNEEEQSSTEKEESNEEEQSSTEKEEESNEEEQSS-TEKEESNEEEEEEE 582
Query: 118 TDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVS 177
+ S + D +DE+ E+ ++ + + + S ++ +E + + + +
Sbjct: 583 ESESNEEESEEEDQ---KDEEEEETGEEDEEEEEESEEQNQE--EEAEEEEDGETDVQEE 637
Query: 178 DSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDG 237
+ +++ + T K + + + +E +E +E ++ E
Sbjct: 638 EEEEEETEKEEEDEEEETEKEEEDEEEETDRVEEEEDAEENEEEEEEESNSEEDEESDGE 697
Query: 238 ASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELRE 289
+S EDE E+ + EE GE + E + EE E E+ EE RE
Sbjct: 698 EEEDEESDSEEDEEEESESEEEEGEESESE--EEEGEESESEEEEGEESDRE 747
Score = 50.1 bits (118), Expect = 3e-04
Identities = 44/244 (18%), Positives = 95/244 (38%), Gaps = 16/244 (6%)
Query: 3 ETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNT 62
E E+ + + + + +E++ +++ S T E+ E + ++ S +N +
Sbjct: 536 EEEQSSTEKEESNEEEQSSTEKEEESNEEEQSSTEKEESNEEEEEEEESESNEEESEEED 595
Query: 63 AS--ENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDT 120
E ET E + + +++E AE + + Q + E+E+ E E + +T
Sbjct: 596 QKDEEEEETGEEDEEEEEESEEQNQEEEAEEEEDGETDVQEEEEEEEETEKEEEDEEEET 655
Query: 121 VK---------DSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNK 171
K D V + D+ NE+E+ E+++ + + E E + + ++
Sbjct: 656 EKEEEDEEEETDRVEEEEDAEENEEEEEEESNSEEDEESDGEEEEDEESDSEEDEEEESE 715
Query: 172 LQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNS----KPVKAPVKASSESSEGVDENHVL 227
+ + + ++ + + S S K K PVK + GV +
Sbjct: 716 SEEEEGEESESEEEEGEESESEEEEGEESDREEEEEIKKKKRPVKLGRQHRSGV-KGQAA 774
Query: 228 EVKE 231
E KE
Sbjct: 775 EPKE 778
Score = 48.9 bits (115), Expect = 6e-04
Identities = 57/298 (19%), Positives = 117/298 (39%), Gaps = 16/298 (5%)
Query: 7 RKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASEN 66
+ +S + ++ R + E + H + S + + P S S + + N
Sbjct: 238 KSSSPSEDSQSVLRAQSEESEAHGASFSNK-SAVSINIQPPASPSETEGSQDKSESVTAN 296
Query: 67 SETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQ-KEGNGEVSDTDTVKDSV 125
E + + ++ EE + K + S+ E+E + + + SD + DS
Sbjct: 297 EEDEKESIQGTEEEEEEEEEEEEDQKEEQKTSADVSEEEEENVTQSSDKASDEEKEGDSD 356
Query: 126 SSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPM 185
+ + + D + E+ +++S+ + E +ERS ++ + + +++ +
Sbjct: 357 TLHPEDESQSDVEDEEEG-ESRSEGQEEEEEEEEEEEERSSTAESESEDEKKGERKERDV 415
Query: 186 NSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQS- 244
+G S V S++ ++ K +SES E +E + E E D S+ +S
Sbjct: 416 EEEEGESEVEESEEGSDH----ESERKKASESEEEGEEEEADSLSEEEEGDSKSDAEESD 471
Query: 245 -------LGSEDERHET-VNAEENGEHEDKAALELKIEEMELRVEKLEEELREVAALE 294
G+E+E+ T E N E + E EE + EK E E ++ E
Sbjct: 472 AESETEGEGAEEEQSSTEKEGESNEEEQSSTEKEESNEEEQSSTEKEESNEEEQSSTE 529
>UniRef100_UPI000024C743 UPI000024C743 UniRef100 entry
Length = 463
Score = 74.7 bits (182), Expect = 1e-11
Identities = 66/298 (22%), Positives = 123/298 (41%), Gaps = 11/298 (3%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
+E+E+ K ++ TK S E RE++ ++S +N+ T+ + + DS+
Sbjct: 98 EESEEEKTVNDTVTKASSENEEREEEEKSNDSE--VNDTMTKHSSEDEEKEKEEKSSDSS 155
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
AS E E D + EE E K + V +AS ++E++E NG+ +
Sbjct: 156 NASSEDEEEEEKNGDSAKASSEDEEE--EEKSGSSV--KASSEDEEEEEENGDSFKASSK 211
Query: 122 KDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQ 181
++S S S N +E+ +K+ +A ++ E + +E + + + +D
Sbjct: 212 EESGDSANASNVNSEEEEQKSKSEATDDLESEEEEDNEDNEEEKSETSASTEDESNDDED 271
Query: 182 KKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNG 241
K + + + N+ S + + + SE E E+ E + E +
Sbjct: 272 KSETSQEEESGEESEGNSESRSERSEEDDEDEESEKEESEVESEAEEDETGENTSEEDDE 331
Query: 242 AQSLGSEDERHETVNAE-----ENGEHEDKAALELKIEEMELRVEKLEEELREVAALE 294
Q SEDE AE G+ ED++ E ++ E + EEE E + +E
Sbjct: 332 EQESSSEDEEKSESEAEGEEEESEGDQEDESDEETSDKDEEEEESENEEEEEEESKVE 389
Score = 67.4 bits (163), Expect = 2e-09
Identities = 63/307 (20%), Positives = 119/307 (38%), Gaps = 25/307 (8%)
Query: 4 TEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTA 63
T RK + + + R++ + S N + + S ++ VG+++
Sbjct: 21 TYNRKEEESDEEESEGSVNERDETEDVEESDGLGNRRDKSEEVEDSDSKTHDKVGENDDD 80
Query: 64 SENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEK---EQKEGNGEVSDTDT 120
SE E + ++ D + EE+ E VN V +S+ E+ E+K + EV+DT T
Sbjct: 81 SEGDEDDDTMI----KDSSEEEESEEEKTVNDTVTKASSENEEREEEEKSNDSEVNDTMT 136
Query: 121 VKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSN 180
S + + + +S+D + + K S + E + K+ S+
Sbjct: 137 KHSSEDEEKEKEEKSSDSSNASSEDEEEEEKNGDSAKASSEDEEEEEKSGSSVKASSEDE 196
Query: 181 QKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVD-----ENHVLEVKEIEIL 235
+++ N + ++ S N+ V + + SE D E E E E
Sbjct: 197 EEEEENGDSFKASSKEESGDSANASNVNSEEEEQKSKSEATDDLESEEEEDNEDNEEEKS 256
Query: 236 DGASNGAQSLGSEDERHETVNAEENGE-------------HEDKAALELKIEEMELRVEK 282
+ +++ ++++ ET EE+GE ED E + EE E+ E
Sbjct: 257 ETSASTEDESNDDEDKSETSQEEESGEESEGNSESRSERSEEDDEDEESEKEESEVESEA 316
Query: 283 LEEELRE 289
E+E E
Sbjct: 317 EEDETGE 323
Score = 66.6 bits (161), Expect = 3e-09
Identities = 58/296 (19%), Positives = 122/296 (40%), Gaps = 14/296 (4%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
++ EK K ++S + + + E++ + D++ + ++ E K+ A++ D
Sbjct: 141 EDEEKEKEEKSSDSSNASSEDEEEEEKNGDSAKASSEDEEEEEKSGSSVKASSE---DEE 197
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
EN ++++ + D + +E + + D E E++E N D +
Sbjct: 198 EEEENGDSFKASSKEESGDSANASNVNSEEEEQKSKSEATDDLESEEEEDN---EDNEEE 254
Query: 122 KDSVSSQGDSFTNEDEKVEKASKDAKSKVKV---SPSESSRGLKERSDRKTNKLQSKVSD 178
K S+ + +N+DE + S++ +S + S S S R ++ D ++ K +S+V
Sbjct: 255 KSETSASTEDESNDDEDKSETSQEEESGEESEGNSESRSERSEEDDEDEESEKEESEVES 314
Query: 179 SNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEV-----KEIE 233
++ + +SS + + ++ + E SEG E+ E +E E
Sbjct: 315 EAEEDETGENTSEEDDEEQESSSEDEEKSESEAEGEEEESEGDQEDESDEETSDKDEEEE 374
Query: 234 ILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELRE 289
+ + E+E E ++ HE K E + E L E+ EE+ E
Sbjct: 375 ESENEEEEEEESKVEEEEEEXXXXSDDESHESKQNGEAEEHEEYLDDEEEEEDEEE 430
Score = 37.4 bits (85), Expect = 1.8
Identities = 27/144 (18%), Positives = 61/144 (41%), Gaps = 4/144 (2%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
+E+EK ++ S+ + E ++ ++ S + +E+ +ES+ + + GD
Sbjct: 303 EESEKEESEVESEAEEDETGENTSEEDDEEQESSSEDEEKSESEAEGEEEESE---GDQE 359
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
S+ ET + + + EE ++++ SD E + + NGE + +
Sbjct: 360 DESDE-ETSDKDEEEEESENEEEEEEESKVEEEEEEXXXXSDDESHESKQNGEAEEHEEY 418
Query: 122 KDSVSSQGDSFTNEDEKVEKASKD 145
D + D E+E+ + K+
Sbjct: 419 LDDEEEEEDEEEEEEEEEHQRGKE 442
>UniRef100_UPI0000247AAF UPI0000247AAF UniRef100 entry
Length = 796
Score = 74.7 bits (182), Expect = 1e-11
Identities = 66/298 (22%), Positives = 123/298 (41%), Gaps = 11/298 (3%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
+E+E+ K ++ TK S E RE++ ++S +N+ T+ + + DS+
Sbjct: 491 EESEEEKTVNDTVTKASSENEEREEEEKSNDSE--VNDTMTKHSSEDEEKEKEEKSSDSS 548
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
AS E E D + EE E K + V +AS ++E++E NG+ +
Sbjct: 549 NASSEDEEEEEKNGDSAKASSEDEEE--EEKSGSSV--KASSEDEEEEEENGDSFKASSK 604
