

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0314.17
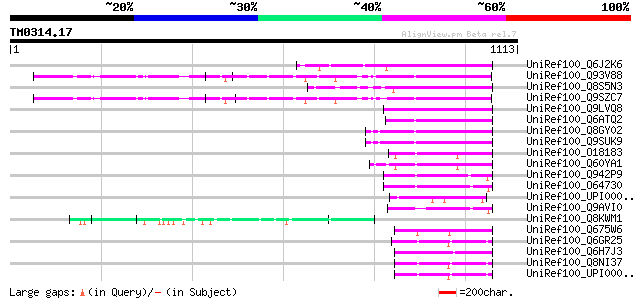
(1113 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6J2K6 BTH-induced protein phosphatase 1 [Oryza sativa] 302 3e-80
UniRef100_Q93V88 Hypothetical protein At4g33500 [Arabidopsis tha... 275 6e-72
UniRef100_Q8S5N3 Hypothetical protein OJ1003C07.6 [Oryza sativa] 273 3e-71
UniRef100_Q9SZC7 Hypothetical protein F17M5.260 [Arabidopsis tha... 263 2e-68
UniRef100_Q9LVQ8 Similarity to unknown protein [Arabidopsis thal... 169 4e-40
UniRef100_Q6ATQ2 Expressed protein [Oryza sativa] 165 6e-39
UniRef100_Q8GY02 Hypothetical protein At4g16580/dl4315c [Arabido... 164 2e-38
UniRef100_Q9SUK9 Hypothetical protein AT4g16580 [Arabidopsis tha... 164 2e-38
UniRef100_O18183 Hypothetical protein W09D10.4 [Caenorhabditis e... 114 2e-23
UniRef100_Q60YA1 Hypothetical protein CBG18323 [Caenorhabditis b... 112 6e-23
UniRef100_Q942P9 Putative senescence-associated protein [Oryza s... 108 9e-22
UniRef100_O64730 Expressed protein [Arabidopsis thaliana] 106 5e-21
UniRef100_UPI000026A9D5 UPI000026A9D5 UniRef100 entry 105 8e-21
UniRef100_Q9AVI0 Putative senescence-associated protein [Pisum s... 103 4e-20
UniRef100_Q8KWM1 Surface protein SdrI [Staphylococcus saprophyti... 99 9e-19
UniRef100_Q675W6 T-cell activation protein phosphatase 2C-like p... 98 1e-18
UniRef100_Q6GR25 MGC81279 protein [Xenopus laevis] 96 5e-18
UniRef100_Q6H7J3 5-azacytidine resistance protein-like [Oryza sa... 96 5e-18
UniRef100_Q8NI37 T-cell activation protein phosphatase 2C [Homo ... 96 8e-18
UniRef100_UPI00001D0928 UPI00001D0928 UniRef100 entry 95 1e-17
>UniRef100_Q6J2K6 BTH-induced protein phosphatase 1 [Oryza sativa]
Length = 569
Score = 302 bits (774), Expect = 3e-80
Identities = 187/449 (41%), Positives = 256/449 (56%), Gaps = 31/449 (6%)
Query: 629 APGEVDEKLDGHEEVADLTSGASGEVEEKLDALEEVVISIVTESDLFSDM-----NSGAS 683
+P E LD T+GA G L++L E + E ++ + N AS
Sbjct: 122 SPSEASSNLD--------TAGAIGGSPLMLESLPETSDTRGCEQEVMPGVVVGSSNRDAS 173
Query: 684 EEVE-EKEGRRTADKGHGVDVATSYLTGSVDADLSELVPASTLLDSEQAVNTEANNLTTS 742
EV E E AD +G+ L SVD +E T + SE+ V+ L
Sbjct: 174 SEVGVESECGSDADGRNGL--GEGELVSSVDGGGAEKSSKVTGVLSEEGVDGMETALEPC 231
Query: 743 LESELVANDEETTHDIVDDSIDASEMEKSTMLHDLPASSDMENKIDVGNTERNDYESSSH 802
+ S E D ++ S+D SE + D +D+E + + E D H
Sbjct: 232 VASVGSITQVEEGVDRMETSLDDSEASDGSTTQDF--DTDVETESSGSSIEEQDMGYGVH 289
Query: 803 LTVPELHSVEMASYGENTSRTE------------LFLVSAAACLPHPSKALTGREDAYFI 850
+ E E+A G +S + L L S AA LPHPSK LTG EDAYFI
Sbjct: 290 IPHTEQAICEVAR-GNKSSEVKSSDRMSSVTLPTLILASGAAMLPHPSKVLTGGEDAYFI 348
Query: 851 SHQNWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENISTIKPAEVLTRSAAETQ 910
+ W VADGVGQWS EG N+G Y RELM+ CK V + ++ EVL ++A E +
Sbjct: 349 ACDGWFGVADGVGQWSFEGINAGLYARELMDGCKKAVMESQGAPEMRTEEVLAKAADEAR 408
Query: 911 TPGSSSVLVAYFDGEALHAANVGNTGFIIIRDGFIFKKSTAKFHEFNFPLHIVKGDDPSE 970
+PGSS+VLVA+FDG+ LHA N+G++GF++IR+G I++KS + FNFPL I KGDDP +
Sbjct: 409 SPGSSTVLVAHFDGQVLHACNIGDSGFLVIRNGEIYQKSKPMTYGFNFPLQIEKGDDPFK 468
Query: 971 IIEGYKVDLCDGDVIIFATNGLFDNLYDQEIASTISKSLQASLGPQEIAEVLAKRAQEVG 1030
+++ Y +DL +GD I+ AT+GLFDN+Y++EIA+ ISKSL+A L P EIAE L RA+EVG
Sbjct: 469 LVQKYTIDLQEGDAIVTATDGLFDNVYEEEIAAVISKSLEAGLKPSEIAEFLVARAKEVG 528
Query: 1031 KSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
+S + RSPF+DAA A+GY+GYSGGKLDD+
Sbjct: 529 RSATCRSPFSDAALAVGYLGYSGGKLDDV 557
>UniRef100_Q93V88 Hypothetical protein At4g33500 [Arabidopsis thaliana]
Length = 724
Score = 275 bits (703), Expect = 6e-72
Identities = 217/652 (33%), Positives = 330/652 (50%), Gaps = 55/652 (8%)
Query: 431 GASGEVEEKIDDHEEASVSSITPESDMFTDISSGALGEVDEKLDVPEEVAVSAVTPESDM 490
G + E+E+ ++ E+A + ++ + +S + +KL V + S E+
Sbjct: 89 GDASEIEKYLEAEEKARCVEVETQNAKIAEEAS-EVSRKQKKL-VSSIIETSTEKEETAA 146
Query: 491 FSDMNN----GSAGEVEEKLDHEEVAVSTVTPEADMFTDVNSGALGEVDEKLDVPEEVAV 546
SD++N V ++ V+ E + D S ++ + + V E + +
Sbjct: 147 PSDLSNVIKIKDRKRVRSPTKKKKETVNVSRSEDKI--DAKSASVSNLSSIVSVAEAIPI 204
Query: 547 SAVTPESDMFSDMNNGSAGEVEEKLDHEEVATVSTVTREADMFTDINNGAPGEVEEK--L 604
S+ E+ + ++ S E L E + VS V + D T+ EV+ + L
Sbjct: 205 SSTEEEAVVEKEITAKSYNV--EPLSSEAMKKVS-VNKIGDCETNGYQENRMEVQARPSL 261
Query: 605 DDKEEVS-VSTVTLEADMFTDMNSSAPGEVDEKLDGHEEVADLTS-----GASGEVEEKL 658
++E++ VST+ ++ ++ + P E +E L D S S +
Sbjct: 262 STQQEITPVSTIEIDDNLDV---TEKPIEAEENLVAEPTATDDLSPDELLSTSEATHRSV 318
Query: 659 DALEEVVISIVTESDLFSDMNSGASEEVEEKEGRRTADKGHGVDVATSYLTG-SVDA--- 714
D + + + +E +L +N+ +EE E TA ++TS T SVD
Sbjct: 319 DEIAQKPVIDTSEENL---LNTFEAEENPVVEPTATAAVSSDELISTSEATRHSVDEIAQ 375
Query: 715 ----DLSELVPASTLLDSEQAVNTEANNLTTSLE--SELVANDEETTHDIVDDSIDASEM 768
D SE P T ++ E AV++ + T L + V ND E +D I
Sbjct: 376 KPIIDTSEKNPMETFVEPE-AVHSSVDESTEKLVVVTSDVENDGENVASTTEDEI----- 429
Query: 769 EKSTMLHDLPASSDMENKIDVGNTERNDYESSSHLTVPELHSVE---MASYGENTSRTEL 825
T+ + S + N D T+ D + L VPE S+E AS E
