

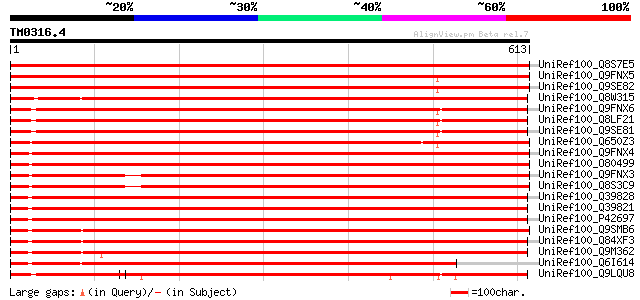
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0316.4
(613 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8S7E5 Putative phragmoplastin [Oryza sativa] 1036 0.0
UniRef100_Q9FNX5 Dynamin-like protein DLP2 [Arabidopsis thaliana] 1022 0.0
UniRef100_Q9SE82 Dynamin-like protein 4 [Arabidopsis thaliana] 984 0.0
UniRef100_Q8W315 Putative GTP-binding protein [Oryza sativa] 971 0.0
UniRef100_Q9FNX6 Dynamin-like protein DLP1 [Arabidopsis thaliana] 962 0.0
UniRef100_Q8LF21 Dynamin, putative [Arabidopsis thaliana] 961 0.0
UniRef100_Q9SE81 Dynamin-like protein 5 [Arabidopsis thaliana] 960 0.0
UniRef100_Q650Z3 Putative phragmoplastin 12 [Oryza sativa] 957 0.0
UniRef100_Q9FNX4 Dynamin-like protein DLP3a [Arabidopsis thaliana] 939 0.0
UniRef100_O80499 Putative phragmoplastin [Arabidopsis thaliana] 935 0.0
UniRef100_Q9FNX3 Dynamin-like protein DLP3b [Arabidopsis thaliana] 898 0.0
UniRef100_Q8S3C9 Dynamin-like protein D [Arabidopsis thaliana] 894 0.0
UniRef100_Q39828 SDL5A [Glycine max] 883 0.0
UniRef100_Q39821 SDL12A [Glycine max] 875 0.0
UniRef100_P42697 Dynamin-like protein [Arabidopsis thaliana] 868 0.0
UniRef100_Q9SMB6 Phragmoplastin [Nicotiana tabacum] 863 0.0
UniRef100_Q84XF3 Dynamin-like protein B [Arabidopsis thaliana] 847 0.0
UniRef100_Q9M362 Dynamin-like protein [Arabidopsis thaliana] 837 0.0
UniRef100_Q6I614 Putative dynamin [Oryza sativa] 764 0.0
UniRef100_Q9LQU8 F10B6.23 [Arabidopsis thaliana] 711 0.0
>UniRef100_Q8S7E5 Putative phragmoplastin [Oryza sativa]
Length = 618
Score = 1036 bits (2678), Expect = 0.0
Identities = 513/615 (83%), Positives = 579/615 (93%), Gaps = 2/615 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME++I LVNRIQRACTVLGDHG S +LWEALP+VAVVGGQSSGKSSVLESIVGR
Sbjct: 4 MENVIVLVNRIQRACTVLGDHGGGDGTASLPTLWEALPSVAVVGGQSSGKSSVLESIVGR 63
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G QEYAEFLHMP+R+F DF+LVR+EIQDETDR+TGK
Sbjct: 64 DFLPRGSGIVTRRPLVLQLHKTEDGVQEYAEFLHMPKRRFNDFALVRKEIQDETDRLTGK 123
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
TKQISP+PIHLSIYSP+VVNLTL+DLPGLTKVAVEGQPESIVQ+IE MVR+YV+KPN II
Sbjct: 124 TKQISPVPIHLSIYSPHVVNLTLIDLPGLTKVAVEGQPESIVQDIENMVRSYVDKPNCII 183
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLAR+VDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW
Sbjct: 184 LAISPANQDIATSDAIKLARDVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 243
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINKN+DMIIARRKE+E+FA+SP+Y HL+++MGSEYLAKLLSQHLE+VIRA
Sbjct: 244 VGIVNRSQADINKNIDMIIARRKEQEFFASSPEYSHLSSRMGSEYLAKLLSQHLEAVIRA 303
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSITSLINK+++ELESEMDH+GRPIA DAGAQLY +LELCRAFE+IF+EHLDGGRPGG
Sbjct: 304 RIPSITSLINKTIDELESEMDHIGRPIASDAGAQLYLVLELCRAFEKIFREHLDGGRPGG 363
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
DRIY VFDNQLP+ALRKLPFDR+LSLQNV++V+SEADGYQPHLIAPEQGYRRLI+SAL+Y
Sbjct: 364 DRIYGVFDNQLPSALRKLPFDRYLSLQNVKRVISEADGYQPHLIAPEQGYRRLIESALNY 423
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASVDAV++VLKELVRKSI ET+ELKRFPT QA LA A ALERFRE+ +KTT+
Sbjct: 424 FRGPAEASVDAVHYVLKELVRKSIGETQELKRFPTLQAELAAACFHALERFREDGRKTTV 483
Query: 481 RLVDMESSYLTVDFFRRLPQEVEK--AGGPATPNVDRYAEGHFRRIASNVSSYIGLVADT 538
RLVDMES+YLTV+FFR+LPQEV+K G P+TP+VDRYA+ HFRRIASNVSSYIG+V+DT
Sbjct: 484 RLVDMESAYLTVEFFRKLPQEVDKTGTGNPSTPSVDRYADAHFRRIASNVSSYIGMVSDT 543
Query: 539 LRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLEL 598
L+NTIPKAVV CQVR+AK+SLLN+FYTQ+G+K+ KQL+++LDEDPALMERRQQC KRLEL
Sbjct: 544 LKNTIPKAVVHCQVREAKRSLLNYFYTQVGRKDAKQLAQLLDEDPALMERRQQCFKRLEL 603
Query: 599 YKAARDEIDSVSWVR 613
YK+ARDEID+VSW R
Sbjct: 604 YKSARDEIDAVSWSR 618
>UniRef100_Q9FNX5 Dynamin-like protein DLP2 [Arabidopsis thaliana]
Length = 624
Score = 1022 bits (2643), Expect = 0.0
Identities = 500/621 (80%), Positives = 574/621 (91%), Gaps = 8/621 (1%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLIGLVNRIQRACTVLGD+G N+F+SLWEALPTVAVVGGQSSGKSSVLESIVGR
Sbjct: 4 MESLIGLVNRIQRACTVLGDYGGGTGSNAFNSLWEALPTVAVVGGQSSGKSSVLESIVGR 63
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK + G++EYAEFLH+P+++FTDF+LVR+EIQDETDR+TGK
Sbjct: 64 DFLPRGSGIVTRRPLVLQLHKTDDGTEEYAEFLHLPKKQFTDFALVRREIQDETDRITGK 123
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
KQISP+PIHLSIYSPNVVNLTL+DLPGLTKVAVEGQPE+I ++IE+MVRTYV+KPN II
Sbjct: 124 NKQISPVPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQPETIAEDIESMVRTYVDKPNCII 183
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLA++VDPTGERTFGVLTKLDLMDKGTNAL+VLEGRSYRLQHPW
Sbjct: 184 LAISPANQDIATSDAIKLAKDVDPTGERTFGVLTKLDLMDKGTNALEVLEGRSYRLQHPW 243
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINKNVDM++ARRKEREYF SPDYGHLA+KMGSEYLAKLLS+HLESVIR
Sbjct: 244 VGIVNRSQADINKNVDMMLARRKEREYFDTSPDYGHLASKMGSEYLAKLLSKHLESVIRT 303
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSI SLINKS+EELE E+D +GRP+A+DAGAQLYTILE+CRAF++IFKEHLDGGRPGG
Sbjct: 304 RIPSILSLINKSIEELERELDRMGRPVAVDAGAQLYTILEMCRAFDKIFKEHLDGGRPGG 363
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
DRIY VFDNQLPAAL+KLPFDRHLSLQ+V+K+VSEADGYQPHLIAPEQGYRRLI+ AL Y
Sbjct: 364 DRIYGVFDNQLPAALKKLPFDRHLSLQSVKKIVSEADGYQPHLIAPEQGYRRLIEGALGY 423
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASVDAV++VLKELVRKSI+ET+ELKRFP+ Q LA AAN +LE+FREESKK+ +
Sbjct: 424 FRGPAEASVDAVHYVLKELVRKSISETEELKRFPSLQVELAAAANSSLEKFREESKKSVI 483
Query: 481 RLVDMESSYLTVDFFRRLPQEVEK--------AGGPATPNVDRYAEGHFRRIASNVSSYI 532
RLVDMES+YLT +FFR+LPQE+E+ P++ +D+Y +GHFRRIASNVS+Y+
Sbjct: 484 RLVDMESAYLTAEFFRKLPQEIERPVTNSKNQTASPSSATLDQYGDGHFRRIASNVSAYV 543
Query: 533 GLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQC 592
+V+DTLRNTIPKA V+CQVRQAK +LLN+FY+QI K+EGKQL ++LDEDPALM+RR +C
Sbjct: 544 NMVSDTLRNTIPKACVYCQVRQAKLALLNYFYSQISKREGKQLGQLLDEDPALMDRRLEC 603