Query: 122 KDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQ 181
++S S S N +E+ +K+ +A ++ E + +E + + + +D
Sbjct: 605 EESGDSANASNVNSEEEEQKSKSEATDDLESEEEEDNEDNEEEKSETSASTEDESNDDED 664
Query: 182 KKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNG 241
K + + + N+ S + + + SE E E+ E + E +
Sbjct: 665 KSETSQEEESGEESEGNSESRSERSEEDDEDEESEKEESEVESEAEEDETGENTSEEDDE 724
Query: 242 AQSLGSEDERHETVNAE-----ENGEHEDKAALELKIEEMELRVEKLEEELREVAALE 294
Q SEDE AE G+ ED++ E ++ E + EEE E + +E
Sbjct: 725 EQESSSEDEEKSESEAEGEEEESEGDQEDESDEETSDKDEEEEESENEEEEEEESKVE 782
Score = 67.4 bits (163), Expect = 2e-09
Identities = 63/307 (20%), Positives = 119/307 (38%), Gaps = 25/307 (8%)
Query: 4 TEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTA 63
T RK + + + R++ + S N + + S ++ VG+++
Sbjct: 414 TYNRKEEESDEEESEGSVNERDETEDVEESDGLGNRRDKSEEVEDSDSKTHDKVGENDDD 473
Query: 64 SENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEK---EQKEGNGEVSDTDT 120
SE E + ++ D + EE+ E VN V +S+ E+ E+K + EV+DT T
Sbjct: 474 SEGDEDDDTMI----KDSSEEEESEEEKTVNDTVTKASSENEEREEEEKSNDSEVNDTMT 529
Query: 121 VKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSN 180
S + + + +S+D + + K S + E + K+ S+
Sbjct: 530 KHSSEDEEKEKEEKSSDSSNASSEDEEEEEKNGDSAKASSEDEEEEEKSGSSVKASSEDE 589
Query: 181 QKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVD-----ENHVLEVKEIEIL 235
+++ N + ++ S N+ V + + SE D E E E E
Sbjct: 590 EEEEENGDSFKASSKEESGDSANASNVNSEEEEQKSKSEATDDLESEEEEDNEDNEEEKS 649
Query: 236 DGASNGAQSLGSEDERHETVNAEENGE-------------HEDKAALELKIEEMELRVEK 282
+ +++ ++++ ET EE+GE ED E + EE E+ E
Sbjct: 650 ETSASTEDESNDDEDKSETSQEEESGEESEGNSESRSERSEEDDEDEESEKEESEVESEA 709
Query: 283 LEEELRE 289
E+E E
Sbjct: 710 EEDETGE 716
Score = 61.2 bits (147), Expect = 1e-07
Identities = 60/279 (21%), Positives = 110/279 (38%), Gaps = 30/279 (10%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
++ EK K ++S + + + E++ + D++ + ++ E K+ G S
Sbjct: 534 EDEEKEKEEKSSDSSNASSEDEEEEEKNGDSAKASSEDEEEEEKS-----------GSSV 582
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEV---SDT 118
AS E E + D S + + NA N + +K + E ++ +
Sbjct: 583 KASSEDEEEEE---ENGDSFKASSKEESGDSANASNVNSEEEEQKSKSEATDDLESEEEE 639
Query: 119 DTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSD 178
D + S + EDE + K S+ + S ES + RS+R S+ D
Sbjct: 640 DNEDNEEEKSETSASTEDESNDDEDKSETSQEEESGEESEGNSESRSER------SEEDD 693
Query: 179 SNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGA 238
+++ ++ S T N S+ ++SSE E + E E +
Sbjct: 694 EDEESEKEESEVESEAEEDETGENTSEEDDEEQESSSEDEEKSE-------SEAEGEEEE 746
Query: 239 SNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEME 277
S G Q S++E + EE E+E++ E K+EE E
Sbjct: 747 SEGDQEDESDEETSDKDEEEEESENEEEEEEESKVEEEE 785
Score = 43.5 bits (101), Expect = 0.025
Identities = 37/171 (21%), Positives = 77/171 (44%), Gaps = 12/171 (7%)
Query: 5 EKRKASRNSQTKGSRRTERREDKLHQDNSSKTL-------NEKGTESKTPQDRSAANNLV 57
E+ + S++ T E +++ +++ S+T N+ +S+T Q+ +
Sbjct: 620 EEEQKSKSEATDDLESEEEEDNEDNEEEKSETSASTEDESNDDEDKSETSQEEESGEESE 679
Query: 58 GDSNTASENSETYENVVIDYVDDVNRSE-EALAEMKVNAMVANQASDTEKEQKEGNGEVS 116
G+S + SE SE + D + SE E+ AE ++ D E+E + E S
Sbjct: 680 GNSESRSERSEEDDE---DEESEKEESEVESEAEEDETGENTSEEDDEEQESSSEDEEKS 736
Query: 117 DTDTVKDSVSSQGDSFTNEDEKV-EKASKDAKSKVKVSPSESSRGLKERSD 166
+++ + S+GD DE+ +K ++ +S+ + E S+ +E +
Sbjct: 737 ESEAEGEEEESEGDQEDESDEETSDKDEEEEESENEEEEEEESKVEEEEEE 787
Score = 42.4 bits (98), Expect = 0.055
Identities = 28/163 (17%), Positives = 66/163 (40%)
Query: 3 ETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNT 62
E+E+ + + +++ + S + ED+ + D +++ + + S + + + +
Sbjct: 634 ESEEEEDNEDNEEEKSETSASTEDESNDDEDKSETSQEEESGEESEGNSESRSERSEEDD 693
Query: 63 ASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVK 122
E SE E+ V ++ E E +++ + + + EG E S+ D
Sbjct: 694 EDEESEKEESEVESEAEEDETGENTSEEDDEEQESSSEDEEKSESEAEGEEEESEGDQED 753
Query: 123 DSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERS 165
+S D E+E + ++ +SKV+ E + S
Sbjct: 754 ESDEETSDKDEEEEESENEEEEEEESKVEEEEEEXXXXSDDES 796
>UniRef100_UPI00001A1808 UPI00001A1808 UniRef100 entry
Length = 290
Score = 69.7 bits (169), Expect = 3e-10
Identities = 67/284 (23%), Positives = 108/284 (37%), Gaps = 20/284 (7%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
K +K+K ++ + + ++ ED+ + N + EK E K +++ V + N
Sbjct: 14 KRIKKKKKEKDKKEEKKKKKNEEEDEKEEKNEEE---EKKEEEKEDEEKEGKKKNVKEDN 70
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
E + E + + EE E K M + +T++E+KE E V
Sbjct: 71 KEEEKKKEEE--------EKEKEEEKEEEEKEKKMEEEEEEETKEEEKEEKKEEEKMKEV 122
Query: 122 KDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQ 181
K + D +DEK EK K + K E KE ++K K + + +
Sbjct: 123 K-----KVDKEVEKDEKKEKKKKKKEKMKKEKREEKKEKKKEEEEKKKKKREEEEDKKEE 177
Query: 182 KKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNG 241
K KG + + K K K +E + E E +
Sbjct: 178 VKKEEEKKGEKKKEEEEKKEEKKKDEKKEEKKRRRKRRRREEEAPPQGGEKEE-EKKKKE 236
Query: 242 AQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEE 285
+ E+E+ E EEN E E+K E KIEE E EK+EE
Sbjct: 237 EEKENKEEEKKEEKEEEENKE-EEKKKEENKIEEEE--EEKMEE 277
Score = 57.0 bits (136), Expect = 2e-06
Identities = 47/230 (20%), Positives = 86/230 (36%), Gaps = 15/230 (6%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
KE EK+K + + + E +E K+ ++ +T E+ E K + + +
Sbjct: 71 KEEEKKKEEEEKEKEEEKEEEEKEKKMEEEEEEETKEEEKEEKKEEEKMKEVKKV----D 126
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
E E E + +E + + K + + EK++K+ E + V
Sbjct: 127 KEVEKDEKKEKK--------KKKKEKMKKEKREEKKEKKKEEEEKKKKKREEEEDKKEEV 178
Query: 122 KDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQ 181
K +G+ E+EK E+ KD K + K + R +E + K + K +
Sbjct: 179 KKEEEKKGEKKKEEEEKKEEKKKDEKKEEKKRRRKRRRREEEAPPQGGEKEEEKKKKEEE 238
Query: 182 KKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKE 231
K+ K + +N K K E E ++EN + KE
Sbjct: 239 KENKEEEKKEEKEEEENKEEEKKKEEN---KIEEEEEEKMEENEEKKKKE 285
Score = 40.4 bits (93), Expect = 0.21
Identities = 32/154 (20%), Positives = 63/154 (40%), Gaps = 5/154 (3%)
Query: 136 DEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQ---KKPMNSTKGPS 192
D++ EK K K ++K E + KE +K N+ + + + N+ KK
Sbjct: 2 DKEDEKGEKSKKKRIKKKKKEKDK--KEEKKKKKNEEEDEKEEKNEEEEKKEEEKEDEEK 59
Query: 193 RVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERH 252
KN +N + K + E E +E + E E + + E+E+
Sbjct: 60 EGKKKNVKEDNKEEEKKKEEEEKEKEEEKEEEEKEKKMEEEEEEETKEEEKEEKKEEEKM 119
Query: 253 ETVNAEENGEHEDKAALELKIEEMELRVEKLEEE 286
+ V + +D+ + K ++ +++ EK EE+
Sbjct: 120 KEVKKVDKEVEKDEKKEKKKKKKEKMKKEKREEK 153
Score = 40.0 bits (92), Expect = 0.27
Identities = 39/162 (24%), Positives = 61/162 (37%), Gaps = 15/162 (9%)
Query: 135 EDEKVEKASK---DAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGP 191
EDEK EK+ K K K K E + E D K K ++ +KK
Sbjct: 4 EDEKGEKSKKKRIKKKKKEKDKKEEKKKKKNEEEDEKEEK-----NEEEEKKEEEKEDEE 58
Query: 192 SRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDER 251
KN +N + K + E E +E + E E + + E+E+
Sbjct: 59 KEGKKKNVKEDNKEEEKKKEEEEKEKEEEKEEEEKEKKMEEEEEEETKEEEKEEKKEEEK 118