Sbjct: 430 ---TVRDTITDSGSISNNDD---TKVEDLQ----LPVPETASLEPIKAASGREELVSKAF 479
Query: 826 FLVSAAACLPHPSKALTGREDAYFISHQNWVAVADGVGQWSPEGNNSGPYIRELMEKCKN 885
+L S A L P KAL GREDAYFISH NW+ +ADGV QWS EG N G Y +ELM C+
Sbjct: 480 YLDSGFASLQSPFKALAGREDAYFISHHNWIGIADGVSQWSFEGINKGMYAQELMSNCEK 539
Query: 886 IVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEALHAANVGNTGFIIIRDGFI 945
I+S+ E P +VL RS ET++ GSS+ L+A+ D LH AN+G++GF++IRDG +
Sbjct: 540 IISN-ETAKISDPVQVLHRSVNETKSSGSSTALIAHLDNNELHIANIGDSGFMVIRDGTV 598
Query: 946 FKKSTAKFHEFNFPLHIVKGDDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYDQEIASTI 1005
+ S+ FH F FPLHI +G D ++ E Y V+L +GDV+I AT+GLFDNLY++EI S +
Sbjct: 599 LQNSSPMFHHFCFPLHITQGCDVLKLAEVYHVNLEEGDVVIAATDGLFDNLYEKEIVSIV 658
Query: 1006 SKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLD 1057
SL+ SL PQ+IAE++A +AQEVG+S + R+PFADAA+ GY G+ GGKLD
Sbjct: 659 CGSLKQSLEPQKIAELVAAKAQEVGRSKTERTPFADAAKEEGYNGHKGGKLD 710
Score = 58.5 bits (140), Expect = 1e-06
Identities = 103/461 (22%), Positives = 198/461 (42%), Gaps = 70/461 (15%)
Query: 52 SNASSSS---SSVPDDVDILSSTECSDGSFVFQFAKASEIRDKLAELEREKLACEVVEGG 108
S+ASSSS +S P+ D++SST+ DGS VF+F ASEI +K E E + EV
Sbjct: 55 SSASSSSPPENSAPEKFDLVSSTQLKDGSHVFRFGDASEI-EKYLEAEEKARCVEVETQN 113
Query: 109 GKVGIQA--LLSDSIEILNNEVDDNLQEESKSSSIVVVGVADQNPQIPINEKESVVLDNE 166
K+ +A + +++++ ++ + ++E ++ +D + I I +++ V +
Sbjct: 114 AKIAEEASEVSRKQKKLVSSIIETSTEKEETAAP------SDLSNVIKIKDRKRVRSPTK 167
Query: 167 SERTVINESQKIDRHLKLDSIEDGDAQHGILSEDGAAECNGVPSVCEEESQADDHKEVVV 226
++ +N S+ D K+D+ + + + SV E + +E VV
Sbjct: 168 KKKETVNVSRSED---KIDA-----------KSASVSNLSSIVSVAEAIPISSTEEEAVV 213
Query: 227 SRTVTAVSDVFSDINGGASEEVEEKVDGQEEVVVSAVNPELDMFGDPVSSAFDGHKEVVS 286
+ +TA S ++ A ++V G E N +++ P S +E+
Sbjct: 214 EKEITAKSYNVEPLSSEAMKKVSVNKIGDCETNGYQEN-RMEVQARPSLST---QQEITP 269
Query: 287 AVTPELDVFGGLNSGASGEVEEEVDGLAVSAATQELDMFGDLNSGGA----LGEVEEK-- 340
T E+D L+ +E E + +A AT +L L++ A + E+ +K
Sbjct: 270 VSTIEID--DNLDV-TEKPIEAEENLVAEPTATDDLSPDELLSTSEATHRSVDEIAQKPV 326
Query: 341 LDGDEDDAISTVTPESDMFSDLNNGASGEVEEKLDGDEEDAISTVTPESDMFSDMNNGAS 400
+D E++ ++T E + + A+ +E IST S+ +
Sbjct: 327 IDTSEENLLNTFEAEENPVVEPTATAAVSSDE--------LIST--------SEATRHSV 370
Query: 401 GEVEEK--LDFHEADALSTDTPGSNVNSDMNNGASGEVEEKID-DHEEASVSSITPES-- 455
E+ +K +D E + + T V+S ++ V D +++ +V+S T +
Sbjct: 371 DEIAQKPIIDTSEKNPMETFVEPEAVHSSVDESTEKLVVVTSDVENDGENVASTTEDEIT 430
Query: 456 --DMFTDISSGALGEVDE------KLDVPEEVAVSAVTPES 488
D TD SG++ D+ +L VPE ++ + S
Sbjct: 431 VRDTITD--SGSISNNDDTKVEDLQLPVPETASLEPIKAAS 469
>UniRef100_Q8S5N3 Hypothetical protein OJ1003C07.6 [Oryza sativa]
Length = 496
Score = 273 bits (697), Expect = 3e-71
Identities = 166/438 (37%), Positives = 246/438 (55%), Gaps = 65/438 (14%)
Query: 655 EEKLDALEEVVISIVTESDLFSDMNSGASEEVEEK--EGRRTADKGHGVDVATSYLTGSV 712
+ + DA+ ++V ES L D + A E + EG TA G+D +G
Sbjct: 79 QRRADAVAPEAAAVV-ESGLDGDAAAAAEPEARDGGGEGEVTAT-ATGLDAEEVVASGGA 136
Query: 713 DADLSELVPASTLLDSEQAVNTEANNLTTSLESELVANDEETTHDIVDDSIDASEMEKST 772
+A T + EA++ +T+ +S+ + E + DD + +
Sbjct: 137 EA---------TATSGLEDAGEEASDGSTARDSDTDVDTESSASTAADDD------QPAE 181
Query: 773 MLHDLPASSDMENKIDVGNTERNDYESSSHLTVPELHSVEMASYGENTSRTELFLVSAAA 832
P + ++ NK+D +E + V +AS + L L S AA
Sbjct: 182 FAVPPPPAEEVCNKVD--------WEKDTSEVKNTDRMVPVAS-------STLVLASGAA 226
Query: 833 CLPHPSKAL-------------------------------TGREDAYFISHQNWVAVADG 861
LPHPSK L TG EDAYFI+ W VADG
Sbjct: 227 ILPHPSKVLIIALRVLFYAVYLWTLVYLDPITEANSFKAATGGEDAYFIACDGWFGVADG 286
Query: 862 VGQWSPEGNNSGPYIRELMEKCKNIVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAY 921
VGQWS EG N+G Y RELM+ CK + ++ + IKP +VL+++A E +PGSS+VLVA+
Sbjct: 287 VGQWSFEGINAGLYARELMDGCKKFIMENQGAADIKPEQVLSKAADEAHSPGSSTVLVAH 346
Query: 922 FDGEALHAANVGNTGFIIIRDGFIFKKSTAKFHEFNFPLHIVKGDDPSEIIEGYKVDLCD 981
FDG+ L+A+N+G++GF++IR+G +++KS + FNFPL I KGD+P ++++ Y ++L D
Sbjct: 347 FDGQFLNASNIGDSGFLVIRNGEVYQKSKPMVYGFNFPLQIEKGDNPLKLVQNYTIELED 406
Query: 982 GDVIIFATNGLFDNLYDQEIASTISKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFAD 1041
GDVI+ A++GLFDN+Y+QE+A+ +SKSLQA L P EIAE LA +AQEVG+S + +PF+D
Sbjct: 407 GDVIVTASDGLFDNVYEQEVATMVSKSLQADLKPTEIAEHLAAKAQEVGRSAAGSTPFSD 466
Query: 1042 AAQAIGYVGYSGGKLDDL 1059
AA A+GY+G+SGGKLDD+
Sbjct: 467 AALAVGYLGFSGGKLDDI 484
>UniRef100_Q9SZC7 Hypothetical protein F17M5.260 [Arabidopsis thaliana]
Length = 1066
Score = 263 bits (672), Expect = 2e-68
Identities = 210/649 (32%), Positives = 326/649 (49%), Gaps = 66/649 (10%)
Query: 431 GASGEVEEKIDDHEEASVSSITPESDMFTDISSGALGEVDEKLDVPEEVAVSAVTPESDM 490
G + E+E+ ++ E+A + ++ + +S + +KL V + S E+
Sbjct: 89 GDASEIEKYLEAEEKARCVEVETQNAKIAEEAS-EVSRKQKKL-VSSIIETSTEKEETAA 146
Query: 491 FSDMNN----GSAGEVEEKLDHEEVAVSTVTPEADMFTDVNSGALGEVDEKLDVPEEVAV 546
SD++N V ++ V+ E + D S ++ + + V E + +
Sbjct: 147 PSDLSNVIKIKDRKRVRSPTKKKKETVNVSRSEDKI--DAKSASVSNLSSIVSVAEAIPI 204