Query: 593 AKRLELYKAARDEIDSVSWVR 613
AKRLELYK ARDEID+V+WVR
Sbjct: 604 AKRLELYKKARDEIDAVAWVR 624
>UniRef100_Q9SE82 Dynamin-like protein 4 [Arabidopsis thaliana]
Length = 626
Score = 984 bits (2545), Expect = 0.0
Identities = 489/623 (78%), Positives = 562/623 (89%), Gaps = 10/623 (1%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLIGLVNRIQRACTVLGD+G N+F+SLWEALPTVAVVGGQSSGKSSVLESIVGR
Sbjct: 4 MESLIGLVNRIQRACTVLGDYGGGTGSNAFNSLWEALPTVAVVGGQSSGKSSVLESIVGR 63
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK + G++EYAEFLH+P+++FTDF+LVR+EIQDETDR+TGK
Sbjct: 64 DFLPRGSGIVTRRPLVLQLHKTDDGTEEYAEFLHLPKKQFTDFALVRREIQDETDRITGK 123
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
KQISP+PIHLSIYSPNVVNLTL+DLPGLTKVAVEGQPE+I ++IE+MVRTYV+KPN II
Sbjct: 124 NKQISPVPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQPETIAEDIESMVRTYVDKPNCII 183
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLA++VDPTGERTFGVLTKLDLMDKGTNAL+VLEGRSYRLQHPW
Sbjct: 184 LAISPANQDIATSDAIKLAKDVDPTGERTFGVLTKLDLMDKGTNALEVLEGRSYRLQHPW 243
Query: 241 VGIVNR-SQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIR 299
VGI ++ DINKNVDM++ARRKEREYF SPDYGHLA+KMGSEYLAKLLS+HLESVIR
Sbjct: 244 VGISEPFNKQDINKNVDMMLARRKEREYFDTSPDYGHLASKMGSEYLAKLLSKHLESVIR 303
Query: 300 ARIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPG 359
RIPSI SLINKS+EELE E+D +GRP+A+DAGAQLYTILE+CRAF++IFKEHLDGGRPG
Sbjct: 304 TRIPSILSLINKSIEELERELDRMGRPVAVDAGAQLYTILEMCRAFDKIFKEHLDGGRPG 363
Query: 360 GDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALS 419
GDRIY VFDNQLPAAL+KLPFDRHLSLQ+V+K+VSEADGYQ LIAPEQGYRRLI+ AL
Sbjct: 364 GDRIYGVFDNQLPAALKKLPFDRHLSLQSVKKIVSEADGYQLTLIAPEQGYRRLIEGALG 423
Query: 420 YFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTT 479
YFRGPAEASVDAV++VLKELVRKSI+ET+ELKRFP+ Q LA AAN +LE+FREESKK+
Sbjct: 424 YFRGPAEASVDAVHYVLKELVRKSISETEELKRFPSLQVELAAAANSSLEKFREESKKSV 483
Query: 480 LRLVDMESSYLTVDFFRRLPQEVE---------KAGGPATPNVDRYAEGHFRRIASNVSS 530
+RLVDMES+YLT +FFR+LPQE+E K ++ +D+Y +GHFRRIASNVS+
Sbjct: 484 IRLVDMESAYLTAEFFRKLPQEIERPVTSKQKPKRASLSSATLDQYGDGHFRRIASNVSA 543
Query: 531 YIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQ 590
Y+ LRNTIPKA V+CQVRQAK +LLN+FY+QI K+EGKQL ++LDEDPALM+RR
Sbjct: 544 YVKWFRTLLRNTIPKACVYCQVRQAKLALLNYFYSQISKREGKQLGQLLDEDPALMDRRL 603
Query: 591 QCAKRLELYKAARDEIDSVSWVR 613
+CAKRLELYK ARDEID+V+WVR
Sbjct: 604 ECAKRLELYKKARDEIDAVAWVR 626
>UniRef100_Q8W315 Putative GTP-binding protein [Oryza sativa]
Length = 611
Score = 971 bits (2511), Expect = 0.0
Identities = 484/611 (79%), Positives = 550/611 (89%), Gaps = 4/611 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
M SLIGLVNRIQRACTVLGDHG +G S LWEALP+VAVVGGQSSGKSSVLESIVGR
Sbjct: 4 MGSLIGLVNRIQRACTVLGDHGGGGEGGS---LWEALPSVAVVGGQSSGKSSVLESIVGR 60
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G QEYAEFLH PR++FTDF+ VR+EI DETDR+TGK
Sbjct: 61 DFLPRGSGIVTRRPLVLQLHKTE-GGQEYAEFLHAPRKRFTDFAAVRKEIADETDRITGK 119
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
TK IS IPIHLSIYSP+VVNLTL+DLPGLTKVAVEGQ ESIVQ+IE MVR+YV+KPNSII
Sbjct: 120 TKAISNIPIHLSIYSPHVVNLTLIDLPGLTKVAVEGQQESIVQDIENMVRSYVDKPNSII 179
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLAR+VDP+G+RTFGVLTKLDLMDKGTNA+DVLEGR YRLQHPW
Sbjct: 180 LAISPANQDIATSDAIKLARDVDPSGDRTFGVLTKLDLMDKGTNAVDVLEGRQYRLQHPW 239
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADIN+NVDM+ ARRKE+EYF +SPDYGHLA+KMG+EYLAKLLSQHLE+VIRA
Sbjct: 240 VGIVNRSQADINRNVDMLAARRKEKEYFESSPDYGHLAHKMGAEYLAKLLSQHLEAVIRA 299
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
+IPSI ++INK+++E+E+E+D LGRPI DAGAQLYTIL++CRAF+R+FKEHLDGGRPGG
Sbjct: 300 KIPSIIAMINKTIDEIEAELDRLGRPIGGDAGAQLYTILDMCRAFDRVFKEHLDGGRPGG 359
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
DRIY VFD+QLPAAL+KLPFD+HLSLQNVRKV+SEADGYQPHLIAPEQGYRRLIDS+L Y
Sbjct: 360 DRIYGVFDHQLPAALKKLPFDKHLSLQNVRKVISEADGYQPHLIAPEQGYRRLIDSSLHY 419
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASVDAV+ VLKELVR+SIA T+ELKRFPT Q +A AANE+LERFRE+ +KT +
Sbjct: 420 FRGPAEASVDAVHLVLKELVRRSIAATEELKRFPTLQTDIAAAANESLERFREDGRKTVI 479
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLR 540
RLV+ME+SYLTV+FFR+LP E +K TP DRY + H RRI SNVSSYI +V +TLR
Sbjct: 480 RLVEMEASYLTVEFFRKLPTEPDKGANNNTPANDRYQDNHLRRIGSNVSSYINMVCETLR 539
Query: 541 NTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
NTIPKAVV CQV++AK++LLN FY +G KE KQLS MLDEDPALME+R KRLELYK
Sbjct: 540 NTIPKAVVHCQVKEAKRNLLNRFYAHVGSKEKKQLSAMLDEDPALMEKRDSLVKRLELYK 599
Query: 601 AARDEIDSVSW 611
+AR+EIDSV+W
Sbjct: 600 SARNEIDSVAW 610
>UniRef100_Q9FNX6 Dynamin-like protein DLP1 [Arabidopsis thaliana]
Length = 614
Score = 962 bits (2488), Expect = 0.0
Identities = 481/616 (78%), Positives = 551/616 (89%), Gaps = 11/616 (1%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
M+SLIGL+N+IQRACTVLGDHG SLWEALPTVAVVGGQSSGKSSVLES+VGR
Sbjct: 4 MKSLIGLINKIQRACTVLGDHGGEG-----MSLWEALPTVAVVGGQSSGKSSVLESVVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+ EYAEFLH P+++F DF+ VR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTEDGTTEYAEFLHAPKKRFADFAAVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+KQIS IPI LSIYSPNVVNLTL+DLPGLTKVAV+GQPESIVQ+IE MVR+YVEKPN II
Sbjct: 119 SKQISNIPIQLSIYSPNVVNLTLIDLPGLTKVAVDGQPESIVQDIENMVRSYVEKPNCII 178
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLAREVDPTGERTFGV TKLD+MDKGT+ LDVLEGRSYRLQHPW
Sbjct: 179 LAISPANQDIATSDAIKLAREVDPTGERTFGVATKLDIMDKGTDCLDVLEGRSYRLQHPW 238
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINK VDMI ARRKE+EYF SP+YGHLA++MGSEYLAKLLSQHLE+VIR
Sbjct: 239 VGIVNRSQADINKRVDMIAARRKEQEYFETSPEYGHLASRMGSEYLAKLLSQHLETVIRQ 298
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
+IPSI +LINKS++E+ +E+D +GRPIA+D+GAQLYTILELCRAF+R+FKEHLDGGRPGG