Query: 252 -------HETVNAEENGEHEDKAALELKIEEMELRVEKLEEE 286
+ V +E E + K ++K E+ E + EK +EE
Sbjct: 119 MKEVKKVDKEVEKDEKKEKKKKKKEKMKKEKREEKKEKKKEE 160
Score = 37.4 bits (85), Expect = 1.8
Identities = 38/198 (19%), Positives = 74/198 (37%), Gaps = 16/198 (8%)
Query: 103 DTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLK 162
D ++E+K+ E D K+ + + EDE+ E K+ K K + K
Sbjct: 24 DKKEEKKKKKNEEEDEKEEKNEEEEKKEE-EKEDEEKEGKKKNVKEDNKEEEKKKEEEEK 82
Query: 163 ERSDRKTNKLQSK--------------VSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVK 208
E+ + K + + K + +++ M K + K+ K K
Sbjct: 83 EKEEEKEEEEKEKKMEEEEEEETKEEEKEEKKEEEKMKEVKKVDKEVEKDEKKEKKKKKK 142
Query: 209 APVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAA 268
+K + E ++ E K+ + + + + + E+E+ EE + E+K
Sbjct: 143 EKMK-KEKREEKKEKKKEEEEKKKKKREEEEDKKEEVKKEEEKKGEKKKEEEEKKEEKKK 201
Query: 269 LELKIEEMELRVEKLEEE 286
E K E+ R + EE
Sbjct: 202 DEKKEEKKRRRKRRRREE 219
Score = 35.0 bits (79), Expect = 8.8
Identities = 30/122 (24%), Positives = 46/122 (37%), Gaps = 3/122 (2%)
Query: 177 SDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILD 236
+D +K S K + K K K + + + +E E KE E +
Sbjct: 1 NDKEDEKGEKSKKKRIKKKKKEKDKKEEKKKKKNEEEDEKEEKNEEEEKKEEEKEDEEKE 60
Query: 237 GASNGAQSLGSEDER---HETVNAEENGEHEDKAALELKIEEMELRVEKLEEELREVAAL 293
G + E+E+ E EE E E+K + EE E + E+ EE+ E
Sbjct: 61 GKKKNVKEDNKEEEKKKEEEEKEKEEEKEEEEKEKKMEEEEEEETKEEEKEEKKEEEKMK 120
Query: 294 EV 295
EV
Sbjct: 121 EV 122
>UniRef100_UPI00003AEAEA UPI00003AEAEA UniRef100 entry
Length = 686
Score = 67.8 bits (164), Expect = 1e-09
Identities = 63/301 (20%), Positives = 134/301 (43%), Gaps = 21/301 (6%)
Query: 1 MKETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDS 60
MK+ ++K ++ T + E E+K + S + +N + +T +++ V +
Sbjct: 153 MKDVTEKKPKEDTVTDMEVKLEGSEEKT--ETSVEKVNSEEHGPQTAVEKNTXKAAVEEK 210
Query: 61 NTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNG----EVS 116
E +E + + N +E L E A+ ++A++ E+K G G E +
Sbjct: 211 AVEKEQKTKHEKA--EATEGRNSMKEVLGEEAKAAVEEDEAAEAPAEKKGGAGEQTAEAT 268
Query: 117 DTDTV---KDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQ 173
+T+T +S+ Q + E ++ + ++ V E++ E +++K + +
Sbjct: 269 ETETTVEKGESIEGQAEVAVEEKAMSQEGRAEGQAAVTAEQKEAAEEQAEAAEKKVEEKE 328
Query: 174 SKVSDSNQK----KPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEG--VDENHVL 227
S + QK K M+++K P + +++ + + K E + ++E+
Sbjct: 329 SAETAPEQKPDESKGMDTSKEPVSAVGEGPANDTGEKTEVAAKVEKEEKKDDLMEESEGA 388
Query: 228 EVKEIEILDGASNGAQSLGSEDERHETVNAEENGEH----EDKAALELKIEEMELRVEKL 283
+V++ E D G ++ SE+E E AE++ E+ EDK + E +I +EL + L
Sbjct: 389 KVEKEEKDDQMEEGEETEESEEEDKENDKAEDDKENELAVEDKESEEEEIGNLELAWDML 448
Query: 284 E 284
E
Sbjct: 449 E 449
>UniRef100_Q95XW8 Hypothetical protein Y55B1BR.3 [Caenorhabditis elegans]
Length = 679
Score = 67.8 bits (164), Expect = 1e-09
Identities = 62/265 (23%), Positives = 109/265 (40%), Gaps = 21/265 (7%)
Query: 29 HQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASENSETYENVVIDYVDDVNRSEEAL 88
HQD S + + E K+P+ RS+ + + +S++ D ++ +
Sbjct: 187 HQDTSDDSEESEDEEEKSPRKRSSKKRRAASVSDSDSDSDS----------DFDKKKWKK 236
Query: 89 AEMKVNAMVANQASDTEKEQKEGNGEVSD-------TDTVKDSVSSQGDSFTNEDEKVEK 141
+ K + + D+E E++ + S ++ K +V+ +E+EK EK
Sbjct: 237 NKAKRSKRDDSSDDDSEMERRRKKSKKSKKSKKFKKSEKRKRAVNDSSSDDEDEEEKPEK 296
Query: 142 ASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSS 201
SK +K V S SE +E+S +K +K K SD Q+ + ++ KN+ S
Sbjct: 297 RSKKSKKAVIDSSSEDEE--EEKSSKKRSKKSKKESDEEQQA--SDSEEEVVEVKKNSKS 352
Query: 202 NNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENG 261
P K VK SE S G +E V++ K+ ++ S E+E E+ +
Sbjct: 353 PKKTPKKTAVKEESEESSGDEEEEVVKKKKSSKINKRKAKESSSDEEEEVEESPKKKTKS 412
Query: 262 EHEDKAALELKIEEMELRVEKLEEE 286
+ K E E + EEE
Sbjct: 413 PRKSSKKSAAKEESEEESSDNEEEE 437
Score = 57.8 bits (138), Expect = 1e-06
Identities = 60/278 (21%), Positives = 112/278 (39%), Gaps = 26/278 (9%)
Query: 5 EKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTAS 64
+K K+ R S K + + E E+ + + + K+P+ S +S S
Sbjct: 408 KKTKSPRKSSKKSAAKEESEEESSDNEEEEEVDYSPKKKVKSPKKSSKKPAAKVESEEPS 467
Query: 65 ENSETYENVVIDYV--DDVNR--SEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDT 120
+N E E V + D R S ++ A+++ N+ + +E + G+ +
Sbjct: 468 DNEEEEEEVEESPIKKDKTPRKYSRKSAAKVESTESSGNEEEEEVEESPKKKGKTPRKSS 527
Query: 121 VKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSN 180
K + + D NE+E VE++ K S K S R KE S+ ++ +V DS
Sbjct: 528 KKSAAVEESD---NEEEDVEESPKKRTSPRK---SSKKRAAKEESEESSDNEVEEVEDSP 581
Query: 181 QKKPMNSTKGPSRVTNKNTSSNN-------------SKPVKAPVKASSESS---EGVDEN 224
+K K P + K S + K K P K+S +S+ E + +
Sbjct: 582 KKNDKTLRKSPRKPAAKVESEESFGNEEEEEVEESPKKKGKTPRKSSKKSAAKEESEESS 641
Query: 225 HVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGE 262
E ++ E +G+ + + DE + +++E+ E
Sbjct: 642 DDEEEEQAEETNGSHTESPTKSKTDESDNSTSSDEDDE 679
Score = 48.9 bits (115), Expect = 6e-04
Identities = 63/318 (19%), Positives = 130/318 (40%), Gaps = 41/318 (12%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
+ ++KR+A+ S + ++ + K ++ + ++ + ++ + +R + +
Sbjct: 208 RSSKKRRAASVSDSDSDSDSDFDKKKWKKNKAKRSKRDDSSDDDSEMERRRKKSKKSKKS 267
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
+ SE + V D D EE + + A S +E E++E + + +
Sbjct: 268 KKFKKSEKRKRAVNDSSSDDEDEEEKPEKRSKKSKKAVIDSSSEDEEEEKSSKKRSKKSK 327
Query: 122 KDSVSSQGDSFTNEDE-KVEKASKDAKSKVKVSP----SESSRG------LKERSDRKTN 170
K+S Q S + E+ +V+K SK K K + SE S G +K++ K N
Sbjct: 328 KESDEEQQASDSEEEVVEVKKNSKSPKKTPKKTAVKEESEESSGDEEEEVVKKKKSSKIN 387
Query: 171 KLQSKVSDSNQKK-----PMNSTKGPSRVTNKNTSSNNSKPV--------------KAPV 211
K ++K S S++++ P TK P + + K+ + S+ K V
Sbjct: 388 KRKAKESSSDEEEEVEESPKKKTKSPRKSSKKSAAKEESEEESSDNEEEEEVDYSPKKKV 447
Query: 212 KASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALEL 271
K+ +SS+ ++E + + N + E+E E+ ++ +
Sbjct: 448 KSPKKSSK-------KPAAKVESEEPSDNEEE----EEEVEESPIKKDKTPRKYSRKSAA 496
Query: 272 KIEEMELRVEKLEEELRE 289
K+E E + EEE+ E
Sbjct: 497 KVESTESSGNEEEEEVEE 514
Score = 42.4 bits (98), Expect = 0.055
Identities = 48/208 (23%), Positives = 89/208 (42%), Gaps = 22/208 (10%)
Query: 2 KETEKRKASRNSQTK-GSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDS 60
K+ RK SR S K S + E++ + S K +KG KTP+ S + V +S
Sbjct: 483 KDKTPRKYSRKSAAKVESTESSGNEEEEEVEESPK---KKG---KTPRKSSKKSAAVEES 536
Query: 61 NTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDT 120
+ E+ E S ++ + + ++SD E E+ E + + +D
Sbjct: 537 DNEEEDVEESPKK--------RTSPRKSSKKRAAKEESEESSDNEVEEVEDSPKKNDKTL 588
Query: 121 VKD------SVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQS 174
K V S+ +SF NE+E+ + S K K S+ S +E + ++ +
Sbjct: 589 RKSPRKPAAKVESE-ESFGNEEEEEVEESPKKKGKTPRKSSKKSAAKEESEESSDDEEEE 647
Query: 175 KVSDSNQKKPMNSTKGPSRVTNKNTSSN 202
+ ++N + TK + ++ +TSS+
Sbjct: 648 QAEETNGSHTESPTKSKTDESDNSTSSD 675
>UniRef100_UPI000045323E UPI000045323E UniRef100 entry
Length = 972
Score = 67.0 bits (162), Expect = 2e-09
Identities = 71/310 (22%), Positives = 137/310 (43%), Gaps = 28/310 (9%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKT------LNEKGTESKTPQDRSAANN 55