Query: 547 SAVTPESDMFSDMNNGSAGEVEEKLDHEEVATVSTVTREADMFTDINNGAPGEVEEK--L 604
S+ E+ + ++ S E L E + VS V + D T+ EV+ + L
Sbjct: 205 SSTEEEAVVEKEITAKSYNV--EPLSSEAMKKVS-VNKIGDCETNGYQENRMEVQARPSL 261
Query: 605 DDKEEVS-VSTVTLEADMFTDMNSSAPGEVDEKLDGHEEVADLTS-----GASGEVEEKL 658
++E++ VST+ ++ ++ + P E +E L D S S +
Sbjct: 262 STQQEITPVSTIEIDDNLDV---TEKPIEAEENLVAEPTATDDLSPDELLSTSEATHRSV 318
Query: 659 DALEEVVISIVTESDLFSDMNSGASEEVEEKEGRRTADKGHGVDVATSYLTG-SVDA--- 714
D + + + +E +L +N+ +EE E TA ++TS T SVD
Sbjct: 319 DEIAQKPVIDTSEENL---LNTFEAEENPVVEPTATAAVSSDELISTSEATRHSVDEIAQ 375
Query: 715 ----DLSELVPASTLLDSEQAVNTEANNLTTSLE--SELVANDEETTHDIVDDSIDASEM 768
D SE P T ++ E AV++ + T L + V ND E +D I
Sbjct: 376 KPIIDTSEKNPMETFVEPE-AVHSSVDESTEKLVVVTSDVENDGENVASTTEDEI----- 429
Query: 769 EKSTMLHDLPASSDMENKIDVGNTERNDYESSSHLTVPELHSVEMASYGENTSRTELFLV 828
T+ + S + N D T+ D + L VPE S+E +
Sbjct: 430 ---TVRDTITDSGSISNNDD---TKVEDLQ----LPVPETASLEP--------------I 465
Query: 829 SAAACLPHPSKALTGREDAYFISHQNWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVS 888
AA+ + +AL GREDAYFISH NW+ +ADGV QWS EG N G Y +ELM C+ I+S
Sbjct: 466 KAASGRNNDVQALAGREDAYFISHHNWIGIADGVSQWSFEGINKGMYAQELMSNCEKIIS 525
Query: 889 SHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEALHAANVGNTGFIIIRDGFIFKK 948
+ E P +VL RS ET++ GSS+ L+A+ D LH AN+G++GF++IRDG + +
Sbjct: 526 N-ETAKISDPVQVLHRSVNETKSSGSSTALIAHLDNNELHIANIGDSGFMVIRDGTVLQN 584
Query: 949 STAKFHEFNFPLHIVKGDDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYDQEIASTISKS 1008
S+ FH F FPLHI +G D ++ E Y V+L +GDV+I AT+GLFDNLY++EI S + S
Sbjct: 585 SSPMFHHFCFPLHITQGCDVLKLAEVYHVNLEEGDVVIAATDGLFDNLYEKEIVSIVCGS 644
Query: 1009 LQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLD 1057
L+ SL PQ+IAE++A +AQEVG+S + R+PFADAA+ GY G+ GGKLD
Sbjct: 645 LKQSLEPQKIAELVAAKAQEVGRSKTERTPFADAAKEEGYNGHKGGKLD 693
Score = 59.7 bits (143), Expect = 5e-07
Identities = 104/467 (22%), Positives = 201/467 (42%), Gaps = 70/467 (14%)
Query: 52 SNASSSS---SSVPDDVDILSSTECSDGSFVFQFAKASEIRDKLAELEREKLACEVVEGG 108
S+ASSSS +S P+ D++SST+ DGS VF+F ASEI +K E E + EV
Sbjct: 55 SSASSSSPPENSAPEKFDLVSSTQLKDGSHVFRFGDASEI-EKYLEAEEKARCVEVETQN 113
Query: 109 GKVGIQA--LLSDSIEILNNEVDDNLQEESKSSSIVVVGVADQNPQIPINEKESVVLDNE 166
K+ +A + +++++ ++ + ++E ++ +D + I I +++ V +
Sbjct: 114 AKIAEEASEVSRKQKKLVSSIIETSTEKEETAAP------SDLSNVIKIKDRKRVRSPTK 167
Query: 167 SERTVINESQKIDRHLKLDSIEDGDAQHGILSEDGAAECNGVPSVCEEESQADDHKEVVV 226
++ +N S+ D K+D+ + + + SV E + +E VV
Sbjct: 168 KKKETVNVSRSED---KIDA-----------KSASVSNLSSIVSVAEAIPISSTEEEAVV 213
Query: 227 SRTVTAVSDVFSDINGGASEEVEEKVDGQEEVVVSAVNPELDMFGDPVSSAFDGHKEVVS 286
+ +TA S ++ A ++V G E N +++ P S +E+
Sbjct: 214 EKEITAKSYNVEPLSSEAMKKVSVNKIGDCETNGYQEN-RMEVQARPSLST---QQEITP 269
Query: 287 AVTPELDVFGGLNSGASGEVEEEVDGLAVSAATQELDMFGDLNSGGA----LGEVEEK-- 340
T E+D L+ +E E + +A AT +L L++ A + E+ +K
Sbjct: 270 VSTIEID--DNLDV-TEKPIEAEENLVAEPTATDDLSPDELLSTSEATHRSVDEIAQKPV 326
Query: 341 LDGDEDDAISTVTPESDMFSDLNNGASGEVEEKLDGDEEDAISTVTPESDMFSDMNNGAS 400
+D E++ ++T E + + A+ +E IST S+ +
Sbjct: 327 IDTSEENLLNTFEAEENPVVEPTATAAVSSDE--------LIST--------SEATRHSV 370
Query: 401 GEVEEK--LDFHEADALSTDTPGSNVNSDMNNGASGEVEEKID-DHEEASVSSITPES-- 455
E+ +K +D E + + T V+S ++ V D +++ +V+S T +
Sbjct: 371 DEIAQKPIIDTSEKNPMETFVEPEAVHSSVDESTEKLVVVTSDVENDGENVASTTEDEIT 430
Query: 456 --DMFTDISSGALGEVDE------KLDVPEEVAVSAVTPESDMFSDM 494
D TD SG++ D+ +L VPE ++ + S +D+
Sbjct: 431 VRDTITD--SGSISNNDDTKVEDLQLPVPETASLEPIKAASGRNNDV 475
>UniRef100_Q9LVQ8 Similarity to unknown protein [Arabidopsis thaliana]
Length = 414
Score = 169 bits (428), Expect = 4e-40
Identities = 93/243 (38%), Positives = 141/243 (57%), Gaps = 4/243 (1%)
Query: 820 TSRTELFLVSAAACLPHPSKALTGREDAYFI-SHQNWVAVADGVGQWSPEGNNSGPYIRE 878
TS L LVS + LPHP K TG EDA+FI + + VADGVG W+ G N+G + RE
Sbjct: 163 TSLKSLRLVSGSCYLPHPEKEATGGEDAHFICDEEQAIGVADGVGGWAEVGVNAGLFSRE 222
Query: 879 LMEKCKNIVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEALHAANVGNTGFI 938
LM + + S+I P VL ++ ++T+ GSS+ + + LHA N+G++GF
Sbjct: 223 LMSYSVSAIQEQHKGSSIDPLVVLEKAHSQTKAKGSSTACIIVLKDKGLHAINLGDSGFT 282
Query: 939 IIRDGFIFKKSTAKFHEFNFPLHIVKGDDPSEIIEG--YKVDLCDGDVIIFATNGLFDNL 996
++R+G +S + H FNF + G+ G + +D+ GDVI+ T+G++DNL
Sbjct: 283 VVREGTTVFQSPVQQHGFNFTYQLESGNSADVPSSGQVFTIDVQSGDVIVAGTDGVYDNL 342
Query: 997 YDQEIASTISKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKL 1056
Y++EI + S++A L P+ A+ +A+ A++ +SPFA AAQ GY Y GGKL
Sbjct: 343 YNEEITGVVVSSVRAGLDPKGTAQKIAELARQRAVDKKRQSPFATAAQEAGY-RYYGGKL 401
Query: 1057 DDL 1059
DD+
Sbjct: 402 DDI 404
>UniRef100_Q6ATQ2 Expressed protein [Oryza sativa]
Length = 479
Score = 165 bits (418), Expect = 6e-39
Identities = 96/239 (40%), Positives = 140/239 (58%), Gaps = 7/239 (2%)
Query: 825 LFLVSAAACLPHPSKALTGREDAYFIS-HQNWVAVADGVGQWSPEGNNSGPYIRELMEKC 883
L LVS + LPHP+K TG ED +FI + + VADGVG W+ G ++G Y +ELM
