

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.14
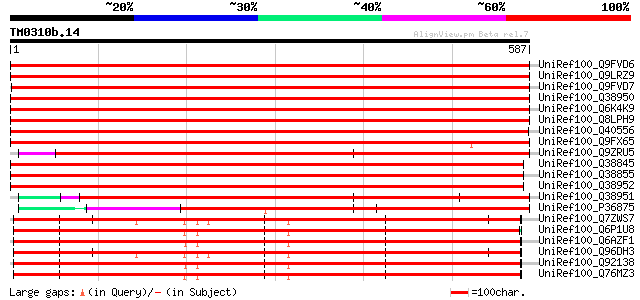
(587 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FVD6 Ser/Thr specific protein phosphatase 2A A regul... 1102 0.0
UniRef100_Q9LRZ9 Protein phosphotase 2a 65kd regulatory subunit ... 1049 0.0
UniRef100_Q9FVD7 Ser/Thr specific protein phosphatase 2A A regul... 1046 0.0
UniRef100_Q38950 Protein phosphatase 2A 65 kDa regulatory subuni... 1045 0.0
UniRef100_Q6K4K9 Phosphatase 2A regulatory A subunit [Oryza sativa] 1041 0.0
UniRef100_Q8LPH9 Protein phosphatase 2A regulatory subunit, puta... 1034 0.0
UniRef100_Q40556 Protein phosphatase 2A [Nicotiana tabacum] 1025 0.0
UniRef100_Q9FX65 Hypothetical protein T6J4.8 [Arabidopsis thaliana] 1019 0.0
UniRef100_Q9ZRU5 Protein phosphatase [Cicer arietinum] 1004 0.0
UniRef100_Q38845 Phosphoprotein phosphatase 2A, regulatory subun... 994 0.0
UniRef100_Q38855 Serine/threonine protein phosphatase type 2A re... 989 0.0
UniRef100_Q38952 65 kDa regulatory subunit of protein phosphatas... 983 0.0
UniRef100_Q38951 Protein phosphatase 2A 65 kDa regulatory subuni... 902 0.0
UniRef100_P36875 Protein phosphatase PP2A regulatory subunit A [... 720 0.0
UniRef100_Q7ZWS7 Ppp2r1a-prov protein [Xenopus laevis] 667 0.0
UniRef100_Q6P1U8 Hypothetical protein MGC76072 [Xenopus tropicalis] 666 0.0
UniRef100_Q6AZF1 Ppp2r1a-B-prov protein [Xenopus laevis] 664 0.0
UniRef100_Q96DH3 Alpha isoform of regulatory subunit A, protein ... 664 0.0
UniRef100_Q92138 Phosphorylase phosphatase [Xenopus laevis] 663 0.0
UniRef100_Q76MZ3 Serine/threonine protein phosphatase 2A, 65 kDa... 663 0.0
>UniRef100_Q9FVD6 Ser/Thr specific protein phosphatase 2A A regulatory subunit beta
isoform [Medicago varia]
Length = 587
Score = 1102 bits (2851), Expect = 0.0
Identities = 560/587 (95%), Positives = 579/587 (98%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD
Sbjct: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLA AEELGVF+PYVGGV+HA+VLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ
Sbjct: 61 DEVLLATAEELGVFVPYVGGVDHASVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVEHFIPLVKRLA+GEWFTARVSSCGLFHIAYPSAPEA K ELRA+YGQLCQDDMPMVR
Sbjct: 121 DLVEHFIPLVKRLASGEWFTARVSSCGLFHIAYPSAPEALKTELRAIYGQLCQDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGP+ T+TELVPAYVRLLRDNEAEVRIAA
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPDSTKTELVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKFSRILSPELAI+HILPCVKELS DSSQHVRSALASVIMGMAPVLGKDATIEQLL
Sbjct: 301 AGKVTKFSRILSPELAIQHILPCVKELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAA N+KRLAEEFGP+WAMQHIIPQV
Sbjct: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAATNIKRLAEEFGPEWAMQHIIPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
LDM+NDPHYLYRMTIL AISLLAPVLGSEIT++ LLPLVVNA+KDRVPNIKFNVAKVLQS
Sbjct: 481 LDMVNDPHYLYRMTILHAISLLAPVLGSEITTTNLLPLVVNAAKDRVPNIKFNVAKVLQS 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
LIPIV+ESVVE+T++PCLVELSEDPDVDVRFFASQALQSSDQVKMSS
Sbjct: 541 LIPIVDESVVESTIKPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
>UniRef100_Q9LRZ9 Protein phosphotase 2a 65kd regulatory subunit [Arabidopsis
thaliana]
Length = 587
Score = 1049 bits (2712), Expect = 0.0
Identities = 526/587 (89%), Positives = 570/587 (96%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+M+D+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD
Sbjct: 1 MSMIDEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIPYVGGVE+A+VLLPPLETL TVEETCVR+K+VESLCR+G+QMRE
Sbjct: 61 DEVLLAMAEELGVFIPYVGGVEYAHVLLPPLETLSTVEETCVREKAVESLCRVGSQMRES 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLV+HFI LVKRLAAGEWFTARVS+CG+FHIAYPSAP+ K ELR++Y QLCQDDMPMVR
Sbjct: 121 DLVDHFISLVKRLAAGEWFTARVSACGVFHIAYPSAPDMLKTELRSLYTQLCQDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AATNLGKFAATVE+AHLK+D+MS+F+DLTQDDQDSVRLLAVEGCAALGKLLEPQ+CV
Sbjct: 181 RAAATNLGKFAATVESAHLKTDVMSMFEDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVQ 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RIL+PE+AI+HILPCVKELS+DSSQHVRSALASVIMGMAPVLGKDATIE LL