K E ++S++ K + + + ++NS+K+ N+ + S+ Q+ A+
Sbjct: 195 KSLESVESSKDDLKKEDKSEDNSAKESTEENSAKSSDSEADANKDFSSSEVAQEEDLASE 254
Query: 56 LVGDSNTASENSETYENVVIDYVDDVN--RSEEALAEMKVNAMVANQASDTEKEQKEGNG 113
+ N + NS+ EN + + N +S + + N+ V N E + + +
Sbjct: 255 STDNLNDQTGNSDGAENTSEEKSSEENDEKSMKTDTNDQSNSDVKNDEVKNEDSKNDASL 314
Query: 114 EVSDTDTVKDSVSSQGDSFTNEDEK----VEKASKDAKSKVKVSPSESSRG--------L 161
+ DT K+S S+ + N D+K VEK ++ K K+ V P + + +
Sbjct: 315 SDASKDTSKES-SNNSEIKANGDDKLEDNVEKTIQEMKDKLGVKPEDETIKRVDDQFIVI 373
Query: 162 KERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKN---TSSNNSKPVKAPVKASSESS 218
++ +K+N++ + D K + K PS N+ +S N ++P K K + S
Sbjct: 374 VKKLKQKSNEVPTDTKDVEVKSHDTTDKNPSEKNNEEKSLSSENQTEPTKEENKDEKKES 433
Query: 219 EGVDENHVLEVKE--IEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEM 276
E ENH + E IE L SN ++ ++++ + N E E ++K +E I+E
Sbjct: 434 ETETENHEQKSNEELIEKLKSLSNNDKNEAADEKIKKINNLLEVSESDEK--MEKLIQEK 491
Query: 277 ELRVEKLEEE 286
+ K EE+
Sbjct: 492 KDYDAKKEEK 501
Score = 42.4 bits (98), Expect = 0.055
Identities = 72/330 (21%), Positives = 135/330 (40%), Gaps = 57/330 (17%)
Query: 2 KETEKRKASR--NSQTKGSRRTERREDKLHQDNSSKTL-NEKGTESKTPQDRSAANNLVG 58
KE K +S+ + Q K ++ E K + N KT E + P++ +
Sbjct: 53 KEFRKIMSSKPNDEQAKEQSSSKSGESKPEESNPDKTKPKEPKPDENKPEESKSGEAKPR 112
Query: 59 DSNTAS---ENSETYENVVIDYVDDVNRSEEALAEMKV--------NAMVANQASDTEKE 107
+SN+ E S++ E+ + D + EEA +E N ++ +T+ E
Sbjct: 113 ESNSDEAKPEESKSDESKPEETKPDETKPEEAKSEEAKPEESKPDENKPKESKPDETKPE 172
Query: 108 Q---------KEGNGEVSDTDTVK-DSVSSQGDSFTNEDEKVEKASKDA--KSKVKVSPS 155
+ +E +G+ + T+ +SV S D ED+ + ++K++ ++ K S S
Sbjct: 173 ETKPEETKHDEENSGKGNSTEPKSLESVESSKDDLKKEDKSEDNSAKESTEENSAKSSDS 232
Query: 156 E-------SSRGLKERSD---RKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSK 205
E SS + + D T+ L + NS + K++ N+ K
Sbjct: 233 EADANKDFSSSEVAQEEDLASESTDNLNDQTG--------NSDGAENTSEEKSSEENDEK 284
Query: 206 PVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEE---NGE 262
+K S S D E++ D ++ + S S+D E+ N E NG+
Sbjct: 285 SMKTDTNDQSNSDVKND--------EVKNEDSKNDASLSDASKDTSKESSNNSEIKANGD 336
Query: 263 HEDKAALELKIEEM--ELRVEKLEEELREV 290
+ + +E I+EM +L V+ +E ++ V
Sbjct: 337 DKLEDNVEKTIQEMKDKLGVKPEDETIKRV 366
Score = 38.5 bits (88), Expect = 0.80
Identities = 35/196 (17%), Positives = 73/196 (36%), Gaps = 3/196 (1%)
Query: 104 TEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKE 163
++KE ++ + + K+ SS+ E+ +K + P ES G +
Sbjct: 51 SQKEFRKIMSSKPNDEQAKEQSSSKSGESKPEESNPDKTKPKEPKPDENKPEESKSGEAK 110
Query: 164 RSDRKTNKLQSKVSDSNQKKPMNSTKG---PSRVTNKNTSSNNSKPVKAPVKASSESSEG 220
+ +++ + + S S++ KP + P ++ SKP + K S
Sbjct: 111 PRESNSDEAKPEESKSDESKPEETKPDETKPEEAKSEEAKPEESKPDENKPKESKPDETK 170
Query: 221 VDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRV 280
+E E K E G N + E + ++ + ED +A E E
Sbjct: 171 PEETKPEETKHDEENSGKGNSTEPKSLESVESSKDDLKKEDKSEDNSAKESTEENSAKSS 230
Query: 281 EKLEEELREVAALEVS 296
+ + ++ ++ EV+
Sbjct: 231 DSEADANKDFSSSEVA 246
Score = 37.7 bits (86), Expect = 1.4
Identities = 28/188 (14%), Positives = 76/188 (39%), Gaps = 8/188 (4%)
Query: 79 DDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEK 138
D+ + + K + + + E+ +++ + + ++ + + DE
Sbjct: 40 DEFGEVDTGIVSQKEFRKIMSSKPNDEQAKEQSSSKSGESKPEESNPDKTKPKEPKPDEN 99
Query: 139 VEKASKDAKSKVKVSPSESSRGLKERSDR------KTNKLQSKVSDSNQKKPMNSTKGPS 192
+ SK ++K + S S+ ++ + +SD K ++ + + + S + KP S P
Sbjct: 100 KPEESKSGEAKPRESNSDEAKPEESKSDESKPEETKPDETKPEEAKSEEAKPEESK--PD 157
Query: 193 RVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERH 252
K + + +KP + + + E + + E K +E ++ + + + ++
Sbjct: 158 ENKPKESKPDETKPEETKPEETKHDEENSGKGNSTEPKSLESVESSKDDLKKEDKSEDNS 217
Query: 253 ETVNAEEN 260
+ EEN
Sbjct: 218 AKESTEEN 225
>UniRef100_Q8I492 Phoshoprotein 300 [Plasmodium falciparum]
Length = 1434
Score = 67.0 bits (162), Expect = 2e-09
Identities = 73/317 (23%), Positives = 132/317 (41%), Gaps = 34/317 (10%)
Query: 1 MKETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTP---------QDRS 51
+KE E+ K + K + ++ ++ + K + EK TE+K + +
Sbjct: 949 VKEKEEVKEKEEVKEKDTESKDKEIEQEKEKEEVKEVKEKDTENKDKVIGQEIIIEEIKK 1008
Query: 52 AANNLVGDSNTASENSETYENVVIDYV--DDVNRSEEA-----------LAEMKVNAMVA 98
V N +EN + NV++ + +DVN + A E+K V
Sbjct: 1009 EVKKRVKKRNNKNENKD---NVIVQEIMNEDVNEKDTANKDKVIEQEKEKEEVKEKEEVK 1065
Query: 99 NQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDS-----FTNED-EKVEKASKDAKSKVKV 152
+ EKE+ + EV + + VK+ + D+ NED + + SKD +V
Sbjct: 1066 EKEEVKEKEEVKEKEEVKEKEEVKEKDTESKDNVIVQEIMNEDVNEKDTESKDKMIGKEV 1125
Query: 153 SPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVK 212
E +K+R +++ NK ++ + N++K + + S N+ + NN K + VK
Sbjct: 1126 IIEEVKEEVKKRVNKEVNKRVNRRNRKNERKDVIEQEIVSEEVNEKDTKNNDKKIGKRVK 1185
Query: 213 ASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELK 272
+ + E V E E E + + ++ SE+E E E E E+++ E +
Sbjct: 1186 KPIDDCK--KEREVQEESEEESEEESEEESEE-ESEEESEEESEEESEEESEEESEEESE 1242
Query: 273 IEEMELRVEKLEEELRE 289
E E E+ EEE E
Sbjct: 1243 EESEEESEEESEEESEE 1259
Score = 60.5 bits (145), Expect = 2e-07
Identities = 52/291 (17%), Positives = 121/291 (40%), Gaps = 30/291 (10%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTP-------------- 47
+E ++++ + + + TE +++ + Q+ ++ +NEK TESK
Sbjct: 1074 EEVKEKEEVKEKEEVKEKDTESKDNVIVQEIMNEDVNEKDTESKDKMIGKEVIIEEVKEE 1133
Query: 48 ---QDRSAANNLVGDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDT 104
+ N V N +E + E ++ ++VN + + K+ V D
Sbjct: 1134 VKKRVNKEVNKRVNRRNRKNERKDVIEQEIVS--EEVNEKDTKNNDKKIGKRVKKPIDDC 1191
Query: 105 EKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKER 164
+KE++ +++ + S + +E+E E++ ++++ + S ES +E
Sbjct: 1192 KKEREVQEESEEESEEESEEESEEESEEESEEESEEESEEESEEE---SEEESEEESEEE 1248
Query: 165 SDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVK---ASSESSEGV 221
S+ ++ + + S+ ++ + K S + ++ K ++ ++ E
Sbjct: 1249 SEEESEEESEEESEEESEEESDEEKNTSGLVHRRNCKKEKKYNNGELEEYYKEKQNEEYF 1308
Query: 222 DENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELK 272
DE ++++ KE L+ N A + E H ++ HED +ELK
Sbjct: 1309 DEEYIIQSKEHNTLNTFPNMALNEDFRREFHNILSI-----HEDTDLMELK 1354
Score = 60.1 bits (144), Expect = 3e-07
Identities = 60/293 (20%), Positives = 121/293 (40%), Gaps = 21/293 (7%)
Query: 5 EKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTAS 64
+ +K + + +E +D ++N K N+K ++ K + + ++G + +
Sbjct: 388 DPKKLTEQEENGTKESSEETKDDKPEENEKKADNKKKSKKK----KKSFFQMLGCNFLCN 443
Query: 65 ENSETY-ENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKE-GNGEVSDTDTVK 122
+N ET E + DD + + L E + +D EK+ K G G+ D VK
Sbjct: 444 KNIETDDEEETLVVKDDAKKKHKFLREANT------EKNDNEKKDKLLGEGDKED---VK 494
Query: 123 DSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKL------QSKV 176
+ Q D E +K + K+ + K KV +KE++D K +K+ Q ++
Sbjct: 495 EKNDEQKDKVLGEGDKEDVKEKNDEQKDKVLGEGDKEDVKEKNDGKKDKVIGSEKTQKEI 554
Query: 177 SDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILD 236
+ +K+ K + K + + VK P E E + + +KE +
Sbjct: 555 KEKVEKRVKKKCKKKVKKGIKENDTEGNDKVKGPEIIIEEVKEEIKKQVEDGIKENDTEG 614