Sbjct: 231 LKLVSGSCYLPHPAKEATGGEDGHFICVDEQAIGVADGVGGWADHGVDAGLYAKELMSNS 290
Query: 884 KNIVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEALHAANVGNTGFIIIRDG 943
+ + E TI P+ VL ++ T+ GSS+ + + +HA N+G++GFII+RDG
Sbjct: 291 MSAIKD-EPQGTIDPSRVLEKAYTCTKARGSSTACIVALKEQGIHAVNLGDSGFIIVRDG 349
Query: 944 FIFKKSTAKFHEFNFPLHIVKG---DDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYDQE 1000
+S + H+FNF + G D PS + + + GDVII T+GLFDNLY E
Sbjct: 350 RTVLRSPVQQHDFNFTYQLESGGGSDLPSSA-QTFHFPVAPGDVIIAGTDGLFDNLYSNE 408
Query: 1001 IASTISKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
I++ + ++L+ L P+ A+ +A AQ+ + +SPFA AAQ GY Y GGKLDD+
Sbjct: 409 ISAIVVEALRTGLEPEATAKKIAALAQQKAMDRNRQSPFAAAAQEAGY-RYFGGKLDDI 466
>UniRef100_Q8GY02 Hypothetical protein At4g16580/dl4315c [Arabidopsis thaliana]
Length = 300
Score = 164 bits (414), Expect = 2e-38
Identities = 99/282 (35%), Positives = 157/282 (55%), Gaps = 8/282 (2%)
Query: 781 SDMENKIDVGNTERNDYESSSHLTVPELHSVEMASYGENTSRTELFLVSAAACLPHPSKA 840
S + N++ GN D + +T ++ S L LVS + LPHP K
Sbjct: 8 SSLSNRLSAGNAP--DVSLDNSVTDEQVRD-SSDSVAAKLCTKPLKLVSGSCYLPHPDKE 64
Query: 841 LTGREDAYFI-SHQNWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENISTIKPA 899
TG EDA+FI + + + VADGVG W+ G ++G Y RELM N + E +I PA
Sbjct: 65 ATGGEDAHFICAEEQALGVADGVGGWAELGIDAGYYSRELMSNSVNAIQD-EPKGSIDPA 123
Query: 900 EVLTRSAAETQTPGSSSVLVAYFDGEALHAANVGNTGFIIIRDGFIFKKSTAKFHEFNFP 959
VL ++ T++ GSS+ + + LHA N+G++GF+++R+G +S + H+FNF
Sbjct: 124 RVLEKAHTCTKSQGSSTACIIALTNQGLHAINLGDSGFMVVREGHTVFRSPVQQHDFNFT 183
Query: 960 LHIVKGDDPSEIIEG--YKVDLCDGDVIIFATNGLFDNLYDQEIASTISKSLQASLGPQE 1017
+ G + G + V + GDVII T+GLFDNLY+ EI + + +++A++ PQ
Sbjct: 184 YQLESGRNGDLPSSGQVFTVAVAPGDVIIAGTDGLFDNLYNNEITAIVVHAVRANIDPQV 243
Query: 1018 IAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
A+ +A A++ + + ++PF+ AAQ G+ Y GGKLDD+
Sbjct: 244 TAQKIAALARQRAQDKNRQTPFSTAAQDAGF-RYYGGKLDDI 284
>UniRef100_Q9SUK9 Hypothetical protein AT4g16580 [Arabidopsis thaliana]
Length = 335
Score = 164 bits (414), Expect = 2e-38
Identities = 99/282 (35%), Positives = 157/282 (55%), Gaps = 8/282 (2%)
Query: 781 SDMENKIDVGNTERNDYESSSHLTVPELHSVEMASYGENTSRTELFLVSAAACLPHPSKA 840
S + N++ GN D + +T ++ S L LVS + LPHP K
Sbjct: 43 SSLSNRLSAGNAP--DVSLDNSVTDEQVRD-SSDSVAAKLCTKPLKLVSGSCYLPHPDKE 99
Query: 841 LTGREDAYFI-SHQNWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENISTIKPA 899
TG EDA+FI + + + VADGVG W+ G ++G Y RELM N + E +I PA
Sbjct: 100 ATGGEDAHFICAEEQALGVADGVGGWAELGIDAGYYSRELMSNSVNAIQD-EPKGSIDPA 158
Query: 900 EVLTRSAAETQTPGSSSVLVAYFDGEALHAANVGNTGFIIIRDGFIFKKSTAKFHEFNFP 959
VL ++ T++ GSS+ + + LHA N+G++GF+++R+G +S + H+FNF
Sbjct: 159 RVLEKAHTCTKSQGSSTACIIALTNQGLHAINLGDSGFMVVREGHTVFRSPVQQHDFNFT 218
Query: 960 LHIVKGDDPSEIIEG--YKVDLCDGDVIIFATNGLFDNLYDQEIASTISKSLQASLGPQE 1017
+ G + G + V + GDVII T+GLFDNLY+ EI + + +++A++ PQ
Sbjct: 219 YQLESGRNGDLPSSGQVFTVAVAPGDVIIAGTDGLFDNLYNNEITAIVVHAVRANIDPQV 278
Query: 1018 IAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
A+ +A A++ + + ++PF+ AAQ G+ Y GGKLDD+
Sbjct: 279 TAQKIAALARQRAQDKNRQTPFSTAAQDAGF-RYYGGKLDDI 319
>UniRef100_O18183 Hypothetical protein W09D10.4 [Caenorhabditis elegans]
Length = 330
Score = 114 bits (285), Expect = 2e-23
Identities = 81/254 (31%), Positives = 125/254 (48%), Gaps = 27/254 (10%)
Query: 831 AACLPHPSKALTGR----------EDAYFISHQN---WVAVADGVGQWSPEGNNSGPYIR 877
A+C P L G +DA+FIS V VADGVG W G + + R
Sbjct: 71 ASCAGFPKDMLNGPSTVLDKGVFGDDAWFISRFKNTFVVGVADGVGGWRKYGIDPSAFSR 130
Query: 878 ELMEKCKNIVSSHENISTIKPAEVLT---RSAAETQTP-GSSSVLVAYFDGEALHAANVG 933
LM++C+ V + KP +L R++AE P GSS+ V E L++AN+G
Sbjct: 131 RLMKECEKRVQKGD-FDPQKPESLLDYAFRASAEAPRPVGSSTACVLVVHQEKLYSANLG 189
Query: 934 NTGFIIIRDGFIFKKSTAKFHEFNFPLHIVKGDDPSEIIEGYKVDLCD--------GDVI 985
++GF+++R+G I KS + H FN P + + + G K D+ D GD+I
Sbjct: 190 DSGFMVVRNGKIVSKSREQVHYFNAPFQLTLPPEGYQGFIGDKADMADKDEMAVKKGDII 249
Query: 986 IFATNGLFDNLYDQEIASTISKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQA 1045
+ AT+G++DNL +Q++ + QE+ LA A+ + + SPFA A+
Sbjct: 250 LLATDGVWDNLSEQQVLDQLKALDAGKSNVQEVCNALALTARRLAFDSKHNSPFAMKARE 309
Query: 1046 IGYVGYSGGKLDDL 1059
G++ GGK DD+
Sbjct: 310 HGFLA-PGGKPDDI 322
>UniRef100_Q60YA1 Hypothetical protein CBG18323 [Caenorhabditis briggsae]
Length = 330
Score = 112 bits (280), Expect = 6e-23
Identities = 88/294 (29%), Positives = 142/294 (47%), Gaps = 32/294 (10%)
Query: 791 NTERNDYESSSHLTVPELHSVEMASYGENTSRTELFLVSAAACLPHPSKALTGR------ 844
+T R SSS + P S E +S +++ E + A+C P L G
Sbjct: 36 STGRRGLSSSS--SKPAKPSNEGSSSPASSAHVENVI---ASCAGFPKDMLNGPSTVLDK 90
Query: 845 ----EDAYFISHQN---WVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENISTIK 897
+DA+FIS V VADGVG W G + + R LM++C+ V E +
Sbjct: 91 GVYGDDAWFISRFKNTFVVGVADGVGGWRKYGIDPSAFSRRLMKECEKRVQGGE-FDPKR 149
Query: 898 PAEVLT---RSAAETQTP-GSSSVLVAYFDGEALHAANVGNTGFIIIRDGFIFKKSTAKF 953