Sbjct: 299 KIPSIVALINKSIDEINAELDRIGRPIAVDSGAQLYTILELCRAFDRVFKEHLDGGRPGG 358
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
DRIY VFD+QLPAAL+KLPFDRHLS +NV+KVVSEADGYQPHLIAPEQGYRRLID ++SY
Sbjct: 359 DRIYGVFDHQLPAALKKLPFDRHLSTKNVQKVVSEADGYQPHLIAPEQGYRRLIDGSISY 418
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
F+GPAEA+VDAV+FVLKELVRKSI+ET+ELKRFPT + +A AANEALERFR+ES+KT L
Sbjct: 419 FKGPAEATVDAVHFVLKELVRKSISETEELKRFPTLASDIAAAANEALERFRDESRKTVL 478
Query: 481 RLVDMESSYLTVDFFRRLPQEVEK-----AGGPATPNVDRYAEGHFRRIASNVSSYIGLV 535
RLVDMESSYLTV+FFR+L E EK PA PN D Y++ HFR+I SNVS+YI +V
Sbjct: 479 RLVDMESSYLTVEFFRKLHLEPEKEKPNPRNAPA-PNADPYSDNHFRKIGSNVSAYINMV 537
Query: 536 ADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKR 595
DTLRN++PKAVV+CQVR+AK+SLLN FY Q+G+KE ++L MLDEDP LMERR AKR
Sbjct: 538 CDTLRNSLPKAVVYCQVREAKRSLLNFFYAQVGRKEKEKLGAMLDEDPQLMERRGTLAKR 597
Query: 596 LELYKAARDEIDSVSW 611
LELYK ARD+ID+V+W
Sbjct: 598 LELYKQARDDIDAVAW 613
>UniRef100_Q8LF21 Dynamin, putative [Arabidopsis thaliana]
Length = 614
Score = 961 bits (2483), Expect = 0.0
Identities = 480/616 (77%), Positives = 550/616 (88%), Gaps = 11/616 (1%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
M+SLIGL+N+IQRACTVLGDHG SLWEALPTVAVVGGQSSGKSSVLES+VGR
Sbjct: 4 MKSLIGLINKIQRACTVLGDHGGEG-----MSLWEALPTVAVVGGQSSGKSSVLESVVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+ EYAEFLH P+++F DF+ VR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTEDGTTEYAEFLHAPKKRFADFAAVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+KQIS IPI LSIYSPNVVNLTL+DLPGLTKVAV+GQPESIVQ+IE MVR+YVEKPN II
Sbjct: 119 SKQISNIPIQLSIYSPNVVNLTLIDLPGLTKVAVDGQPESIVQDIENMVRSYVEKPNCII 178
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLAREVDPTGERTFGV TKLD+MDKGT+ LDVLEGRSYRLQHPW
Sbjct: 179 LAISPANQDIATSDAIKLAREVDPTGERTFGVATKLDIMDKGTDCLDVLEGRSYRLQHPW 238
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINK VDMI ARRKE+EYF SP+YGHLA++MGSEYLAKLLSQHLE+VIR
Sbjct: 239 VGIVNRSQADINKRVDMIAARRKEQEYFETSPEYGHLASRMGSEYLAKLLSQHLETVIRQ 298
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
+IPSI +LINKS++E+ +E+D +GRPIA+D+GAQLYTILELCRAF+R+FKEHLDGGRPGG
Sbjct: 299 KIPSIVALINKSIDEINAELDRIGRPIAVDSGAQLYTILELCRAFDRVFKEHLDGGRPGG 358
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
DRIY VFD+QLPAAL+KLPFDRHLS +NV+KVVSEADGYQPHLIAPEQGYRRLID ++SY
Sbjct: 359 DRIYGVFDHQLPAALKKLPFDRHLSTKNVQKVVSEADGYQPHLIAPEQGYRRLIDGSISY 418
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
F+GPAEA+VDAV+FVLKELVRKSI+ET+ELKRFPT + +A AANEALERFR+ES+KT L
Sbjct: 419 FKGPAEATVDAVHFVLKELVRKSISETEELKRFPTLASDIAAAANEALERFRDESRKTVL 478
Query: 481 RLVDMESSYLTVDFFRRLPQEVEK-----AGGPATPNVDRYAEGHFRRIASNVSSYIGLV 535
RLVDMESSYLTV+FFR+L E EK PA PN D Y++ HFR+I SN S+YI +V
Sbjct: 479 RLVDMESSYLTVEFFRKLHLEPEKEKPNPRNAPA-PNADPYSDNHFRKIGSNXSAYINMV 537
Query: 536 ADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKR 595
DTLRN++PKAVV+CQVR+AK+SLLN FY Q+G+KE ++L MLDEDP LMERR AKR
Sbjct: 538 CDTLRNSLPKAVVYCQVREAKRSLLNFFYAQVGRKEKEKLGAMLDEDPQLMERRGTLAKR 597
Query: 596 LELYKAARDEIDSVSW 611
LELYK ARD+ID+V+W
Sbjct: 598 LELYKQARDDIDAVAW 613
>UniRef100_Q9SE81 Dynamin-like protein 5 [Arabidopsis thaliana]
Length = 614
Score = 960 bits (2481), Expect = 0.0
Identities = 480/616 (77%), Positives = 550/616 (88%), Gaps = 11/616 (1%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
M+SLIGL+N+IQRACTVLG HG SLWEALPTVAVVGGQSSGKSSVLES+VGR
Sbjct: 4 MKSLIGLINKIQRACTVLGHHGGEG-----MSLWEALPTVAVVGGQSSGKSSVLESVVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+ EYAEFLH P+++F DF+ VR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTEDGTTEYAEFLHAPKKRFADFAAVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+KQIS IPI LSIYSPNVVNLTL+DLPGLTKVAV+GQPESIVQ+IE MVR+YVEKPN II
Sbjct: 119 SKQISNIPIQLSIYSPNVVNLTLIDLPGLTKVAVDGQPESIVQDIENMVRSYVEKPNCII 178
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLAREVDPTGERTFGV TKLD+MDKGT+ LDVLEGRSYRLQHPW
Sbjct: 179 LAISPANQDIATSDAIKLAREVDPTGERTFGVATKLDIMDKGTDCLDVLEGRSYRLQHPW 238
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINK VDMI ARRKE+EYF SP+YGHLA++MGSEYLAKLLSQHLE+VIR
Sbjct: 239 VGIVNRSQADINKRVDMIAARRKEQEYFETSPEYGHLASRMGSEYLAKLLSQHLETVIRQ 298
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
+IPSI +LINKS++E+ +E+D +GRPIA+D+GAQLYTILELCRAF+R+FKEHLDGGRPGG
Sbjct: 299 KIPSIVALINKSIDEINAELDRIGRPIAVDSGAQLYTILELCRAFDRVFKEHLDGGRPGG 358
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
DRIY VFD+QLPAAL+KLPFDRHLS +NV+KVVSEADGYQPHLIAPEQGYRRLID ++SY
Sbjct: 359 DRIYGVFDHQLPAALKKLPFDRHLSTKNVQKVVSEADGYQPHLIAPEQGYRRLIDGSISY 418
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
F+GPAEA+VDAV+FVLKELVRKSI+ET+ELKRFPT + +A AANEALERFR+ES+KT L
Sbjct: 419 FKGPAEATVDAVHFVLKELVRKSISETEELKRFPTLASDIAAAANEALERFRDESRKTVL 478
Query: 481 RLVDMESSYLTVDFFRRLPQEVEK-----AGGPATPNVDRYAEGHFRRIASNVSSYIGLV 535
RLVDMESSYLTV+FFR+L E EK PA PN D Y++ HFR+I SNVS+YI +V
Sbjct: 479 RLVDMESSYLTVEFFRKLHLEPEKEKPNPRNAPA-PNADPYSDNHFRKIGSNVSAYINMV 537
Query: 536 ADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKR 595
DTLRN++PKAVV+CQVR+AK+SLLN FY Q+G+KE ++L MLDEDP LMERR AKR
Sbjct: 538 CDTLRNSLPKAVVYCQVREAKRSLLNFFYAQVGRKEKEKLGAMLDEDPQLMERRGTLAKR 597
Query: 596 LELYKAARDEIDSVSW 611
LELYK ARD+ID+V+W
Sbjct: 598 LELYKQARDDIDAVAW 613
>UniRef100_Q650Z3 Putative phragmoplastin 12 [Oryza sativa]
Length = 626
Score = 957 bits (2474), Expect = 0.0
Identities = 474/625 (75%), Positives = 551/625 (87%), Gaps = 14/625 (2%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LIGL+NRIQRACT LGDHG +G + +LWE+LPT+AVVGGQSSGKSSVLESIVGR
Sbjct: 4 MEGLIGLMNRIQRACTALGDHGGG-EGANLPTLWESLPTIAVVGGQSSGKSSVLESIVGR 62
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLH+++ G+ +YAEFLH+P+ +F+DF+LVRQEI DETDRVTGK
Sbjct: 63 DFLPRGSGIVTRRPLVLQLHQIDKGAHDYAEFLHLPKTRFSDFALVRQEIADETDRVTGK 122