Query: 237 GASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELRE 289
+ +E+ + E E G E+ K++ E+ E+++EE+++
Sbjct: 615 NDKVKGPEIITEEVKEEIKKQVEEGIKENDTEGNDKVKGPEIITEEVKEEIKK 667
Score = 42.4 bits (98), Expect = 0.055
Identities = 60/328 (18%), Positives = 126/328 (38%), Gaps = 49/328 (14%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNL-VGDS 60
KET + K + S+ G + + + + E G +T + + + G+S
Sbjct: 238 KETGESKETGESKETGESKETGESKETGESKETGESKETGESKETGESKETGESKETGES 297
Query: 61 NTASENSET--YENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEG---NGEV 115
E+ ET YE + + R E N + ++ D E + G N E
Sbjct: 298 KETGESKETRIYEETKYNKITSEFRETE-------NVKITEESKDREGNKVSGPYENSEN 350
Query: 116 SDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSK 175
S+ + + + NE EK+ + D S+ P + + + + + + +
Sbjct: 351 SNVTSESEETKKLAEKEENEGEKLGENVNDGASENSEDPKKLTEQEENGTKESSEETKDD 410
Query: 176 VSDSNQKKPMNSTK------------GPSRVTNKNTSSNNSKP---VKAPVK-------- 212
+ N+KK N K G + + NKN +++ + VK K
Sbjct: 411 KPEENEKKADNKKKSKKKKKSFFQMLGCNFLCNKNIETDDEEETLVVKDDAKKKHKFLRE 470
Query: 213 ASSESSEGVDENHVL-EVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALEL 271
A++E ++ ++ +L E + ++ + + E ++ + E+N E +DK E
Sbjct: 471 ANTEKNDNEKKDKLLGEGDKEDVKEKNDEQKDKVLGEGDKEDV--KEKNDEQKDKVLGEG 528
Query: 272 KIEEME----------LRVEKLEEELRE 289
E+++ + EK ++E++E
Sbjct: 529 DKEDVKEKNDGKKDKVIGSEKTQKEIKE 556
Score = 41.6 bits (96), Expect = 0.094
Identities = 65/312 (20%), Positives = 130/312 (40%), Gaps = 22/312 (7%)
Query: 1 MKETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESK-------TPQDRSAA 53
+K+ K+K + + ++DKL + + + EK E K +D
Sbjct: 457 VKDDAKKKHKFLREANTEKNDNEKKDKLLGEGDKEDVKEKNDEQKDKVLGEGDKEDVKEK 516
Query: 54 NNLVGDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNG 113
N+ D + E + D V SE+ E+K + +K+ K+G
Sbjct: 517 NDEQKDKVLGEGDKEDVKEKNDGKKDKVIGSEKTQKEIK-EKVEKRVKKKCKKKVKKGIK 575
Query: 114 EVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQ 173
E +DT+ D V E+++K +D + ++ +G + ++ +++
Sbjct: 576 E-NDTEG-NDKVKGPEIIIEEVKEEIKKQVEDGIKENDTEGNDKVKGPEIITEEVKEEIK 633
Query: 174 SKVSD---SNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKAS-SESSEGVDENHVL-- 227
+V + N + + KGP +T + K V+ +K + +ES + + ++
Sbjct: 634 KQVEEGIKENDTEGNDKVKGPEIIT-EEVKEEIKKQVEEGIKENDTESKDKLIGQEIITE 692
Query: 228 EVKEIEILDGASNGAQSLGSE---DERHETVNAEENGEHEDKA-ALELKIEEMELRVEKL 283
EVKE + N + +G E +E E + E + E++DK E+ EE++ +EK
Sbjct: 693 EVKEGIKENDTENKDKVIGQEIITEEVKEGIK-ENDTENKDKVIGQEIITEEVKKEIEKQ 751
Query: 284 EEELREVAALEV 295
EE+ + LE+
Sbjct: 752 EEKGNKENILEI 763
Score = 40.0 bits (92), Expect = 0.27
Identities = 72/339 (21%), Positives = 133/339 (38%), Gaps = 58/339 (17%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
++ +++ + + G E ++K + + K L E E ++ + ++G
Sbjct: 491 EDVKEKNDEQKDKVLGEGDKEDVKEK-NDEQKDKVLGEGDKEDVKEKNDGKKDKVIGSEK 549
Query: 62 TASENSETYENVV----------------IDYVDDVNRSEEALAEMKVNAMVANQASDTE 105
T E E E V + D V E + E+K + Q D
Sbjct: 550 TQKEIKEKVEKRVKKKCKKKVKKGIKENDTEGNDKVKGPEIIIEEVKEE--IKKQVEDGI 607
Query: 106 KEQK-EGNGEVSD----TDTVKDSVSSQ-----GDSFTNEDEKV---EKASKDAKSKVKV 152
KE EGN +V T+ VK+ + Q ++ T ++KV E +++ K ++K
Sbjct: 608 KENDTEGNDKVKGPEIITEEVKEEIKKQVEEGIKENDTEGNDKVKGPEIITEEVKEEIKK 667
Query: 153 SPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVK 212
E G+KE +KL + + + K +N + N K + +
Sbjct: 668 QVEE---GIKENDTESKDKLIGQEIITEEVK---------EGIKENDTENKDKVIGQEI- 714
Query: 213 ASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELK 272
+ E EG+ EN E K+ I Q + +E+ + E EE G E+ L+
Sbjct: 715 ITEEVKEGIKENDT-ENKDKVI-------GQEIITEEVKKEIEKQEEKGNKENI----LE 762
Query: 273 IEEMELRVEKLEEELREVAALEVSLYSVVPEHGSSAHKV 311
I+++ + E + EE+++V +V + H S KV
Sbjct: 763 IKDIVIGQEVIIEEVKKVIKKKVE-KGIKENHTESKDKV 800
Score = 39.3 bits (90), Expect = 0.47
Identities = 59/297 (19%), Positives = 124/297 (40%), Gaps = 24/297 (8%)
Query: 5 EKRKASRNSQTKGSRR--TERREDKLHQD----NSSKTLNEKGTESKTPQDRSAANNLVG 58
E +K + KG + TE ++ + Q+ + + ++ E D + + ++G
Sbjct: 776 EVKKVIKKKVEKGIKENHTESKDKVIGQEIIVEEVKEEIEKQVEEGIKENDTESKDKVIG 835
Query: 59 DSNTASE-NSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSD 117
+ N E EN D V + +E E+K + + + E+K+ V
Sbjct: 836 QEVIKGDVNEEGPENK--DKVTKQEKVKEVKKEVKKKVKKRVKKRNNKNERKDN---VIG 890
Query: 118 TDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVS 177
+ +K+ V+ + + N+D+++E+ K K +V E + +E +++ K + +V
Sbjct: 891 KEIMKEDVNEKDTA--NKDKEIEQE----KEKEEVKEKEEVKEKEEVKEKEEVKEKEEVK 944
Query: 178 DSNQKKPMNSTKGPSRVTNKNTSSNN----SKPVKAPVKASSE-SSEGVDENHVLEVKEI 232
+ + K K V K+T S + + K VK E +E D+ E+
Sbjct: 945 EKEEVKEKEEVKEKEEVKEKDTESKDKEIEQEKEKEEVKEVKEKDTENKDKVIGQEIIIE 1004
Query: 233 EILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELRE 289
EI + +++E + V +E +ED + ++ + EK +EE++E
Sbjct: 1005 EIKKEVKKRVKKRNNKNENKDNVIVQEI-MNEDVNEKDTANKDKVIEQEKEKEEVKE 1060
Score = 37.4 bits (85), Expect = 1.8
Identities = 61/305 (20%), Positives = 112/305 (36%), Gaps = 50/305 (16%)
Query: 1 MKETEKRKASRNSQTKGSRRTERRE--------DKLHQDNSSKTLNEKGTESKTPQDRSA 52
M+E+E+RK K ++E +++ + + K E + R
Sbjct: 139 MQESERRKQQEEENAKDIEEIRKKEKEYLMKELEEMDESDVEKAFRELQFIKLRDRTRPR 198
Query: 53 AN-NLVGDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEG 111
+ N++G+S E+ ET E+ + S+E E KE
Sbjct: 199 KHVNVMGESKETDESKETDESKETGESKETGESKET------------------GESKE- 239
Query: 112 NGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNK 171
GE +T K++ G+S + K SK+ + S+ + KE + K
Sbjct: 240 TGESKETGESKET----GESKETGESKETGESKETGESKETGESKETGESKETGESKETG 295
Query: 172 LQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKE 231
+ +S + + TK +++T++ + N K + S+ EG + E E
Sbjct: 296 ESKETGESKETRIYEETKY-NKITSEFRETENVK-----ITEESKDREGNKVSGPYENSE 349
Query: 232 IEILDGASNGAQSLGSEDERH-----ETVN--AEENGEHEDKAALELKIEEMELRVEKLE 284
+ S + L ++E E VN A EN E K E+ E ++
Sbjct: 350 NSNVTSESEETKKLAEKEENEGEKLGENVNDGASENSEDPKKLT-----EQEENGTKESS 404
Query: 285 EELRE 289
EE ++
Sbjct: 405 EETKD 409
>UniRef100_Q8I7C4 NAD(+) ADP-ribosyltransferase-3 [Dictyostelium discoideum]
Length = 2536
Score = 65.9 bits (159), Expect = 5e-09
Identities = 53/237 (22%), Positives = 99/237 (41%), Gaps = 20/237 (8%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
KE EK K + + + +++ + +E + + NS +K +SK +D + +S
Sbjct: 196 KEKEKAKKQKEKEKEKAKKLKEKEKEKKKKNSESEKPKKSNKSKKQEDTENESESESESE 255
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
+ S +SE+ + + + ++ + K ++ +TE E++ G+ E S D
Sbjct: 256 SESSSSESEKE---ESSKKSSSKKKTTTKEKSKPTKKSKKQETEDEEESGSDESSGDDDE 312
Query: 122 KDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSE---------SSRGLKERSDRKTNKL 172
K S ++EDE+ +K+SK + + P E R E SD +
Sbjct: 313 KSS--------SDEDEREKKSSKKSSTAANGKPQEIKPIVKKSTGKRKKNEDSDASSGDE 364
Query: 173 QSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEV 229
+ + D K G S+ K + +KP KA +KA ++ VLEV
Sbjct: 365 EEEEEDKTTTKKSKGKGGESKKVVKEKVTKVAKPTKASIKAQTKKGNPNPAIPVLEV 421
Score = 57.8 bits (138), Expect = 1e-06
Identities = 62/286 (21%), Positives = 104/286 (35%), Gaps = 48/286 (16%)