P +L R++AE P GSS+ V E L++AN+G++GF+++R+G + KS +
Sbjct: 150 PDSLLDFAFRASAEAPRPVGSSTACVLVVHQEKLYSANLGDSGFMVVRNGKVISKSREQV 209
Query: 954 HEFNFPLHIVKGDDPSEIIEGYKVDLCD--------GDVIIFATNGLFDNLYDQEIASTI 1005
H FN P + + + G + D+ D GD+I+ AT+G++DNL + ++ +
Sbjct: 210 HYFNAPFQLTLPPEGYQGFIGDRADMADKEEMDVKKGDIILLATDGVWDNLSENQVLDQL 269
Query: 1006 SKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
+ QE+ LA A+ + + SPFA A+ G++ GGK DD+
Sbjct: 270 KALNEGKGNVQEVCNALALTARRLAFDSKHNSPFAMKAREHGFLA-PGGKPDDI 322
>UniRef100_Q942P9 Putative senescence-associated protein [Oryza sativa]
Length = 331
Score = 108 bits (270), Expect = 9e-22
Identities = 78/254 (30%), Positives = 130/254 (50%), Gaps = 21/254 (8%)
Query: 821 SRTELFLVSAAACLPHPSKALTGREDAYFISHQNW--VAVADGVGQWSPEGNNSGPYIRE 878
++ E L +PHP KA TG EDA+F++ + AVADGV W+ + N + RE
Sbjct: 38 AKLEAVLTIGTHLIPHPRKAETGGEDAFFVNGDDGGVFAVADGVSGWAEKDVNPALFSRE 97
Query: 879 LMEKCKNIVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEA-LHAANVGNTGF 937
LM + E P +L ++ A T + GS++V++A + L A+VG+ G
Sbjct: 98 LMAHTSTFLKDEE--VNHDPQLLLMKAHAATTSVGSATVIIAMLEKTGILKIASVGDCGL 155
Query: 938 IIIRDGFIFKKSTAKFHEFNFPLHIVKGDDPSEIIEGY--KVDLCDGDVIIFATNGLFDN 995
+IR G + + + H F+ P + ++ V+L +GD+I+ ++G FDN
Sbjct: 156 KVIRKGQVMFSTCPQEHYFDCPYQLSSEAIGQTYLDALVCTVNLMEGDMIVSGSDGFFDN 215
Query: 996 LYDQEIASTISKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIG-------- 1047
++DQEI S IS+ S G E A+ LA+ A++ + SP++ A++ G
Sbjct: 216 IFDQEIVSVISE----SPGVDEAAKALAELARKHSVDVTFDSPYSMEARSRGFDVPSWKK 271
Query: 1048 YVG--YSGGKLDDL 1059
++G GGK+DD+
Sbjct: 272 FIGGKLIGGKMDDI 285
>UniRef100_O64730 Expressed protein [Arabidopsis thaliana]
Length = 298
Score = 106 bits (264), Expect = 5e-21
Identities = 74/253 (29%), Positives = 124/253 (48%), Gaps = 21/253 (8%)
Query: 822 RTELFLVSAAACLPHPSKALTGREDAYFISHQNW--VAVADGVGQWSPEGNNSGPYIREL 879
R EL L +PHP K G EDA+F+S +AVADGV W+ + + + +EL
Sbjct: 42 RPELSLSVGIHAIPHPDKVEKGGEDAFFVSSYRGGVMAVADGVSGWAEQDVDPSLFSKEL 101
Query: 880 MEKCKNIVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEA-LHAANVGNTGFI 938
M +V E P ++ ++ T + GS+++++A + L NVG+ G
Sbjct: 102 MANASRLVDDQE--VRYDPGFLIDKAHTATTSRGSATIILAMLEEVGILKIGNVGDCGLK 159
Query: 939 IIRDGFIFKKSTAKFHEFNFPLHIVKGDDPSEIIEGYK--VDLCDGDVIIFATNGLFDNL 996
++R+G I + + H F+ P + ++ V++ GDVI+ ++GLFDN+
Sbjct: 160 LLREGQIIFATAPQEHYFDCPYQLSSEGSAQTYLDASFSIVEVQKGDVIVMGSDGLFDNV 219
Query: 997 YDQEIASTISKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGY-------- 1048
+D EI S ++K + E + +LA+ A + T SP+A A+A G+
Sbjct: 220 FDHEIVSIVTKHTDVA----ESSRLLAEVASSHSRDTEFESPYALEARAKGFDVPLWKKV 275
Query: 1049 --VGYSGGKLDDL 1059
+GGKLDD+
Sbjct: 276 LGKKLTGGKLDDV 288
>UniRef100_UPI000026A9D5 UPI000026A9D5 UniRef100 entry
Length = 250
Score = 105 bits (262), Expect = 8e-21
Identities = 69/236 (29%), Positives = 121/236 (51%), Gaps = 25/236 (10%)
Query: 834 LPHPSKALTGREDAYFISHQNWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENI 893
+PHP K G EDA F S N + VADGVG W+ +G + G Y LM + +
Sbjct: 1 MPHPDKVHKGGEDAAFGS-PNAIGVADGVGGWASKGVDPGLYSEGLMRSAEKEALAKPAP 59
Query: 894 STIKPAEVLTRSAAETQTP---GSSSVLVAYFDGEA-----LHAANVGNTGFIIIRDGFI 945
+T +P E+L ++ + + GS++ ++ + A +H AN+G++G +++R F
Sbjct: 60 NTPRPVELLRKAYDDVEQQRVLGSTTAIIVTLESIARDRALIHIANLGDSGILLVRP-FA 118
Query: 946 FKKSTAKF-------HEFNFPLHIVKGDDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYD 998
+ ++ K H FN+P + D E+ + + + + + D+++ T+GLFDNL+D
Sbjct: 119 WDRAPQKLFRSEEQQHSFNYPYQLGGDSDKPEMAQQHTIPVNENDLVVLGTDGLFDNLFD 178
Query: 999 QEIASTISKSLQASLGPQEIAEV-LAKRAQEVGKSTSTR-------SPFADAAQAI 1046
+EI I + + A G LAK ++ + K+T TR +PFA A+ +
Sbjct: 179 EEINDIIKQHIVACGGIGNCNRAGLAKMSEAIAKATHTRATSRTADTPFAKGARRV 234
>UniRef100_Q9AVI0 Putative senescence-associated protein [Pisum sativum]
Length = 300
Score = 103 bits (256), Expect = 4e-20
Identities = 73/246 (29%), Positives = 121/246 (48%), Gaps = 42/246 (17%)
Query: 829 SAAACL-PHPSKALTGREDAYFISHQNW--VAVADGVGQWSPEGNNSGPYIRELMEKCKN 885
S CL PHP K G EDA+F+S+ N +AVADGV W+ E + + RELM N
Sbjct: 60 SVGTCLIPHPKKVDKGGEDAFFVSNYNGGVIAVADGVSGWAEEDVDPSLFPRELMANAYN 119
Query: 886 IVSSHENISTIKPAEVLTRSAAETQTPGSSSVLVAYFDGEA-LHAANVGNTGFIIIRDGF 944
V S++V++A + L ANVG+ G +IR+G
Sbjct: 120 FVGDD-----------------------SATVIIAMLEKNGNLKIANVGDCGLRVIRNGI 156
Query: 945 IFKKSTAKFHEFNFPLHIVKGD-DPSEIIEGYKVDLCDGDVIIFATNGLFDNLYDQEIAS 1003
+ ++ + H F+ P + + ++ G V+L +GD+I+ ++GL+DN++D EIA
Sbjct: 157 VTFSTSPQEHYFDCPFQLSSERVGQTYLMHGKNVELMEGDIIVMGSDGLYDNVFDHEIAL 216
Query: 1004 TISKSLQASLGPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGY----------VGYSG 1053
T+++ S E A+ LA A + ++ SP++ A++ G+ + +G
Sbjct: 217 TVARYRDVS----EAAKALANLASSHARDSNFDSPYSWEARSKGFEAPLWKKILGMKLTG 272
Query: 1054 GKLDDL 1059
GK DD+
Sbjct: 273 GKPDDI 278
>UniRef100_Q8KWM1 Surface protein SdrI [Staphylococcus saprophyticus]
Length = 1893
Score = 98.6 bits (244), Expect = 9e-19