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
TKQISP+PIHLSIYSPNVVNLTL+DLPGLTKVAVEGQPES+V +IE MVR+YVEKPN II
Sbjct: 123 TKQISPVPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQPESVVHDIENMVRSYVEKPNCII 182
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKL++EVDP+GERTFGVLTKLDLMDKGTNALDVLEGR+YRLQ+PW
Sbjct: 183 LAISPANQDIATSDAIKLSKEVDPSGERTFGVLTKLDLMDKGTNALDVLEGRAYRLQYPW 242
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADIN+ VDMI+AR KEREYF NSPDY HLA+KMGS YLAKLLSQHLE+VI+A
Sbjct: 243 VGIVNRSQADINRKVDMIVAREKEREYFENSPDYAHLASKMGSVYLAKLLSQHLEAVIKA 302
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSITSLINK+++ELESE+D +G+ +A D GAQLYTILELCRAF+R+FKEHLDGGR GG
Sbjct: 303 RIPSITSLINKTIDELESELDTIGKEVAADPGAQLYTILELCRAFDRVFKEHLDGGRSGG 362
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
D+IY VFD++LPAA RKLPFDR+LS+QNV+KVVSEADGYQPHLIAPEQGYRRL+++ L+Y
Sbjct: 363 DKIYGVFDHKLPAAFRKLPFDRYLSVQNVKKVVSEADGYQPHLIAPEQGYRRLVEAGLAY 422
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
F+GPAEA+VDAV+ VL++LVRKSI ET+ L+RFPT QAA+ATAANEALERFRE+ + T L
Sbjct: 423 FKGPAEATVDAVHVVLRDLVRKSIGETEPLRRFPTLQAAIATAANEALERFREDGRSTAL 482
Query: 481 RLVDMESSYLTVDFFRRLPQEVE------------KAGGPATPNVDRYAEGHFRRIASNV 528
RLVDME +YLTV+FFR+LPQ+ + G + VDRY +GH+R IASNV
Sbjct: 483 RLVDME-AYLTVEFFRKLPQDPDSGSKVGNNTNESNGSGSGSVTVDRYGDGHYRNIASNV 541
Query: 529 SSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMER 588
S YI +V D L + IPKAVV CQVR+AK+SLLNHFY IGKKE Q +LDEDPA++ER
Sbjct: 542 SQYIKMVGDQLLHKIPKAVVHCQVREAKRSLLNHFYVHIGKKEASQFGHLLDEDPAMLER 601
Query: 589 RQQCAKRLELYKAARDEIDSVSWVR 613
RQQC KRLELYK+ARDEIDSV+W R
Sbjct: 602 RQQCWKRLELYKSARDEIDSVAWTR 626
>UniRef100_Q9FNX4 Dynamin-like protein DLP3a [Arabidopsis thaliana]
Length = 612
Score = 939 bits (2428), Expect = 0.0
Identities = 467/614 (76%), Positives = 547/614 (89%), Gaps = 3/614 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLI L+N IQRACTV+GDHG D N+ SSLWEALP+VAVVGGQSSGKSSVLESIVGR
Sbjct: 1 MESLIVLINTIQRACTVVGDHGG--DSNALSSLWEALPSVAVVGGQSSGKSSVLESIVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+++ AEFLH+ +KFT+FSLVR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTENGTEDNAEFLHLTNKKFTNFSLVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
KQIS IPIHLSI+SPNVVNLTL+DLPGLTKVAVEGQPE+IV++IE+MVR+YVEKPN +I
Sbjct: 119 NKQISSIPIHLSIFSPNVVNLTLIDLPGLTKVAVEGQPETIVEDIESMVRSYVEKPNCLI 178
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDA+KLA+EVDP G+RTFGVLTKLDLMDKGTNALDV+ GRSY+L++PW
Sbjct: 179 LAISPANQDIATSDAMKLAKEVDPIGDRTFGVLTKLDLMDKGTNALDVINGRSYKLKYPW 238
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINKNVDM++ARRKEREYF SPDYGHLA +MGSEYLAKLLS+ LESVIR+
Sbjct: 239 VGIVNRSQADINKNVDMMVARRKEREYFETSPDYGHLATRMGSEYLAKLLSKLLESVIRS 298
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSI SLIN ++EELE E+D LGRPIA+DAGAQLYTIL +CRAFE+IFKEHLDGGRPGG
Sbjct: 299 RIPSILSLINNNIEELERELDQLGRPIAIDAGAQLYTILGMCRAFEKIFKEHLDGGRPGG 358
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
RIY +FD LP A++KLPFDRHLSLQ+V+++VSE+DGYQPHLIAPE GYRRLI+ +L++
Sbjct: 359 ARIYGIFDYNLPTAIKKLPFDRHLSLQSVKRIVSESDGYQPHLIAPELGYRRLIEGSLNH 418
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASV+A++ +LKELVRK+IAET+ELKRFP+ Q L AAN +L++FREES K+ L
Sbjct: 419 FRGPAEASVNAIHLILKELVRKAIAETEELKRFPSLQIELVAAANSSLDKFREESMKSVL 478
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAG-GPATPNVDRYAEGHFRRIASNVSSYIGLVADTL 539
RLVDMESSYLTVDFFR+L E + T +D+Y +GHFR+IASNV++YI +VA+TL
Sbjct: 479 RLVDMESSYLTVDFFRKLHVESQNMSLSSPTSAIDQYGDGHFRKIASNVAAYIKMVAETL 538
Query: 540 RNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELY 599
NTIPKAVV CQVRQAK SLLN+FY QI + +GK+L ++LDE+PALMERR QCAKRLELY
Sbjct: 539 VNTIPKAVVHCQVRQAKLSLLNYFYAQISQSQGKRLGQLLDENPALMERRMQCAKRLELY 598
Query: 600 KAARDEIDSVSWVR 613
K ARDEID+ WVR
Sbjct: 599 KKARDEIDAAVWVR 612
>UniRef100_O80499 Putative phragmoplastin [Arabidopsis thaliana]
Length = 613
Score = 935 bits (2417), Expect = 0.0
Identities = 467/615 (75%), Positives = 548/615 (88%), Gaps = 4/615 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLI L+N IQRACTV+GDHG D N+ SSLWEALP+VAVVGGQSSGKSSVLESIVGR
Sbjct: 1 MESLIVLINTIQRACTVVGDHGG--DSNALSSLWEALPSVAVVGGQSSGKSSVLESIVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+++ AEFLH+ +KFT+FSLVR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTENGTEDNAEFLHLTNKKFTNFSLVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
KQIS IPIHLSI+SPNVVNLTL+DLPGLTKVAVEGQPE+IV++IE+MVR+YVEKPN +I
Sbjct: 119 NKQISSIPIHLSIFSPNVVNLTLIDLPGLTKVAVEGQPETIVEDIESMVRSYVEKPNCLI 178
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDA+KLA+EVDP G+RTFGVLTKLDLMDKGTNALDV+ GRSY+L++PW
Sbjct: 179 LAISPANQDIATSDAMKLAKEVDPIGDRTFGVLTKLDLMDKGTNALDVINGRSYKLKYPW 238
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINKNVDM++ARRKEREYF SPDYGHLA +MGSEYLAKLLS+ LESVIR+
Sbjct: 239 VGIVNRSQADINKNVDMMVARRKEREYFETSPDYGHLATRMGSEYLAKLLSKLLESVIRS 298
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSI SLIN ++EELE E+D LGRPIA+DAGAQLYTIL +CRAFE+IFKEHLDGGRPGG
Sbjct: 299 RIPSILSLINNNIEELERELDQLGRPIAIDAGAQLYTILGMCRAFEKIFKEHLDGGRPGG 358
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
RIY +FD LP A++KLPFDRHLSLQ+V+++VSE+DGYQPHLIAPE GYRRLI+ +L++
Sbjct: 359 ARIYGIFDYNLPTAIKKLPFDRHLSLQSVKRIVSESDGYQPHLIAPELGYRRLIEGSLNH 418
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASV+A++ +LKELVRK+IAET+ELKRFP+ Q L AAN +L++FREES K+ L
Sbjct: 419 FRGPAEASVNAIHLILKELVRKAIAETEELKRFPSLQIELVAAANSSLDKFREESMKSVL 478
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAG-GPATPNVDRYAEGHFRRIASNVSSYIGLVADTL 539
RLVDMESSYLTVDFFR+L E + T +D+Y +GHFR+IASNV++YI +VA+TL
Sbjct: 479 RLVDMESSYLTVDFFRKLHVESQNMSLSSPTSAIDQYGDGHFRKIASNVAAYIKMVAETL 538
Query: 540 RNTIPKAVVFCQVRQAKQSLLNHFYTQIGK-KEGKQLSEMLDEDPALMERRQQCAKRLEL 598