Query: 46 TPQDRSAANNLVGDSNTASENSETYENVVIDYVDDVNRSEEALAEM------KVNAMVAN 99
T D VG +++S + E++++ N +E+ + +VN +V N
Sbjct: 57 TVSDEDETEYKVGPESSSS--NIPIESIIVTTTLKANSKQESSYRISKIELRRVNTIVEN 114
Query: 100 QASDTEKEQKEGNGEVSDTDTVKDSVSSQGDS---------------------------- 131
++S E+E++E NGE ++S S +S
Sbjct: 115 ESSSEEEEEEESNGEEEKEGNAQESGSESEESESESEVPVKKTKKQLVEEQKKSAAAAKK 174
Query: 132 FTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQK-----KPMN 186
E EK ++ K K K K E ++ KE+ K KL+ K + +K KP
Sbjct: 175 LQKEKEKEKEKEKAKKFKEKEKEKEKAKKQKEKEKEKAKKLKEKEKEKKKKNSESEKPKK 234
Query: 187 STKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLG 246
S K + +N S + S ++ SESS E K +
Sbjct: 235 SNKSKKQEDTENESESES-------ESESESSSSESEKEESSKKSSSKKKTTTKEKSKPT 287
Query: 247 SEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELREVAA 292
+ ++ ET + EE+G E + K E EK + AA
Sbjct: 288 KKSKKQETEDEEESGSDESSGDDDEKSSSDEDEREKKSSKKSSTAA 333
Score = 51.6 bits (122), Expect = 9e-05
Identities = 67/302 (22%), Positives = 123/302 (40%), Gaps = 35/302 (11%)
Query: 4 TEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTA 63
T K + + S + S+ RR + + ++ SS E+ E ++ N +S +
Sbjct: 87 TLKANSKQESSYRISKIELRRVNTIVENESSS--EEEEEEESNGEEEKEGN--AQESGSE 142
Query: 64 SENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKE---QKEGNGEVSDTDT 120
SE SE+ V V ++++ L E + + A + EKE +KE + + +
Sbjct: 143 SEESESESEV------PVKKTKKQLVEEQKKSAAAAKKLQKEKEKEKEKEKAKKFKEKEK 196
Query: 121 VKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVS--PSESSRGLK----ERSDRKTNKLQS 174
K+ Q + + +K+++ K+ K K S P +S++ K E ++ +S
Sbjct: 197 EKEKAKKQKEKEKEKAKKLKEKEKEKKKKNSESEKPKKSNKSKKQEDTENESESESESES 256
Query: 175 KVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVK---------ASSESSEGVDENH 225
+ S S +K +S K S+ K T+ SKP K K S ESS DE
Sbjct: 257 ESSSSESEKEESSKKSSSK--KKTTTKEKSKPTKKSKKQETEDEEESGSDESSGDDDEKS 314
Query: 226 VLEVKEIEILDGASNGAQSLGSEDE-----RHETVNAEENGEHEDKAALELKIEEMELRV 280
+ E E + + G E + T ++N + + + E + EE +
Sbjct: 315 SSDEDEREKKSSKKSSTAANGKPQEIKPIVKKSTGKRKKNEDSDASSGDEEEEEEDKTTT 374
Query: 281 EK 282
+K
Sbjct: 375 KK 376
>UniRef100_Q7RH28 Mature-parasite-infected erythrocyte surface antigen [Plasmodium
yoelii yoelii]
Length = 1003
Score = 65.9 bits (159), Expect = 5e-09
Identities = 69/302 (22%), Positives = 123/302 (39%), Gaps = 46/302 (15%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLH------QDNSSKTLNEKGTESKTPQDRSAANN 55
KE E + +S+ K E E+K + S K++N +G+ S+ + S +
Sbjct: 717 KENENLSSYVDSKNKNEPNLEENENKRKSFSGESESESEKSINGEGSTSEESESESESEK 776
Query: 56 LVGDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVN---AMVANQASDTEKEQKEGN 112
V + SE SE+ + + + E+ +E V + S++EK K+G
Sbjct: 777 SVKKEESTSEESESESEKSVKKEESTSEESESESEKSVKKEESTSEESESESEKSVKKGG 836
Query: 113 GEVSDTDTVKDSVSSQGDSFTNEDE--------KVEKASKDA---------KSKVKVSPS 155
++D+ + + +G S + E E K E S+++ K + K S
Sbjct: 837 STSEESDSESEKSAKKGGSASGESESESEKSVKKEESTSEESETESEESKKKGEFKSKES 896
Query: 156 ESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASS 215
ES S++ K +S +S + + KG S T++ + S + K VK K S
Sbjct: 897 ES------ESEKSVKKEESTSEESESESEKSVKKGGS--TSEESESESEKSVK---KGGS 945
Query: 216 ESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEE 275
S E E+ KE G+ S SE E ++V EE+ E ++ + + E
Sbjct: 946 TSEESESESEKSVKKE---------GSTSEESESESEKSVKKEESTSEESESESDDNVVE 996
Query: 276 ME 277
+
Sbjct: 997 TQ 998
Score = 55.5 bits (132), Expect = 6e-06
Identities = 55/235 (23%), Positives = 104/235 (43%), Gaps = 31/235 (13%)
Query: 1 MKETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDS 60
+K+ E S+++ S + E + + S K++ + G+ S+ S + G S
Sbjct: 796 VKKEESTSEESESESEKSVKKEESTSEESESESEKSVKKGGSTSEESDSESEKSAKKGGS 855
Query: 61 NTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDT 120
+ SE+ ++V + EE+ +E S+TE E+ + GE
Sbjct: 856 ASGESESESEKSV---------KKEESTSE----------ESETESEESKKKGEFKS--- 893
Query: 121 VKDSVSSQGDSFTNEDEKVEKASKDAKSKVKV--SPSESSRGLKERSDRKTNKLQSKVSD 178
K+S S S E+ E++ +++ VK S SE S E+S +K S+ S+
Sbjct: 894 -KESESESEKSVKKEESTSEESESESEKSVKKGGSTSEESESESEKSVKKGGS-TSEESE 951
Query: 179 SNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIE 233
S +K + T++ + S + K VK +++SE SE +++V+E + E
Sbjct: 952 SESEKSVKKEGS----TSEESESESEKSVKKE-ESTSEESESESDDNVVETQSEE 1001
Score = 42.0 bits (97), Expect = 0.072
Identities = 46/184 (25%), Positives = 79/184 (42%), Gaps = 31/184 (16%)
Query: 112 NGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNK 171
N +++D K + D+ N D+K + ++ KS K + ++ +K S + +N
Sbjct: 82 NKKINDVLIKKKKRDTLEDTQKNVDKKNDYLYRNNKSLRKGTEPNNTFLIKSSSPKISNF 141
Query: 172 LQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKE 231
++K K S V+ KN S +P + + SS
Sbjct: 142 QKNK-------KMGTSVISTKPVSTKNNMSLFQRPRRKNIIYSS---------------- 178
Query: 232 IEILDGASNGAQSLGSEDERHETVNAEENGEHED---KAALELKIEEMEL--RVEKLEEE 286
LD + G + + E+E +E V EE E ED + +E + EE+E+ VE++EEE
Sbjct: 179 ---LDSSEEGKEVIEEEEEEYEEVEVEEEEEVEDEYEEVEVEEEYEEVEVEEEVEEVEEE 235
Query: 287 LREV 290
EV
Sbjct: 236 YEEV 239
>UniRef100_Q8MTN7 Glutamic acid-rich protein cNBL1700 [Trichinella spiralis]
Length = 571
Score = 65.9 bits (159), Expect = 5e-09
Identities = 66/313 (21%), Positives = 127/313 (40%), Gaps = 36/313 (11%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKT-LNEKGTESKTPQDRSAANNLVGDS 60
+E ++ AS ++G + +E++ + N ++ + + +E +T ++ +A+ +
Sbjct: 233 EEEDEESASEEQTSEGEEKGASQEEEEDEGNEQESEVESQASEEQTSEEEESASEEEDEE 292
Query: 61 NTASENSETYENVVIDYVDDVNRSE--EALAEMKVNAMVANQASDTEKEQKEGNGEVSDT 118
N + E + E + D+ + SE E A + N++ + E++E E D
Sbjct: 293 NESKEQTTEEEESASEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDE 352
Query: 119 DTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESS----RGLKERSDRKTNKLQS 174
++V + +S+G+ E+ E D +S+V+ SE G E D + N+ +
Sbjct: 353 ESVSEEQTSEGEEKGASQEEEEDEGNDQESEVESQASEEQTSEEEGASEEEDEE-NESEE 411
Query: 175 KVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEI 234
+ ++ + S KN S E EG ++ +E
Sbjct: 412 QTTEEESASEEEDEESASEGEEKNASQEE------------EEDEGNEQESEVE------ 453
Query: 235 LDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELREVAALE 294
S ++ SE+E E + EE+ E+E + E EE E E EE E +A E
Sbjct: 454 ----SQASEEQTSEEEEKEGASQEEDEENESE---EQTSEEEE---EGASEEEDEESAFE 503
Query: 295 VSLYSVVPEHGSS 307
E G+S
Sbjct: 504 EQTSEEEEEKGAS 516
Score = 63.5 bits (153), Expect = 2e-08
Identities = 62/308 (20%), Positives = 118/308 (38%), Gaps = 24/308 (7%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAAN---NLVG 58
KE+E++ + + K +D + NS + + TP+++++ +
Sbjct: 111 KESEEKSSQEKDEDKSESEASEEKDVSQEQNSKEEKGASEEDEDTPEEQNSKEENGSSEE 170
Query: 59 DSNTASENSETYE--------NVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKE 110
D ASE + E N V + + E+ + + V +QAS+ E+ +E
Sbjct: 171 DDEDASEEQASNEEKEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASE-EQTTEE 229
Query: 111 GNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERS----- 165
G E D ++ + +S+G+ E+ E + +S+V+ SE +E S
Sbjct: 230 GASEEEDEESASEEQTSEGEEKGASQEEEEDEGNEQESEVESQASEEQTSEEEESASEEE 289