Identities = 128/638 (20%), Positives = 243/638 (38%), Gaps = 29/638 (4%)
Query: 179 DRHLKLDSIEDGDAQHGILSEDGAAECNGVPSVCEEESQADDHKEVVVSRTVTAVSDVFS 238
D DS D DA ++ A + + +S AD + A SD +
Sbjct: 1057 DADADADSDADADADADSDADADADSDADSDADADADSDADADADADADSDADADSDADA 1116
Query: 239 DINGGASEEVEEKVDGQEEVVVSAVNPELDMFGDPVSSAFDGHKEVVSAVTPELDVFGGL 298
D + A + + D + + D D + A D + + + D
Sbjct: 1117 DADADADSDADADADADSDA-------DADSDSDADADA-DADSDADADADADSDADADA 1168
Query: 299 NSGASGEVEEEVDGLAVSAATQELDMFGDLNSGG-ALGEVEEKLDGDED---DAISTVTP 354
+S A + + + D A S A + D D ++ A + + D D D DA +
Sbjct: 1169 DSDADADADSDADADADSDADSDADADADSDADADADADADSDADADADSDADADADADA 1228
Query: 355 ESDMFSDLNNGASGEVEEKLDGD---EEDAISTVTPESDMFSDMNNGASGEVEEKLDFHE 411
+SD SD ++ A + + D D + DA + ++D +D ++ A + + D +
Sbjct: 1229 DSDADSDADSDADSDADADADSDADADADADADSDADADSDADADSDADADADADAD-SD 1287
Query: 412 ADALSTDTPGSNVNSDMNNGASGEVEEKID-DHEEASVSSITPESDMFTDISSGALGEVD 470
ADA S S+ ++D + A + + D D + S + +SD D + A + D
Sbjct: 1288 ADADSDADADSDADADSDADADADADSDADADADADSDADADADSDSDADADADADSDAD 1347
Query: 471 EKLDVPEEV-AVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVAVSTVTPEADMFTDVNSG 529
D + A S +SD +D + S + + D + A S +AD D ++
Sbjct: 1348 ADADADSDADADSDADADSDADADADADSDADADSDADADADADSDADADADSDADADAD 1407
Query: 530 ALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVATVSTVT-READM 588
A + D D + A ++D +D ++ + + + D + A + +AD
Sbjct: 1408 ADSDADADSDADADADADA-DSDADADADADSDADADADADADADSDADADADSDADADA 1466
Query: 589 FTDINNGAPGEVEEKLD-----DKEEVSVSTVTLEADMFTDMNSSAPGEVDEKLDGHEEV 643
+D ++ A + + D D + + S ++D D ++ A + D D +
Sbjct: 1467 DSDADSDADADADSDADADADADSDADADSDADADSDADADSDADADADADSDADADSD- 1525
Query: 644 ADLTSGASGEVEEKLDALEEVVISIVTESDLFSDMNSGASEEVEEKEGRRTADKGHGVDV 703
AD S A + + DA + +SD +D +S A + + + AD D
Sbjct: 1526 ADADSDADADADADSDADADSDADADADSDADADADSDADAD-SDADADADADADSDAD- 1583
Query: 704 ATSYLTGSVDADLSELVPASTLLDSEQAVNTEANNLTTSLESELVANDEETTHDIVDDSI 763
A S DAD A D++ + +A + + +++ A+ + + D
Sbjct: 1584 ADSDADSDADADADSDSDADADADADSDADADA-DADSDADADSDADADADSDSDADADA 1642
Query: 764 DASEMEKSTMLHDLPASSDMENKIDVGNTERNDYESSS 801
DA + D A +D + D + D +S +
Sbjct: 1643 DADSDADADSDADADADADSDADADADSDADADADSDA 1680
Score = 97.4 bits (241), Expect = 2e-18
Identities = 112/533 (21%), Positives = 207/533 (38%), Gaps = 27/533 (5%)
Query: 179 DRHLKLDSIEDGDAQHGILSEDGAAECNGVPSVCEEESQADDHKEVVVSRTVTAVSDVFS 238
D D+ D DA ++ A + + + ++ AD + A +D S
Sbjct: 1333 DSDADADADADSDADADADADSDADADSDADADSDADADADADSDADADSDADADADADS 1392
Query: 239 DINGGASEEVEEKVDGQEEVVVSAVNPELDMFGDPVSSAFDGHKEVVSAVTPELDVFGGL 298
D + A + + D + + + + D D S A D + S + D
Sbjct: 1393 DADADADSDADADADADSDADADS-DADADADADADSDA-DADADADSDADADADADADA 1450
Query: 299 NSGASGEVEEEVDGLAVSAATQELDMFGDLNSGGALGEVEEKLDGDED-DAISTVTPESD 357
+S A + + + D A S A + D D + A + + D D D DA S +SD
Sbjct: 1451 DSDADADADSDADADADSDADSDADADAD-SDADADADADSDADADSDADADSDADADSD 1509
Query: 358 MFSDLNNGASGEVEEKLDGDEE-----DAISTVTPESDMFSDMNNGASGEVEEKLDFH-- 410
+D + + + + D D + DA S +SD +D ++ A + + D
Sbjct: 1510 ADADADADSDADADSDADADSDADADADADSDADADSDADADADSDADADADSDADADSD 1569
Query: 411 -EADALSTDTPGSNVNSDMNNGASGEVEEKIDDHEEASVSSITPESDMFTDISSGALGEV 469
+ADA + ++ +SD ++ A + + D +A S ++D D S A +
Sbjct: 1570 ADADADADADSDADADSDADSDADADADSDSDADADADADS---DADADADADSDADADS 1626
Query: 470 DEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVAVSTVTPEADMFTDVNSG 529
D D + S ++D SD + S + + D + A + +AD +D ++
Sbjct: 1627 DADADADSD---SDADADADADSDADADSDADADADADSDADADADSDADADADSDADAD 1683
Query: 530 ALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVATVSTVTREADMF 589
A + D D A S ++D SD + S + + D + A S +AD
Sbjct: 1684 ADSDADSDADAD---ADSDADADADADSDADADSDADADADADSDADAD-SDADADADAD 1739
Query: 590 TDINNGAPGEVEEKLDDKEEVSVSTVTLEADMFTDMNSSAPGEVDEKLDGHEEVADLTSG 649
+D + + + + D + + +AD D ++ A + D D + AD S
Sbjct: 1740 SDADADSDADADSDADADSDADADS-DADADSDADADADADSDADADSDADAD-ADADSD 1797
Query: 650 ASGEVEEKLDALEEVVISIVTESDLFSDMNSGASEEVE---EKEGRRTADKGH 699
A + + DA + +SD +D +S A + + +K TADK +
Sbjct: 1798 ADADADSDADADADSDADADADSDADADADSDADADADSDADKYHNDTADKSN 1850
Score = 84.3 bits (207), Expect = 2e-14
Identities = 102/526 (19%), Positives = 205/526 (38%), Gaps = 29/526 (5%)
Query: 279 DGHKEVVSAVTPELDVFGGLNSGASGEVEEEVDGLAVSAATQELDMFGDLNSGGALGEVE 338
+G+ + S + D ++ A + + + D A + + + D D +S A + +
Sbjct: 979 NGYFDEDSDADADSDADADSDADADADADSDADADADADSDADSDADADADSD-ADADAD 1037
Query: 339 EKLDGDED-DAISTVTPESDMFSDLNNGASGEVEEKLDGDEEDAISTVTPESDMFSDMNN 397
D D D DA + +SD +D ++ A + + D D + +SD SD +
Sbjct: 1038 ADADSDSDADADADADADSDADADADSDADADADADSDADAD-------ADSDADSDADA 1090
Query: 398 GASGEVEEKLDFHEADALSTDTPGSNVNSDMNNGASGEVEEKIDDHEEASVSSITPESDM 457
A + + D ADA S S+ ++D + A + + D +A +SD
Sbjct: 1091 DADSDADADAD---ADADSDADADSDADADADADADSDADADADADSDADA-----DSDS 1142
Query: 458 FTDISSGALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVAVSTVT 517