NTIPKAVV CQVRQAK SLLN+FY QI + ++GK+L ++LDE+PALMERR QCAKRLEL
Sbjct: 539 VNTIPKAVVHCQVRQAKLSLLNYFYAQISQSQQGKRLGQLLDENPALMERRMQCAKRLEL 598
Query: 599 YKAARDEIDSVSWVR 613
YK ARDEID+ WVR
Sbjct: 599 YKKARDEIDAAVWVR 613
>UniRef100_Q9FNX3 Dynamin-like protein DLP3b [Arabidopsis thaliana]
Length = 595
Score = 898 bits (2321), Expect = 0.0
Identities = 451/614 (73%), Positives = 530/614 (85%), Gaps = 20/614 (3%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLI L+N IQRACTV+GDHG D N+ SSLWEALP+VAVVGGQSSGKSSVLESIVGR
Sbjct: 1 MESLIVLINTIQRACTVVGDHGG--DSNALSSLWEALPSVAVVGGQSSGKSSVLESIVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+++ AEFLH+ +KFT+FSLVR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTENGTEDNAEFLHLTNKKFTNFSLVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
KQIS IPIHLSI+SPN EGQPE+IV++IE+MVR+YVEKPN +I
Sbjct: 119 NKQISSIPIHLSIFSPN-----------------EGQPETIVEDIESMVRSYVEKPNCLI 161
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDA+KLA+EVDP G+RTFGVLTKLDLMDKGTNALDV+ GRSY+L++PW
Sbjct: 162 LAISPANQDIATSDAMKLAKEVDPIGDRTFGVLTKLDLMDKGTNALDVINGRSYKLKYPW 221
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINKNVDM++ARRKEREYF SPDYGHLA +MGSEYLAKLLS+ LESVIR+
Sbjct: 222 VGIVNRSQADINKNVDMMVARRKEREYFETSPDYGHLATRMGSEYLAKLLSKLLESVIRS 281
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSI SLIN ++EELE E+D LGRPIA+DAGAQLYTIL +CRAFE+IFKEHLDGGRPGG
Sbjct: 282 RIPSILSLINNNIEELERELDQLGRPIAIDAGAQLYTILGMCRAFEKIFKEHLDGGRPGG 341
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
RIY +FD LP A++KLPFDRHLSLQ+V+++VSE+DGYQPHLIAPE GYRRLI+ +L++
Sbjct: 342 ARIYGIFDYNLPTAIKKLPFDRHLSLQSVKRIVSESDGYQPHLIAPELGYRRLIEGSLNH 401
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASV+A++ +LKELVRK+IAET+ELKRFP+ Q L AAN +L++FREES K+ L
Sbjct: 402 FRGPAEASVNAIHLILKELVRKAIAETEELKRFPSLQIELVAAANSSLDKFREESMKSVL 461
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAG-GPATPNVDRYAEGHFRRIASNVSSYIGLVADTL 539
RLVDMESSYLTVDFFR+L E + T +D+Y +GHFR+IASNV++YI +VA+TL
Sbjct: 462 RLVDMESSYLTVDFFRKLHVESQNMSLSSPTSAIDQYGDGHFRKIASNVAAYIKMVAETL 521
Query: 540 RNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELY 599
NTIPKAVV CQVRQAK SLLN+FY QI + +GK+L ++LDE+PALMERR QCAKRLELY
Sbjct: 522 VNTIPKAVVHCQVRQAKLSLLNYFYAQISQSQGKRLGQLLDENPALMERRMQCAKRLELY 581
Query: 600 KAARDEIDSVSWVR 613
K ARDEID+ WVR
Sbjct: 582 KKARDEIDAAVWVR 595
>UniRef100_Q8S3C9 Dynamin-like protein D [Arabidopsis thaliana]
Length = 596
Score = 894 bits (2310), Expect = 0.0
Identities = 451/615 (73%), Positives = 531/615 (86%), Gaps = 21/615 (3%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLI L+N IQRACTV+GDHG D N+ SSLWEALP+VAVVGGQSSGKSSVLESIVGR
Sbjct: 1 MESLIVLINTIQRACTVVGDHGG--DSNALSSLWEALPSVAVVGGQSSGKSSVLESIVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+++ AEFLH+ +KFT+FSLVR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTENGTEDNAEFLHLTNKKFTNFSLVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
KQIS IPIHLSI+SPN EGQPE+IV++IE+MVR+YVEKPN +I
Sbjct: 119 NKQISSIPIHLSIFSPN-----------------EGQPETIVEDIESMVRSYVEKPNCLI 161
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDA+KLA+EVDP G+RTFGVLTKLDLMDKGTNALDV+ GRSY+L++PW
Sbjct: 162 LAISPANQDIATSDAMKLAKEVDPIGDRTFGVLTKLDLMDKGTNALDVINGRSYKLKYPW 221
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINKNVDM++ARRKEREYF SPDYGHLA +MGSEYLAKLLS+ LESVIR+
Sbjct: 222 VGIVNRSQADINKNVDMMVARRKEREYFETSPDYGHLATRMGSEYLAKLLSKLLESVIRS 281
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSI SLIN ++EELE E+D LGRPIA+DAGAQLYTIL +CRAFE+IFKEHLDGGRPGG
Sbjct: 282 RIPSILSLINNNIEELERELDQLGRPIAIDAGAQLYTILGMCRAFEKIFKEHLDGGRPGG 341
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
RIY +FD LP A++KLPFDRHLSLQ+V+++VSE+DGYQPHLIAPE GYRRLI+ +L++
Sbjct: 342 ARIYGIFDYNLPTAIKKLPFDRHLSLQSVKRIVSESDGYQPHLIAPELGYRRLIEGSLNH 401
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASV+A++ +LKELVRK+IAET+ELKRFP+ Q L AAN +L++FREES K+ L
Sbjct: 402 FRGPAEASVNAIHLILKELVRKAIAETEELKRFPSLQIELVAAANSSLDKFREESMKSVL 461
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAG-GPATPNVDRYAEGHFRRIASNVSSYIGLVADTL 539
RLVDMESSYLTVDFFR+L E + T +D+Y +GHFR+IASNV++YI +VA+TL
Sbjct: 462 RLVDMESSYLTVDFFRKLHVESQNMSLSSPTSAIDQYGDGHFRKIASNVAAYIKMVAETL 521
Query: 540 RNTIPKAVVFCQVRQAKQSLLNHFYTQIGK-KEGKQLSEMLDEDPALMERRQQCAKRLEL 598
NTIPKAVV CQVRQAK SLLN+FY QI + ++GK+L ++LDE+PALMERR QCAKRLEL
Sbjct: 522 VNTIPKAVVHCQVRQAKLSLLNYFYAQISQSQQGKRLGQLLDENPALMERRMQCAKRLEL 581
Query: 599 YKAARDEIDSVSWVR 613
YK ARDEID+ WVR
Sbjct: 582 YKKARDEIDAAVWVR 596
>UniRef100_Q39828 SDL5A [Glycine max]
Length = 610
Score = 883 bits (2282), Expect = 0.0
Identities = 420/611 (68%), Positives = 538/611 (87%), Gaps = 3/611 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME+LI LVN+IQRACT LGDHG + ++ +LW++LP +AVVGGQSSGKSSVLES+VG+
Sbjct: 1 MENLISLVNKIQRACTALGDHG---ENSALPTLWDSLPAIAVVGGQSSGKSSVLESVVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK+E GS+EYAEFLH+PR++FTDF VR+EIQDETDR TG+
Sbjct: 58 DFLPRGSGIVTRRPLVLQLHKIEEGSREYAEFLHLPRKRFTDFVAVRKEIQDETDRETGR 117
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
TKQIS +PIHLSIYSPNVVNLTLVDLPGLTKVAVEGQP+SIV++IE MVR+Y+EKPN II
Sbjct: 118 TKQISTVPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPDSIVKDIEDMVRSYIEKPNCII 177
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQD+ATSDAIK++REVDPTG+RT GVLTK+DLMDKGT+A+D+LEGR+YRL+ PW
Sbjct: 178 LAISPANQDLATSDAIKISREVDPTGDRTIGVLTKIDLMDKGTDAVDILEGRAYRLKFPW 237
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DINKNVDMI ARR+EREYF ++P+Y HLAN+MGSE+LAK+LS+HLE+VI++
Sbjct: 238 IGVVNRSQQDINKNVDMIAARRREREYFNSTPEYKHLANRMGSEHLAKMLSKHLETVIKS 297
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
+IP I SLINK++ ELE+E+ LG+P+A DAG +LY I+E+CR+F++IFK+HLDG RPGG
Sbjct: 298 KIPGIQSLINKTIAELEAELTRLGKPVAADAGGKLYAIMEICRSFDQIFKDHLDGVRPGG 357