Query: 166 DRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPV--KAPVKASSESSEGVDE 223
D + + + + S KN S + ++ + + E +E
Sbjct: 290 DEENESKEQTTEEEESASEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESASEE 349
Query: 224 NHVLEVKEIEILDGASNGAQSLGSEDERHETVN-----AEENGEHEDKAALELKIEEMEL 278
V E + +G GA EDE ++ + A E E++ A E + EE E
Sbjct: 350 EDEESVSEEQTSEGEEKGASQEEEEDEGNDQESEVESQASEEQTSEEEGASEEEDEENES 409
Query: 279 RVEKLEEE 286
+ EEE
Sbjct: 410 EEQTTEEE 417
Score = 63.2 bits (152), Expect = 3e-08
Identities = 63/305 (20%), Positives = 121/305 (39%), Gaps = 22/305 (7%)
Query: 2 KETEKR----KASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLV 57
K T K K S+ +++ E+ EDK + S + + E + +++ A+
Sbjct: 96 KPTSKEESGEKTSQEKESEEKSSQEKDEDKSESEASEE--KDVSQEQNSKEEKGASEE-- 151
Query: 58 GDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSD 117
D +T E + EN + D+ E+A E K + N S+ K E E D
Sbjct: 152 -DEDTPEEQNSKEENGSSEEDDEDASEEQASNEEKEASEEKNTVSEERKGASEEEDEEKD 210
Query: 118 TDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKV- 176
+ S + T E+ E+ +++ S+ + S E +E + + N+ +S+V
Sbjct: 211 DGHESEVESQASEEQTTEEGASEEEDEESASEEQTSEGEEKGASQEEEEDEGNEQESEVE 270
Query: 177 SDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILD 236
S +++++ + S ++ S + + E E E + E D
Sbjct: 271 SQASEEQTSEEEESASEEEDEENESKEQTTEEEESASEEEDEESASEREEKNASQEEEED 330
Query: 237 GASNGAQSL------GSEDERHETVNAEENGEHEDKAALELKIE------EMELRVEKLE 284
+ + SE+E E+V+ E+ E E+K A + + E E E+ + E
Sbjct: 331 EGNESKEQTTEEEESASEEEDEESVSEEQTSEGEEKGASQEEEEDEGNDQESEVESQASE 390
Query: 285 EELRE 289
E+ E
Sbjct: 391 EQTSE 395
Score = 59.3 bits (142), Expect = 4e-07
Identities = 53/280 (18%), Positives = 118/280 (41%), Gaps = 14/280 (5%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
++T + + S + + +ER E Q+ NE E T ++ SA+ +S
Sbjct: 297 EQTTEEEESASEEEDEESASEREEKNASQEEEEDEGNES-KEQTTEEEESASEEEDEESV 355
Query: 62 TASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTV 121
+ + SE E ++ EE + V +QAS+ + ++EG E D +
Sbjct: 356 SEEQTSEGEEK-------GASQEEEEDEGNDQESEVESQASEEQTSEEEGASEEEDEENE 408
Query: 122 KDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQ 181
+ +++ +S + E++ E+++ + + K E G ++ S+ ++ + + S+ +
Sbjct: 409 SEEQTTEEESASEEED--EESASEGEEKNASQEEEEDEGNEQESEVESQASEEQTSEEEE 466
Query: 182 KKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASS----ESSEGVDENHVLEVKEIEILDG 237
K+ + + + + TS + S ++SE +E + +E + +
Sbjct: 467 KEGASQEEDEENESEEQTSEEEEEGASEEEDEESAFEEQTSEEEEEKGASQEEEEDEENE 526
Query: 238 ASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEME 277
+ +S SE++ E A E G+ + E + EE E
Sbjct: 527 QESEVESQASEEQTSEEEGASEEGQDASEEEDEDESEEEE 566
Score = 59.3 bits (142), Expect = 4e-07
Identities = 61/295 (20%), Positives = 117/295 (38%), Gaps = 14/295 (4%)
Query: 2 KETEKRKASRNSQTKGSRRTERRE-DKLH------QDNSSKTLNEKGTESKTPQDRSAAN 54
KE + K + + + KG+ E E D H Q + +T E +E + + S
Sbjct: 185 KEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQTTEEGASEEEDEESASEEQ 244
Query: 55 NLVGDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGE 114
G+ AS+ E E + + SEE +E + + A++ D E E KE E
Sbjct: 245 TSEGEEKGASQEEEEDEGNEQESEVESQASEEQTSEEEES---ASEEEDEENESKEQTTE 301
Query: 115 VSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQS 174
++ + ++ S + + E+ + +SK + + E S +E + + + S
Sbjct: 302 EEESASEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESVSEEQTS 361
Query: 175 KVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLE---VKE 231
+ + + +G + + + ++ + + + E E E E E
Sbjct: 362 EGEEKGASQEEEEDEGNDQESEVESQASEEQTSEEEGASEEEDEENESEEQTTEEESASE 421
Query: 232 IEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEE 286
E + AS G + S++E + N E+ E E +A+ E EE E EE+
Sbjct: 422 EEDEESASEGEEKNASQEEEEDEGN-EQESEVESQASEEQTSEEEEKEGASQEED 475
Score = 58.9 bits (141), Expect = 6e-07
Identities = 59/289 (20%), Positives = 118/289 (40%), Gaps = 25/289 (8%)
Query: 3 ETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNT 62
E E S+ S+ + S E ++ ++N SK + ES + ++ + + + N
Sbjct: 264 EQESEVESQASEEQTSEEEESASEEEDEENESKEQTTEEEESASEEEDEESASEREEKNA 323
Query: 63 ASENSETYENVVIDYVDDVNRS------EEALAEMKVNAMVANQASDTEKEQKEGN---- 112
+ E E N + + S EE+++E + + AS E+E+ EGN
Sbjct: 324 SQEEEEDEGNESKEQTTEEEESASEEEDEESVSEEQTSEGEEKGASQ-EEEEDEGNDQES 382
Query: 113 ---GEVSDTDTVKDSVSSQGDSFTNEDEK---VEKASKDAKSKVKVSPSESSRGLKERSD 166
+ S+ T ++ +S+ + NE E+ E+++ + + + S E +E +
Sbjct: 383 EVESQASEEQTSEEEGASEEEDEENESEEQTTEEESASEEEDEESASEGEEKNASQEEEE 442
Query: 167 RKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHV 226
+ N+ +S+V ++ + + + + N S+ + S E EG E
Sbjct: 443 DEGNEQESEVESQASEEQTSEEEEKEGASQEEDEENESEE-----QTSEEEEEGASEE-- 495
Query: 227 LEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEE 275
E +E + S + G+ E E E+ E E +A+ E EE
Sbjct: 496 -EDEESAFEEQTSEEEEEKGASQEEEEDEENEQESEVESQASEEQTSEE 543
Score = 57.0 bits (136), Expect = 2e-06
Identities = 56/290 (19%), Positives = 124/290 (42%), Gaps = 23/290 (7%)
Query: 5 EKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTAS 64
E + + + + E + ++ ++ + KG + +++ + +S AS
Sbjct: 164 ENGSSEEDDEDASEEQASNEEKEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQ-AS 222
Query: 65 ENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDS 124
E +T E + D+ + SEE +E + + + E+E+ EGN + S+ +
Sbjct: 223 EE-QTTEEGASEEEDEESASEEQTSEGE------EKGASQEEEEDEGNEQESEVE----- 270
Query: 125 VSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKP 184
S + T+E+E+ +D +++ K +E E D ++ + + + S +++
Sbjct: 271 -SQASEEQTSEEEESASEEEDEENESKEQTTEEEESASEEEDEESASEREEKNASQEEEE 329
Query: 185 MNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQS 244
+ + T + S++ + ++ + ++SEG ++ E +E E D S +S
Sbjct: 330 DEGNESKEQTTEEEESASEEEDEES--VSEEQTSEGEEKGASQEEEEDEGNDQESE-VES 386
Query: 245 LGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELREVAALE 294
SE++ E A E + E+++ EE E EE E +A E
Sbjct: 387 QASEEQTSEEEGASEEEDEENES------EEQTTEEESASEEEDEESASE 430
Score = 49.3 bits (116), Expect = 5e-04
Identities = 70/313 (22%), Positives = 123/313 (38%), Gaps = 29/313 (9%)
Query: 16 KGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASENSETYENVVI 75
K +R +D +D + T+ EK +E+ +++ + G+ + + SE +
Sbjct: 64 KWKKREGNSDDICTEDET--TVIEKESENGVDKEKPTSKEESGEKTSQEKESEEKSS--- 118
Query: 76 DYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTN- 134
D ++SE +E K + N KE+K + E DT ++S G S +
Sbjct: 119 -QEKDEDKSESEASEEKDVSQEQNS-----KEEKGASEEDEDTPEEQNSKEENGSSEEDD 172
Query: 135 EDEKVEKASKDAK--SKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPS 192
ED E+AS + K S+ K + SE +G E D + + +S + + +G S
Sbjct: 173 EDASEEQASNEEKEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQTTEEGAS 232
Query: 193 RVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSE-DER 251
++ ++S AS E E EV+ + S +S E DE
Sbjct: 233 EEEDEESASEEQTSEGEEKGASQEEEEDEGNEQESEVESQASEEQTSEEEESASEEEDEE 292
Query: 252 HETVNA-------------EENG-EHEDKAALELKIEEMELRVEKLEEELREVAALEVSL 297
+E+ EE+ E E+K A + + E+ ++ E E A+ E
Sbjct: 293 NESKEQTTEEEESASEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDE 352