D + A + D D + A +SD +D ++ + + + D + A +
Sbjct: 1143 DADADADADSDADADADADSDADADA---DSDADADADSDADADADSDADSDADADADSD 1199
Query: 518 PEADMFTDVNSGALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEKLDHEEVA 577
+AD D +S A + D D + + +SD SD ++ + + + D + A
Sbjct: 1200 ADADADADADSDADADADSDADADADADADS-DADSDADSDADSDADADADSDADADADA 1258
Query: 578 TVSTVTREADMFTDINNGAPGEVEEKLDDKEEVSVSTVTLEADMFTDMNSSAPGEVDEKL 637
+ +AD D ++ A + + D + +AD D +S A + D
Sbjct: 1259 DADS-DADADSDADADSDADADADADADSDADADSDA---DADSDADADSDADADADADS 1314
Query: 638 DGHEEVADLTSGASGEVEEKLDALEEVVISIVTESDLFSDMNSGASEEVE-EKEGRRTAD 696
D + AD S A + + DA + ++D +D ++ A + + + + AD
Sbjct: 1315 DADAD-ADADSDADADADSDSDADADADADSDADADADADSDADADSDADADSDADADAD 1373
Query: 697 KGHGVDVATSYLTGSVDADLSELVPASTLLDSEQAVNTEAN-NLTTSLESELVANDEETT 755
D A S DAD A + D++ +++A+ + +++ A+ +
Sbjct: 1374 ADSDAD-ADSDADADADADSDADADADSDADADADADSDADADSDADADADADADSDADA 1432
Query: 756 HDIVDDSIDASEMEKSTMLHDLPASSDMENKIDVGNTERNDYESSS 801
D DA + D A +D + D + +D ++ +
Sbjct: 1433 DADADSDADADADADADADSDADADADSDADADADSDADSDADADA 1478
Score = 50.1 bits (118), Expect = 4e-04
Identities = 154/785 (19%), Positives = 287/785 (35%), Gaps = 143/785 (18%)
Query: 131 NLQEESKSSSIVVVGVADQNPQIP-------INEKESV-----VLDNESERTVINESQKI 178
N Q SK SIV GV D+N +++ +++ +L TV + Q
Sbjct: 513 NTQLRSKEGSIVANGVYDENTTTTTYTFTNYVDQYQNITGSFNLLATPKRETVTTDKQTY 572
Query: 179 DRHLKLDSIEDGDAQHGILSEDGAAECNGV-PSVCEEESQADDHKEVV-VSRTVTAVSDV 236
++ +I + + + + G E + +V ++ + H EVV ++++ + D
Sbjct: 573 PMNV---TIANQEVSENFVVDYGNHEDHLTNAAVVNVDNVNNQHNEVVYLNQSGDRIYDA 629
Query: 237 --FSDINGGASEEVEEKVDG--QEEVVVSAVNPELDMFGDPVSSAFDGHKEVVSAVTPEL 292
FS + G E KV + V+V + NP+L+ G V K+V S TP+
Sbjct: 630 KYFSIVQNGTFIPNEVKVYEVLDDNVLVDSFNPDLN--GPAV-------KDVTSEFTPQY 680
Query: 293 DV----------FGGLNSGASGEVEEEV--DGLAVSAATQELDMFGD------------- 327
+ +N G+ + + V G T EL +G+
Sbjct: 681 SLNNTRVDIDLNRSNMNKGSRYIITQAVKPSGTGNVNTTYELTRYGNEASRYPTGTKSTT 740
Query: 328 ----LNSGGALGE-----------VEEKLDGDEDD------AISTVTPESD--------- 357
S A G+ +++ DG ++D + + +S+
Sbjct: 741 VSYINGSSTAQGDNPTYNLGDYVWLDKNKDGIQNDDEKGISGVYVILKDSNNKELQRATT 800
Query: 358 ------MFSDLNNGASGEVEEKLDGDEEDAISTVTPESDMFSD-MNNGASGEVEEKLDFH 410
F++L NG + VE + + + S + SD +G S V K +
Sbjct: 801 DDTGRYQFNNLQNG-TYNVEFVIPNNYTPSPSNTIDNDTIDSDGQKDGDSNVVVAKGTIN 859
Query: 411 EADALSTDTP---------GSNVNSDMNNGASGEVEEK--------IDDHEEASVSSITP 453
AD ++ DT G V D N + +EK + D + + T
Sbjct: 860 NADNMTVDTGFYETPKYSLGDYVWKDTNKDGVQDSDEKGIQGVTVTLKDKNGNVLKTTTT 919
Query: 454 E---SDMFTDISSGALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVEEK--LDH 508
+ S F ++ SG D + + ++ T SD SD N + GE DH
Sbjct: 920 DENGSYRFDNLDSG-----DYIVHFEKPEGLTQTTTNSD--SDENKDADGEEVHVTITDH 972
Query: 509 EEVAVSTVTPEADMFTDVNSGALGEVDEKLDVPEEVAVSAVTPESDMFSDMNNGSAGEVE 568
++ ++ + D D +S A + D D A S ++D SD ++ + + +
Sbjct: 973 DDFSIDNGYFDEDSDADADSDADADSDADADAD---ADSDADADADADSDADSDADADAD 1029
Query: 569 EKLDHEEVATV-STVTREADMFTDINNGAPGEVEEKLD---------DKEEVSVSTVTLE 618
D + A S +AD D ++ A + + D D + S + +
Sbjct: 1030 SDADADADADADSDSDADADADADADSDADADADSDADADADADSDADADADSDADSDAD 1089
Query: 619 ADMFTDMNSSAPGEVDEKLDGHEEV-ADLTSGASGEVEEKLDALEEVVISIVTESDLFSD 677
AD +D ++ A + D D + AD + A + + DA + +++D +D
Sbjct: 1090 ADADSDADADADADADSDADADSDADADADADADSDADADADADSDADADSDSDADADAD 1149
Query: 678 MNSGASEEVE-EKEGRRTADKGHGVDVATSYLTGSVDADLSELVPASTLLDSEQAVNTEA 736
+S A + + + + AD D A S D+D A D++ + +A
Sbjct: 1150 ADSDADADADADSDADADADSDADAD-ADSDADADADSDADSDADADADSDADADADADA 1208
Query: 737 NNLTTSLESELVANDEETTHDIVDDSIDASEMEKSTMLHDLPASSDMENKIDVGNTERND 796
++ +++ A+ + D DA S D A +D + D +D
Sbjct: 1209 DS-----DADADADSDADADADADADSDADSDADSDADSDADADADSDADADADADADSD 1263
Query: 797 YESSS 801
++ S
Sbjct: 1264 ADADS 1268
>UniRef100_Q675W6 T-cell activation protein phosphatase 2C-like protein [Oikopleura
dioica]
Length = 354
Score = 98.2 bits (243), Expect = 1e-18
Identities = 76/238 (31%), Positives = 122/238 (50%), Gaps = 25/238 (10%)
Query: 845 EDAYFI----SHQNWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHEN-----IST 895
EDA F ++++ +ADGVG W G + + LM CK++ + + I
Sbjct: 105 EDACFALTNSRRKDYIGIADGVGGWRDRGFDPSVFSSSLMRICKDMANKKQEDPMRLIDD 164
Query: 896 IKPAEVLTRSAAETQTPGSSSVLVAYFDGEA--LHAANVGNTGFIIIRDGFIFKKSTAKF 953
+L Q GSS+V + F+ E L AN+G++G++I+R+G I +S +
Sbjct: 165 SYNKLLLLNKKKNFQIVGSSTVCILSFEQETGILTTANLGDSGYLIVRNGEIIDRSEKQT 224
Query: 954 HEFNFPLHIVK--------GDDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYDQEIASTI 1005
H+FN P + D PS+ E K GD+I+ AT+GLFDN+ D+ + +
Sbjct: 225 HKFNIPKQLAYAPPSLRFIADMPSDAHE-KKFVTHPGDLIVTATDGLFDNVPDEVLIQEL 283
Query: 1006 SKSLQAS-LGPQEI---AEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
S A + Q++ A+ LA RA + + S SPFA AA++ G+ Y+GGK+DD+
Sbjct: 284 SYLPHADHIENQDLERSAKRLATRAHKNALNKSYVSPFALAAKSAGF-HYTGGKMDDV 340
>UniRef100_Q6GR25 MGC81279 protein [Xenopus laevis]