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
D+IY+VFDNQLPAAL++L FD+ LS++N+RK+++EADGYQPHLIAPEQGYRRLI+S+L
Sbjct: 358 DKIYNVFDNQLPAALKRLQFDKQLSMENIRKLITEADGYQPHLIAPEQGYRRLIESSLIT 417
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
RGPAEA+VDAV+ +LK+LV K+I+ET +LK++P + + AA ++LER R+ESK+ TL
Sbjct: 418 IRGPAEAAVDAVHSLLKDLVHKAISETLDLKQYPGLRVEVGAAAVDSLERMRDESKRATL 477
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLR 540
+LVDME YLTVDFFR+LPQ+V+K G P DRY + + RRI + + SY+ +V TLR
Sbjct: 478 QLVDMECGYLTVDFFRKLPQDVDKGGNPTHSIFDRYNDSYLRRIGTTILSYVNMVCATLR 537
Query: 541 NTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
N+IPK++V+CQVR+AK+SLL+HF+T++GK E K+LS +L+EDPA+MERR AKRLELY+
Sbjct: 538 NSIPKSIVYCQVREAKRSLLDHFFTELGKMETKRLSSLLNEDPAIMERRSALAKRLELYR 597
Query: 601 AARDEIDSVSW 611
+A+ EID+V+W
Sbjct: 598 SAQAEIDAVAW 608
>UniRef100_Q39821 SDL12A [Glycine max]
Length = 610
Score = 875 bits (2262), Expect = 0.0
Identities = 414/611 (67%), Positives = 538/611 (87%), Gaps = 3/611 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME+LI LVN+IQRACT LGDHG + ++ +LW++LP +AVVGGQSSGKSSVLES+VG+
Sbjct: 1 MENLISLVNKIQRACTALGDHG---ENSALPTLWDSLPAIAVVGGQSSGKSSVLESVVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK++ GS+EYAEFLH+PR++FTDF VR+EIQDETDR TG+
Sbjct: 58 DFLPRGSGIVTRRPLVLQLHKIDEGSREYAEFLHLPRKRFTDFVAVRKEIQDETDRETGR 117
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
TKQIS +PIHLSIYSPNVVNLTL+DLPGLTKVAVEGQP+SIV++IE MVR+Y+EKPN II
Sbjct: 118 TKQISSVPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQPDSIVKDIEDMVRSYIEKPNCII 177
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQD+ATSDAIK++REVDPTG+RT GVLTK+DLMDKGT+A+D+LEGR+YRL+ PW
Sbjct: 178 LAISPANQDLATSDAIKISREVDPTGDRTIGVLTKIDLMDKGTDAVDILEGRAYRLKFPW 237
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DINKNVDMI ARR+EREYF ++P+Y HLAN+MGSE+LAK+LS+HLE+VI++
Sbjct: 238 IGVVNRSQQDINKNVDMIAARRREREYFNSTPEYKHLANRMGSEHLAKMLSKHLETVIKS 297
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
+IP I SLINK++ ELE+E+ LG+P+A DAG +LY I+E+CR+F++IFK+HLDG RPGG
Sbjct: 298 KIPGIQSLINKTIAELEAELTRLGKPVAADAGGKLYAIMEICRSFDQIFKDHLDGVRPGG 357
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
D+IY+VFDNQLPAAL++L FD+ LS++N+RK+++EADGYQPHLIAPEQGYRRLI+S+L
Sbjct: 358 DKIYNVFDNQLPAALKRLQFDKQLSMENIRKLITEADGYQPHLIAPEQGYRRLIESSLIT 417
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
RGPAE++VDAV+ +LK+LV K+++ET +LK++P + + A+ ++LER R+ESK+ TL
Sbjct: 418 IRGPAESAVDAVHSLLKDLVHKAMSETLDLKQYPGLRVEVGAASVDSLERMRDESKRATL 477
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLR 540
+LVDME YLTVDFFR+LPQ+V+K G P DRY + + RRI + + SY+ +V TLR
Sbjct: 478 QLVDMECGYLTVDFFRKLPQDVDKGGNPTHSICDRYNDSYLRRIGTTILSYVNMVCATLR 537
Query: 541 NTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
++IPK++V+CQVR+AK+SLL+HF+T++GK E K+LS +L+EDPA+MERR AKRLELY+
Sbjct: 538 HSIPKSIVYCQVREAKRSLLDHFFTELGKMEIKRLSSLLNEDPAIMERRSALAKRLELYR 597
Query: 601 AARDEIDSVSW 611
+A+ EID+V+W
Sbjct: 598 SAQAEIDAVAW 608
>UniRef100_P42697 Dynamin-like protein [Arabidopsis thaliana]
Length = 610
Score = 868 bits (2242), Expect = 0.0
Identities = 416/611 (68%), Positives = 532/611 (86%), Gaps = 3/611 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME+LI LVN+IQRACT LGDHG D ++ +LW++LP +AVVGGQSSGKSSVLESIVG+
Sbjct: 1 MENLISLVNKIQRACTALGDHG---DSSALPTLWDSLPAIAVVGGQSSGKSSVLESIVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQL K++ G++EYAEFLH+PR+KFTDF+ VR+EIQDETDR TG+
Sbjct: 58 DFLPRGSGIVTRRPLVLQLQKIDDGTREYAEFLHLPRKKFTDFAAVRKEIQDETDRETGR 117
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+K IS +PIHLSIYSPNVVNLTL+DLPGLTKVAV+GQ +SIV++IE MVR+Y+EKPN II
Sbjct: 118 SKAISSVPIHLSIYSPNVVNLTLIDLPGLTKVAVDGQSDSIVKDIENMVRSYIEKPNCII 177
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQD+ATSDAIK++REVDP+G+RTFGVLTK+DLMDKGT+A+++LEGRS++L++PW
Sbjct: 178 LAISPANQDLATSDAIKISREVDPSGDRTFGVLTKIDLMDKGTDAVEILEGRSFKLKYPW 237
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VG+VNRSQADINKNVDMI AR++EREYF+N+ +Y HLANKMGSE+LAK+LS+HLE VI++
Sbjct: 238 VGVVNRSQADINKNVDMIAARKREREYFSNTTEYRHLANKMGSEHLAKMLSKHLERVIKS 297
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIP I SLINK++ ELE+E+ LG+PIA DAG +LY+I+E+CR F++IFKEHLDG R GG
Sbjct: 298 RIPGIQSLINKTVLELETELSRLGKPIAADAGGKLYSIMEICRLFDQIFKEHLDGVRAGG 357
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
+++Y+VFDNQLPAAL++L FD+ L++ N+RK+V+EADGYQPHLIAPEQGYRRLI+S++
Sbjct: 358 EKVYNVFDNQLPAALKRLQFDKQLAMDNIRKLVTEADGYQPHLIAPEQGYRRLIESSIVS 417
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
RGPAEASVD V+ +LK+LV KS+ ET ELK++P + + AA E+L++ RE SKK TL
Sbjct: 418 IRGPAEASVDTVHAILKDLVHKSVNETVELKQYPALRVEVTNAAIESLDKMREGSKKATL 477
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLR 540
+LVDME SYLTVDFFR+LPQ+VEK G P DRY + + RRI SNV SY+ +V LR
Sbjct: 478 QLVDMECSYLTVDFFRKLPQDVEKGGNPTHSIFDRYNDSYLRRIGSNVLSYVNMVCAGLR 537
Query: 541 NTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
N+IPK++V+CQVR+AK+SLL+HF+ ++G + K+LS +L+EDPA+MERR +KRLELY+
Sbjct: 538 NSIPKSIVYCQVREAKRSLLDHFFAELGTMDMKRLSSLLNEDPAIMERRSAISKRLELYR 597
Query: 601 AARDEIDSVSW 611
AA+ EID+V+W
Sbjct: 598 AAQSEIDAVAW 608
>UniRef100_Q9SMB6 Phragmoplastin [Nicotiana tabacum]
Length = 609
Score = 863 bits (2231), Expect = 0.0
Identities = 419/613 (68%), Positives = 531/613 (86%), Gaps = 4/613 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME+LI LVNR+ RACT LGD G + ++ +LW+ALP +AVVGGQSSGKS+VLESIVG+
Sbjct: 1 MENLIQLVNRLHRACTALGDLG---EESALPTLWDALPAIAVVGGQSSGKSAVLESIVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFL RGSGIVTRRPLVLQLH+++ G +EYAEF H+PR++FTDF+ VR+EI DETDR TG+
Sbjct: 58 DFLRRGSGIVTRRPLVLQLHRIDEG-REYAEFGHLPRKRFTDFAAVRKEIADETDRETGR 116
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+KQIS +PI+LSIYSPNVVNLTL+DLPGLTKVAVEGQ E IV +IE MVR+Y+EKPN II