Query: 298 YSVVPEHGSSAHK 310
SV E S +
Sbjct: 353 ESVSEEQTSEGEE 365
Score = 48.1 bits (113), Expect = 0.001
Identities = 48/274 (17%), Positives = 111/274 (39%), Gaps = 17/274 (6%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSN 61
+E ++ AS + S+ E E ++ +++ E +E + + S G+
Sbjct: 308 EEEDEESASEREEKNASQEEEEDEGNESKEQTTEE-EESASEEEDEESVSEEQTSEGEEK 366
Query: 62 TASENSET-----YENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVS 116
AS+ E E+ V + SEE A + + ++ TE+E S
Sbjct: 367 GASQEEEEDEGNDQESEVESQASEEQTSEEEGASEEEDEENESEEQTTEEE--------S 418
Query: 117 DTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKV 176
++ + +S+G+ E+ E + +S+V+ SE +E + + + + +
Sbjct: 419 ASEEEDEESASEGEEKNASQEEEEDEGNEQESEVESQASEEQTSEEEEKEGASQE-EDEE 477
Query: 177 SDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILD 236
++S ++ +G S ++ ++ + K +S+ E +EN + E+E
Sbjct: 478 NESEEQTSEEEEEGASEEEDEESAFEEQTSEEEEEKGASQEEEEDEENE--QESEVESQA 535
Query: 237 GASNGAQSLGSEDERHETVNAEENGEHEDKAALE 270
++ G+ +E + E+ E E++ + E
Sbjct: 536 SEEQTSEEEGASEEGQDASEEEDEDESEEEESDE 569
Score = 47.8 bits (112), Expect = 0.001
Identities = 49/228 (21%), Positives = 87/228 (37%), Gaps = 18/228 (7%)
Query: 77 YVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNED 136
Y D V+R + + ++ + EKE + G + T S G+ + E
Sbjct: 57 YDDGVDRKWKKREGNSDDICTEDETTVIEKESENGVDKEKPT-----SKEESGEKTSQEK 111
Query: 137 EKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSK-----VSDSNQKKPMNSTKGP 191
E EK+S++ S + + + + + K K S+ + N K+ S++
Sbjct: 112 ESEEKSSQEKDEDKSESEASEEKDVSQEQNSKEEKGASEEDEDTPEEQNSKEENGSSEED 171
Query: 192 SRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLE-----VKEIEILDGASNGAQSLG 246
++ +SN K SE +G E E E+E +
Sbjct: 172 DEDASEEQASNEEKEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQTTEEGA 231
Query: 247 SEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELREVAALE 294
SE+E E+ + E+ E E+K A + EE E + E E+ A+ E
Sbjct: 232 SEEEDEESASEEQTSEGEEKGASQ---EEEEDEGNEQESEVESQASEE 276
>UniRef100_Q86BT4 Phoshoprotein 300 [Plasmodium falciparum]
Length = 597
Score = 65.5 bits (158), Expect = 6e-09
Identities = 71/291 (24%), Positives = 125/291 (42%), Gaps = 21/291 (7%)
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQ-DRSAANNLVGDS 60
KE +KR RN+ + E +++ + ++ + +NEK T +K ++ V +
Sbjct: 150 KEVKKRVKKRNN------KNENKDNVIGKEIMKEDVNEKDTANKDKVIEQEKEKEEVKEK 203
Query: 61 NTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDT 120
E E E + ++V EE + ++ D Q+ N EV + DT
Sbjct: 204 EEVKEKEEVKEKEEVKEKEEVKEKEEVKEK-------DTESRDNVIVQEIMNEEVKEKDT 256
Query: 121 V-KDSVSSQGDSFTNED-EKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSD 178
KD+V Q NED + + SKD +V E +K+R +++ NK ++ +
Sbjct: 257 ESKDNVIVQ--EIMNEDVNEKDTESKDKMIGKEVIIEEVKEEVKKRVNKEVNKRVNRRNR 314
Query: 179 SNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGA 238
N++K + + S N+ + NN K + VK + + E V E E E + +
Sbjct: 315 KNERKDVIEQEIVSEEVNEKDTKNNDKKIGKRVKKPIDDCK--KEREVQEESEEESEEES 372
Query: 239 SNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELRE 289
++ SE+E E E E E+++ E + E E E+ EEE E
Sbjct: 373 EEESEE-ESEEESEEESEEESEEESEEESEEESEEESEEESEEESEEESEE 422
Score = 60.5 bits (145), Expect = 2e-07
Identities = 58/294 (19%), Positives = 119/294 (39%), Gaps = 18/294 (6%)
Query: 5 EKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQ-DRSAANNLVGDSNTA 63
E +K + K + + ER+++ + ++ + +NEK T +K + ++ V +
Sbjct: 8 EVKKKVKKRVKKRNNKNERKDNVIGKEIMKEDVNEKDTANKDKEIEQEKEKEEVKEKEEV 67
Query: 64 SENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQK--EGNGEVSDTDTV 121
E E E + ++V EE + +V + DTE + K E E + V
Sbjct: 68 KEKEEVKEKEEVKEKEEVKEKEEVKEKEEVKEKEEVKEKDTESKDKEIEQEKEKEEVKEV 127
Query: 122 KDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRG--------LKERSDRKTNKLQ 173
K+ + D ++ +E+ K+ K +VK +++ +KE + K +
Sbjct: 128 KEKDTENKDKVIGQEIIIEEIKKEVKKRVKKRNNKNENKDNVIGKEIMKEDVNEKDTANK 187
Query: 174 SKVSDSNQKKPMNSTKGPSRVTNK-----NTSSNNSKPVKAPVKASSESSEGVDENHVLE 228
KV + Q+K K V K + VK + + +E D V E
Sbjct: 188 DKVIE--QEKEKEEVKEKEEVKEKEEVKEKEEVKEKEEVKEKEEVKEKDTESRDNVIVQE 245
Query: 229 VKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEK 282
+ E+ + + ++ ++ +E VN ++ + E+ IEE++ V+K
Sbjct: 246 IMNEEVKEKDTESKDNVIVQEIMNEDVNEKDTESKDKMIGKEVIIEEVKEEVKK 299
Score = 59.3 bits (142), Expect = 4e-07
Identities = 57/296 (19%), Positives = 122/296 (40%), Gaps = 36/296 (12%)
Query: 2 KETEKR-----KASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTP--------- 47
K+TE R + N + K + TE +++ + Q+ ++ +NEK TESK
Sbjct: 233 KDTESRDNVIVQEIMNEEVK-EKDTESKDNVIVQEIMNEDVNEKDTESKDKMIGKEVIIE 291
Query: 48 --------QDRSAANNLVGDSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVAN 99
+ N V N +E + E ++ ++VN + + K+ V
Sbjct: 292 EVKEEVKKRVNKEVNKRVNRRNRKNERKDVIEQEIVS--EEVNEKDTKNNDKKIGKRVKK 349
Query: 100 QASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSR 159
D +KE++ +++ + S + +E+E E++ ++++ + S ES
Sbjct: 350 PIDDCKKEREVQEESEEESEEESEEESEEESEEESEEESEEESEEESEEE---SEEESEE 406
Query: 160 GLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVK---ASSE 216
+E S+ ++ + + S+ ++ + K S + ++ K ++ +
Sbjct: 407 ESEEESEEESEEESEEESEEESEEESDEEKNTSGLVHRRNCKKEKKYNNGELEEYYKEKQ 466
Query: 217 SSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVNAEENGEHEDKAALELK 272
+ E DE ++++ KE L+ N A + E H ++ HED +ELK
Sbjct: 467 NEEYFDEEYIIQSKEHNTLNTFPNMALNEDFRREFHNILSI-----HEDTDLMELK 517
Score = 38.5 bits (88), Expect = 0.80
Identities = 52/234 (22%), Positives = 94/234 (39%), Gaps = 30/234 (12%)
Query: 64 SENSETYENVVIDYV--DDVNRSEEA-----LAEMKVNAMVANQASDTEKEQKEGNGEVS 116
+ +E +NV+ + +DVN + A + + K V + EKE+ + EV
Sbjct: 21 NNKNERKDNVIGKEIMKEDVNEKDTANKDKEIEQEKEKEEVKEKEEVKEKEEVKEKEEVK 80
Query: 117 DTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKV 176
+ + VK+ ++E EK K +VK +ES E+ K + K
Sbjct: 81 EKEEVKE-----------KEEVKEKEEVKEKEEVKEKDTESKDKEIEQEKEKEEVKEVKE 129
Query: 177 SDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILD 236
D+ K + + + K VK VK + +E D E+ + ++ +
Sbjct: 130 KDTENKDKV--------IGQEIIIEEIKKEVKKRVKKRNNKNENKDNVIGKEIMKEDVNE 181
Query: 237 -GASNGAQSLGSEDERHETVNAEENGEHEDKAALELKIEEMELRVEKLEEELRE 289
+N + + E E+ E EE E E+ ++ K E E K +EE++E
Sbjct: 182 KDTANKDKVIEQEKEKEEVKEKEEVKEKEE---VKEKEEVKEKEEVKEKEEVKE 232
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.127 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,333,795,852
Number of Sequences: 2790947
Number of extensions: 58158712
Number of successful extensions: 268415
Number of sequences better than 10.0: 8130
Number of HSP's better than 10.0 without gapping: 441
Number of HSP's successfully gapped in prelim test: 8034
Number of HSP's that attempted gapping in prelim test: 227836
Number of HSP's gapped (non-prelim): 28094
length of query: 799
length of database: 848,049,833
effective HSP length: 136
effective length of query: 663
effective length of database: 468,481,041
effective search space: 310602930183
effective search space used: 310602930183
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0318.1