Length = 297
Score = 96.3 bits (238), Expect = 5e-18
Identities = 74/242 (30%), Positives = 115/242 (46%), Gaps = 24/242 (9%)
Query: 839 KALTGREDAYFISHQNW---VAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENIST 895
K + +DA FI+ + VADGVG W G + + LM C+ +V + T
Sbjct: 47 KGMCYGDDACFIARHRTADVLGVADGVGGWRDYGVDPSQFSETLMRTCERLVKEGRFVPT 106
Query: 896 IKPAEVLTRSAAE---TQTP--GSSSVLVAYFD--GEALHAANVGNTGFIIIRDGFIFKK 948
P +LT S E + P GSS+ + D LH AN+G++GF+++R G + +
Sbjct: 107 -NPVGILTSSYRELLQNKVPLLGSSTACLVVLDRTSHRLHTANLGDSGFLVVRAGEVVHR 165
Query: 949 STAKFHEFNFPLHI----------VKGDDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYD 998
S + H FN P + V D P + + D+ GD+I+ AT+GLFDN+ D
Sbjct: 166 SDEQQHYFNTPFQLSIAPPEAEGAVLSDSP-DAADSNSFDVQLGDIILTATDGLFDNMPD 224
Query: 999 QEIASTISKSLQASL-GPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLD 1057
I + K + Q+ A +A++A ++ + SPFA A G + GGK D
Sbjct: 225 YMILQELKKLKNTNYESIQQTARSIAEQAHDLAYDPNYMSPFAQFACDYG-LNVRGGKPD 283
Query: 1058 DL 1059
D+
Sbjct: 284 DI 285
>UniRef100_Q6H7J3 5-azacytidine resistance protein-like [Oryza sativa]
Length = 315
Score = 96.3 bits (238), Expect = 5e-18
Identities = 64/223 (28%), Positives = 114/223 (50%), Gaps = 9/223 (4%)
Query: 845 EDAYFISHQ-NWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIV--SSHENISTIKPAEV 901
ED +F+ + VA+ADGVG + G ++ + R LM +V ++ I P +
Sbjct: 86 EDTHFVRPEAGVVALADGVGGYRAPGVDAAAFARALMYNAFEMVVATTPGGAGGICPYAL 145
Query: 902 LT---RSAAETQTPGSSSVLVAYFDGEALHAANVGNTGFIIIRDGFIFKKSTAKFHEFNF 958
L A +T G+S+ ++ G L A +G++ F + RDG +F +S A+ H FN+
Sbjct: 146 LGWAYEQAVSARTQGASTAVILSLAGATLKYAYIGDSAFAVFRDGKLFFRSEAQVHSFNY 205
Query: 959 P--LHIVKGDDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYDQEIASTISKSLQASLGPQ 1016
P L + G+ + G V++ +GDV++ T+GLFDN+ +E+ ++ L P+
Sbjct: 206 PFQLSVKNGNSVTSAARG-GVEVKEGDVVVAGTDGLFDNVTSEELQRIVAMGRALGLSPK 264
Query: 1017 EIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
+ A+V+A A E + +PF+ ++ + GK DD+
Sbjct: 265 QTADVVAGFAYEASTTMGRDTPFSLESRKKQGTIFRRGKRDDI 307
>UniRef100_Q8NI37 T-cell activation protein phosphatase 2C [Homo sapiens]
Length = 304
Score = 95.5 bits (236), Expect = 8e-18
Identities = 72/236 (30%), Positives = 115/236 (48%), Gaps = 24/236 (10%)
Query: 845 EDAYFISHQ---NWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENISTIKPAEV 901
+DA F++ + + VADGVG W G + + LM C+ +V + + P +
Sbjct: 60 DDACFVARHRSADVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGRFVPS-NPIGI 118
Query: 902 LTRSAAE---TQTP--GSSSVLVAYFD--GEALHAANVGNTGFIIIRDGFIFKKSTAKFH 954
LT S E + P GSS+ + D LH AN+G++GF+++R G + +S + H
Sbjct: 119 LTTSYCELLQNKVPLLGSSTACIVVLDRTSHRLHTANLGDSGFLVVRGGEVVHRSDEQQH 178
Query: 955 EFNFPLH----------IVKGDDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYDQEIAST 1004
FN P +V D P + + D+ GD+I+ AT+GLFDN+ D I
Sbjct: 179 YFNTPFQLSIAPPEAEGVVLSDSP-DAADSTSFDVQLGDIILTATDGLFDNMPDYMILQE 237
Query: 1005 ISKSLQASL-GPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
+ K ++ Q+ A +A++A E+ + SPFA A G + GGK DD+
Sbjct: 238 LKKLKNSNYESIQQTARSIAEQAHELAYDPNYMSPFAQFACDNG-LNVRGGKPDDI 292
>UniRef100_UPI00001D0928 UPI00001D0928 UniRef100 entry
Length = 307
Score = 95.1 bits (235), Expect = 1e-17
Identities = 72/236 (30%), Positives = 115/236 (48%), Gaps = 24/236 (10%)
Query: 845 EDAYFISHQ---NWVAVADGVGQWSPEGNNSGPYIRELMEKCKNIVSSHENISTIKPAEV 901
+DA F++ + + VADGVG W G + + LM C+ +V + + P +
Sbjct: 63 DDACFVARHRSADVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGRFVPS-NPVGI 121
Query: 902 LTRSAAE---TQTP--GSSSVLVAYFDGEA--LHAANVGNTGFIIIRDGFIFKKSTAKFH 954
LT S E + P GSS+ + D + LH AN+G++GF+++R G + +S + H
Sbjct: 122 LTTSYCELLQNKVPLLGSSTACIVVLDRSSHRLHTANLGDSGFLVVRGGEVVHRSDEQQH 181
Query: 955 EFNFPLH----------IVKGDDPSEIIEGYKVDLCDGDVIIFATNGLFDNLYDQEIAST 1004
FN P +V D P + + D+ GD+I+ AT+GLFDN+ D I
Sbjct: 182 YFNTPFQLSIAPPEAEGVVLSDSP-DAADSTSFDVQLGDIILTATDGLFDNMPDYMILQE 240
Query: 1005 ISKSLQASL-GPQEIAEVLAKRAQEVGKSTSTRSPFADAAQAIGYVGYSGGKLDDL 1059
+ K ++ Q A +A++A E+ + SPFA A G + GGK DD+
Sbjct: 241 LKKLKNSNYESIQRTARSIAEQAHELAYDPNYMSPFAQFACDNG-LNVRGGKPDDI 295
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.307 0.127 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,885,167,551
Number of Sequences: 2790947
Number of extensions: 85279795
Number of successful extensions: 254365
Number of sequences better than 10.0: 2471
Number of HSP's better than 10.0 without gapping: 93
Number of HSP's successfully gapped in prelim test: 2499
Number of HSP's that attempted gapping in prelim test: 240676
Number of HSP's gapped (non-prelim): 7735
length of query: 1113
length of database: 848,049,833
effective HSP length: 138
effective length of query: 975
effective length of database: 462,899,147
effective search space: 451326668325
effective search space used: 451326668325
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0314.17