Sbjct: 117 SKQISSVPIYLSIYSPNVVNLTLIDLPGLTKVAVEGQSEGIVADIENMVRSYIEKPNCII 176
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA+SPANQD+ATSDAIK++REVDP GERTFGVLTK+DLMDKGT+A+D+LEGR+Y+LQ PW
Sbjct: 177 LAVSPANQDLATSDAIKISREVDPKGERTFGVLTKIDLMDKGTDAVDILEGRAYKLQFPW 236
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DINKNVDMI ARR+E+EYF+++P+Y H+AN+MGSE+L K++S+HLESVI++
Sbjct: 237 IGVVNRSQQDINKNVDMIAARRREKEYFSSTPEYRHMANRMGSEHLGKVMSKHLESVIKS 296
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIP + SLI+K++ ELE+E+ LG+PIA DAG +LY I+E+CR F+ IFKEHLDG RPGG
Sbjct: 297 RIPGLQSLISKTIIELETELSRLGKPIATDAGGKLYMIMEVCRTFDGIFKEHLDGVRPGG 356
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
D+IY+VFDNQLPAAL++L FD+ LS+ NVRK+++EADGYQPHLIAPEQGYRRLI+S+L+
Sbjct: 357 DKIYNVFDNQLPAALKRLQFDKQLSMDNVRKLITEADGYQPHLIAPEQGYRRLIESSLTS 416
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
+GPAEA+VDAV+ +LKELV KSI+ET ELK++P+ + + AA E+LER R+ESKK TL
Sbjct: 417 MKGPAEAAVDAVHAILKELVHKSISETAELKQYPSLRVEVNGAAVESLERMRDESKKATL 476
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLR 540
+LV+ME SYLTVDFFR+LPQ++EK G P DRY + + RRI SNV SY+ +V TLR
Sbjct: 477 QLVEMECSYLTVDFFRKLPQDIEKGGNPTHSIFDRYNDSYLRRIGSNVLSYVNMVCATLR 536
Query: 541 NTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
N+IPK+VV+CQVR+AK+SLL+HF+T +GKKEGKQL +LDEDPA+M+R AKRLELY+
Sbjct: 537 NSIPKSVVYCQVREAKRSLLDHFFTDLGKKEGKQLGTLLDEDPAIMQRCISLAKRLELYR 596
Query: 601 AARDEIDSVSWVR 613
AA+ EIDSV+W +
Sbjct: 597 AAQAEIDSVAWAK 609
>UniRef100_Q84XF3 Dynamin-like protein B [Arabidopsis thaliana]
Length = 610
Score = 847 bits (2189), Expect = 0.0
Identities = 407/612 (66%), Positives = 525/612 (85%), Gaps = 5/612 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLI LVN+IQRACT LGDHG +G+S +LW++LP +AVVGGQSSGKSSVLES+VG+
Sbjct: 1 MESLIALVNKIQRACTALGDHG---EGSSLPTLWDSLPAIAVVGGQSSGKSSVLESVVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRG+GIVTRRPLVLQLH+++ G +EYAEF+H+P++KFTDF+ VRQEI DETDR TG+
Sbjct: 58 DFLPRGAGIVTRRPLVLQLHRIDEG-KEYAEFMHLPKKKFTDFAAVRQEISDETDRETGR 116
Query: 121 T-KQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSI 179
+ K IS +PIHLSI+SPNVVNLTLVDLPGLTKVAV+GQPESIVQ+IE MVR+++EKPN I
Sbjct: 117 SSKVISTVPIHLSIFSPNVVNLTLVDLPGLTKVAVDGQPESIVQDIENMVRSFIEKPNCI 176
Query: 180 ILAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHP 239
ILAISPANQD+ATSDAIK++REVDP G+RTFGVLTK+DLMD+GTNA+D+LEGR Y+L++P
Sbjct: 177 ILAISPANQDLATSDAIKISREVDPKGDRTFGVLTKIDLMDQGTNAVDILEGRGYKLRYP 236
Query: 240 WVGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIR 299
WVG+VNRSQADINK+VDMI ARR+ER+YF SP+Y HL +MGSEYL K+LS+HLE VI+
Sbjct: 237 WVGVVNRSQADINKSVDMIAARRRERDYFQTSPEYRHLTERMGSEYLGKMLSKHLEVVIK 296
Query: 300 ARIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPG 359
+RIP + SLI K++ ELE+E+ LG+P+A DAG +LY I+E+CRAF++ FKEHLDG R G
Sbjct: 297 SRIPGLQSLITKTISELETELSRLGKPVAADAGGKLYMIMEICRAFDQTFKEHLDGTRSG 356
Query: 360 GDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALS 419
G++I VFDNQ PAA+++L FD+HLS+ NVRK+++EADGYQPHLIAPEQGYRRLI+S L
Sbjct: 357 GEKINSVFDNQFPAAIKRLQFDKHLSMDNVRKLITEADGYQPHLIAPEQGYRRLIESCLV 416
Query: 420 YFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTT 479
RGPAEA+VDAV+ +LK+L+ KS+ ET ELK++PT + ++ AA ++L+R R+ES+K T
Sbjct: 417 SIRGPAEAAVDAVHSILKDLIHKSMGETSELKQYPTLRVEVSGAAVDSLDRMRDESRKAT 476
Query: 480 LRLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTL 539
L LVDMES YLTV+FFR+LPQ+ EK G P DRY + + RRI SNV SY+ +V L
Sbjct: 477 LLLVDMESGYLTVEFFRKLPQDSEKGGNPTHSIFDRYNDAYLRRIGSNVLSYVNMVCAGL 536
Query: 540 RNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELY 599
RN+IPK++V+CQVR+AK+SLL+ F+T++G+KE +LS++LDEDPA+ +RR AKRLELY
Sbjct: 537 RNSIPKSIVYCQVREAKRSLLDIFFTELGQKEMSKLSKLLDEDPAVQQRRTSIAKRLELY 596
Query: 600 KAARDEIDSVSW 611
++A+ +I++V+W
Sbjct: 597 RSAQTDIEAVAW 608
>UniRef100_Q9M362 Dynamin-like protein [Arabidopsis thaliana]
Length = 627
Score = 837 bits (2163), Expect = 0.0
Identities = 407/629 (64%), Positives = 525/629 (82%), Gaps = 22/629 (3%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLI LVN+IQRACT LGDHG +G+S +LW++LP +AVVGGQSSGKSSVLES+VG+
Sbjct: 1 MESLIALVNKIQRACTALGDHG---EGSSLPTLWDSLPAIAVVGGQSSGKSSVLESVVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLV-------------- 106
DFLPRG+GIVTRRPLVLQLH+++ G +EYAEF+H+P++KFTDF +V
Sbjct: 58 DFLPRGAGIVTRRPLVLQLHRIDEG-KEYAEFMHLPKKKFTDFGIVWEEFMLLSFPVTAC 116
Query: 107 ---RQEIQDETDRVTGKT-KQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIV 162
RQEI DETDR TG++ K IS +PIHLSI+SPNVVNLTLVDLPGLTKVAV+GQPESIV
Sbjct: 117 TAVRQEISDETDRETGRSSKVISTVPIHLSIFSPNVVNLTLVDLPGLTKVAVDGQPESIV 176
Query: 163 QEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKG 222
Q+IE MVR+++EKPN IILAISPANQD+ATSDAIK++REVDP G+RTFGVLTK+DLMD+G
Sbjct: 177 QDIENMVRSFIEKPNCIILAISPANQDLATSDAIKISREVDPKGDRTFGVLTKIDLMDQG 236
Query: 223 TNALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMG 282
TNA+D+LEGR Y+L++PWVG+VNRSQADINK+VDMI ARR+ER+YF SP+Y HL +MG
Sbjct: 237 TNAVDILEGRGYKLRYPWVGVVNRSQADINKSVDMIAARRRERDYFQTSPEYRHLTERMG 296
Query: 283 SEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELC 342
SEYL K+LS+HLE VI++RIP + SLI K++ ELE+E+ LG+P+A DAG +LY I+E+C
Sbjct: 297 SEYLGKMLSKHLEVVIKSRIPGLQSLITKTISELETELSRLGKPVAADAGGKLYMIMEIC 356
Query: 343 RAFERIFKEHLDGGRPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPH 402
RAF++ FKEHLDG R GG++I VFDNQ PAA+++L FD+HLS+ NVRK+++EADGYQPH
Sbjct: 357 RAFDQTFKEHLDGTRSGGEKINSVFDNQFPAAIKRLQFDKHLSMDNVRKLITEADGYQPH 416
Query: 403 LIAPEQGYRRLIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALAT 462
LIAPEQGYRRLI+S L RGPAEA+VDAV+ +LK+L+ KS+ ET ELK++PT + ++
Sbjct: 417 LIAPEQGYRRLIESCLVSIRGPAEAAVDAVHSILKDLIHKSMGETSELKQYPTLRVEVSG 476
Query: 463 AANEALERFREESKKTTLRLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFR 522
AA ++L+R R+ES+K TL LVDMES YLTV+FFR+LPQ+ EK G P DRY + + R
Sbjct: 477 AAVDSLDRMRDESRKATLLLVDMESGYLTVEFFRKLPQDSEKGGNPTHSIFDRYNDAYLR 536
Query: 523 RIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDED 582
RI SNV SY+ +V LRN+IPK++V+CQVR+AK+SLL+ F+T++G+KE +LS++LDED
Sbjct: 537 RIGSNVLSYVNMVCAGLRNSIPKSIVYCQVREAKRSLLDIFFTELGQKEMSKLSKLLDED 596
Query: 583 PALMERRQQCAKRLELYKAARDEIDSVSW 611
PA+ +RR AKRLELY++A+ +I++V+W
Sbjct: 597 PAVQQRRTSIAKRLELYRSAQTDIEAVAW 625
>UniRef100_Q6I614 Putative dynamin [Oryza sativa]
Length = 540
Score = 764 bits (1972), Expect = 0.0
Identities = 366/527 (69%), Positives = 461/527 (87%), Gaps = 4/527 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME+LI LVN++QRACT LGDHG + ++ +LW++LP +AVVGGQSSGKSSVLES+VG+
Sbjct: 1 MENLISLVNKLQRACTALGDHG---EESALPTLWDSLPAIAVVGGQSSGKSSVLESVVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLH+++ G +EYAEF+H+PR++FTDF+LVR+EI DETDR TG+
Sbjct: 58 DFLPRGSGIVTRRPLVLQLHRID-GDREYAEFMHLPRKRFTDFALVRKEIADETDRETGR 116
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+KQIS +PIHLSIYSPNVVNLTL+DLPGLTKVAVEGQP+SIVQ+IE MVR+++EKPN II
Sbjct: 117 SKQISSVPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQPDSIVQDIENMVRSFIEKPNCII 176
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA+SPANQD+ATSDAIK++REVDP GERTFGVLTK+DLMDKGT+A+D+LEGRSYRLQ W
Sbjct: 177 LAVSPANQDLATSDAIKISREVDPKGERTFGVLTKIDLMDKGTDAVDILEGRSYRLQQQW 236
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DINKNVDMI ARR+EREYF+ +P+Y HLA++MGSE+LAK LS+HLE+VI++
Sbjct: 237 IGVVNRSQQDINKNVDMIAARRREREYFSTTPEYKHLAHRMGSEHLAKSLSKHLETVIKS 296
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIP + SLI K++ ELE+E++ LG+PIA DAG +LYTI+E+CR F+ I+KEHLDG RPGG
Sbjct: 297 RIPGLQSLITKTIAELETELNRLGKPIATDAGGKLYTIMEICRMFDGIYKEHLDGVRPGG 356
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
++IYHVFDNQ P A+++L FD+ L+++NV+K+++EADGYQPHLIAPEQGYRRLI+S L
Sbjct: 357 EKIYHVFDNQFPVAIKRLQFDKQLAMENVKKLITEADGYQPHLIAPEQGYRRLIESCLVS 416
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
RGPAEA+VDAV+ +LKELV K+I ET ELK+FPT + + AA E+L+R R+ESKK TL
Sbjct: 417 IRGPAEAAVDAVHAILKELVHKAINETHELKQFPTLRVEVGNAAFESLDRMRDESKKNTL 476
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASN 527
+LVDME SYLTVDFFR+LPQ+VEK G P+ DRY + + RRI +
Sbjct: 477 KLVDMECSYLTVDFFRKLPQDVEKGGNPSHSIFDRYNDSYLRRIGKS 523
>UniRef100_Q9LQU8 F10B6.23 [Arabidopsis thaliana]
Length = 749
Score = 711 bits (1836), Expect = 0.0
Identities = 376/563 (66%), Positives = 436/563 (76%), Gaps = 83/563 (14%)
Query: 130 HLSIYSPNVVNLTLVDLPGLTKVAV-------------------EGQPESIVQEIETMVR 170
H +++S VVNLTL+DLPGLTKVAV +GQPESIVQ+IE MVR
Sbjct: 188 HSTLFS-TVVNLTLIDLPGLTKVAVVTDMNLVLKLVTDMNIIRVDGQPESIVQDIENMVR 246
Query: 171 TYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLE 230
+YVEKPN IILAISPANQDIATSDAIKLAREVDPTGERTFGV TKLD+MDKGT+ LDVLE
Sbjct: 247 SYVEKPNCIILAISPANQDIATSDAIKLAREVDPTGERTFGVATKLDIMDKGTDCLDVLE 306
Query: 231 GRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLL 290
GRSYRLQHPWVGIVNRSQADINK VDMI ARRKE+EYF SP+YGHLA++MGSEYLAKLL
Sbjct: 307 GRSYRLQHPWVGIVNRSQADINKRVDMIAARRKEQEYFETSPEYGHLASRMGSEYLAKLL 366
Query: 291 SQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFK 350
SQHLE+VIR +IPSI +LINKS++E+ +E+D +GRPIA+D+GAQLYTILELCRAF+R+FK
Sbjct: 367 SQHLETVIRQKIPSIVALINKSIDEINAELDRIGRPIAVDSGAQLYTILELCRAFDRVFK 426
Query: 351 EHLDGGRPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGY 410
EHLDGGRPGGDRIY VFD+QLPAAL+KLPFDRHLS +NV+KVVSEADGYQPHLIAPEQGY
Sbjct: 427 EHLDGGRPGGDRIYGVFDHQLPAALKKLPFDRHLSTKNVQKVVSEADGYQPHLIAPEQGY 486
Query: 411 RRLIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETK---------------------- 448
RRLID ++SYF+GPAEA+VDAV+FVLKELVRKSI+ET+
Sbjct: 487 RRLIDGSISYFKGPAEATVDAVHFVLKELVRKSISETEVRTDKMPLVLRTLPSSLRSVLV 546
Query: 449 ---------ELKRFPTFQAALATAANEALERFREESKKTTLRLVDMESSYLTVDFFRRLP 499
ELKRFPT + +A AANEALERFR+ES+KT LRLVDMESSYLTV+FFR+L
Sbjct: 547 LTYHCCFFQELKRFPTLASDIAAAANEALERFRDESRKTVLRLVDMESSYLTVEFFRKLH 606
Query: 500 QEVEKA-----GGPATPNVDRYAEGHFRRIA--------------------------SNV 528
E EK PA PN D Y++ HFR+I SNV
Sbjct: 607 LEPEKEKPNPRNAPA-PNADPYSDNHFRKIGTCLSRLPLLRNDTLSSLCLLTLDNSGSNV 665
Query: 529 SSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMER 588
S+YI +V DTLRN++PKAVV+CQVR+AK+SLLN FY Q+G+KE ++L MLDEDP LMER
Sbjct: 666 SAYINMVCDTLRNSLPKAVVYCQVREAKRSLLNFFYAQVGRKEKEKLGAMLDEDPQLMER 725
Query: 589 RQQCAKRLELYKAARDEIDSVSW 611
R AKRLELYK ARD+ID+V+W
Sbjct: 726 RGTLAKRLELYKQARDDIDAVAW 748
Score = 215 bits (547), Expect = 4e-54
Identities = 107/137 (78%), Positives = 120/137 (87%), Gaps = 5/137 (3%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
M+SLIGL+N+IQRACTVLGDHG SLWEALPTVAVVGGQSSGKSSVLES+VGR
Sbjct: 4 MKSLIGLINKIQRACTVLGDHGGEG-----MSLWEALPTVAVVGGQSSGKSSVLESVVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+ EYAEFLH P+++F DF+ VR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTEDGTTEYAEFLHAPKKRFADFAAVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPN 137
+KQIS IPI LSIYSPN
Sbjct: 119 SKQISNIPIQLSIYSPN 135
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 968,884,943
Number of Sequences: 2790947
Number of extensions: 40305800
Number of successful extensions: 125416
Number of sequences better than 10.0: 467
Number of HSP's better than 10.0 without gapping: 409
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 123806
Number of HSP's gapped (non-prelim): 777
length of query: 613
length of database: 848,049,833
effective HSP length: 133
effective length of query: 480
effective length of database: 476,853,882
effective search space: 228889863360
effective search space used: 228889863360
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0316.4