Sbjct: 301 AGKVTKFCRILNPEIAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLGVGFFDDKLGALCMQWL+DKV+SIRDAAANN+KRLAEEFGP+WAMQHI+PQV
Sbjct: 421 IPLLASQLGVGFFDDKLGALCMQWLQDKVHSIRDAAANNLKRLAEEFGPEWAMQHIVPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L+M+N+PHYLYRMTIL+A+SLLAPV+GSEIT SKLLP+V+ ASKDRVPNIKFNVAKVLQS
Sbjct: 481 LEMVNNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPVVMTASKDRVPNIKFNVAKVLQS 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
LIPIV++SVVE T+RP LVELSEDPDVDVRFFA+QALQS D V MSS
Sbjct: 541 LIPIVDQSVVEKTIRPGLVELSEDPDVDVRFFANQALQSIDNVMMSS 587
>UniRef100_Q9FVD7 Ser/Thr specific protein phosphatase 2A A regulatory subunit alpha
isoform [Medicago varia]
Length = 585
Score = 1046 bits (2706), Expect = 0.0
Identities = 526/585 (89%), Positives = 563/585 (95%)
Query: 3 MVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDDE 62
M D+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFL+ENNDDDDE
Sbjct: 1 MADEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLTENNDDDDE 60
Query: 63 VLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDL 122
VLLAMAEELGVFIPYVGGVEHA+VLLPPLE C+VEETCVRDK+VESLCRIG+QMRE DL
Sbjct: 61 VLLAMAEELGVFIPYVGGVEHASVLLPPLEAFCSVEETCVRDKAVESLCRIGSQMRESDL 120
Query: 123 VEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRS 182
VE+FIPLVKRLAAGEWFTARVS+CGLFHIAYPSAPEA K ELR++Y QLCQDDMPMVRRS
Sbjct: 121 VEYFIPLVKRLAAGEWFTARVSACGLFHIAYPSAPEASKTELRSIYSQLCQDDMPMVRRS 180
Query: 183 AATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHI 242
AATNLGKFAATVE HLK+DIMS+FDDLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVAHI
Sbjct: 181 AATNLGKFAATVEYTHLKADIMSIFDDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVAHI 240
Query: 243 LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAG 302
LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTR ELVPAYVRLLRDNEAEVRIAAAG
Sbjct: 241 LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRAELVPAYVRLLRDNEAEVRIAAAG 300
Query: 303 KVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPI 362
KVTKF RILSP+LAI+HILPCVKELS+DSSQHVRSALASVIMGMAPVLGK+ATIEQLL I
Sbjct: 301 KVTKFCRILSPDLAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKEATIEQLLHI 360
Query: 363 FLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIP 422
FLSLLKDEFPDVR+NIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIP
Sbjct: 361 FLSLLKDEFPDVRINIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIP 420
Query: 423 LLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLD 482
LLASQLGVGFFDDKLGALCMQWL+DKV+SIR+AAANN+KRLAEEFGP+WAMQHIIPQVL+
Sbjct: 421 LLASQLGVGFFDDKLGALCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIIPQVLE 480
Query: 483 MINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLI 542
MI +PHYLYRMTIL+AISLLAPV+GSE+T SKLLP VV ASKDRVPNIKFNVAKVL+S+
Sbjct: 481 MITNPHYLYRMTILRAISLLAPVMGSEVTCSKLLPAVVAASKDRVPNIKFNVAKVLESIF 540
Query: 543 PIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
PIV++SVVE T+RPCLVEL EDPDVDVRFF++QALQ+ D V MSS
Sbjct: 541 PIVDQSVVEKTIRPCLVELGEDPDVDVRFFSNQALQAIDHVMMSS 585
>UniRef100_Q38950 Protein phosphatase 2A 65 kDa regulatory subunit [Arabidopsis
thaliana]
Length = 587
Score = 1045 bits (2703), Expect = 0.0
Identities = 525/587 (89%), Positives = 569/587 (96%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+M+D+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD
Sbjct: 1 MSMIDEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIPYVGGVE+A+VLLPPLETL TVEETCVR+K+VESLCR+G+QMRE
Sbjct: 61 DEVLLAMAEELGVFIPYVGGVEYAHVLLPPLETLSTVEETCVREKAVESLCRVGSQMRES 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLV+HFI LVKRLAAGEWFTARVS+CG+FHIAYPSAP+ K ELR++Y QLCQDDMPMVR
Sbjct: 121 DLVDHFISLVKRLAAGEWFTARVSACGVFHIAYPSAPDMLKTELRSLYTQLCQDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AATNLGKFAATVE+AHLK+D+MS+F+DLTQDDQDSVRLLAVEGCAALGKLLEPQ+CV
Sbjct: 181 RAAATNLGKFAATVESAHLKTDVMSMFEDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVQ 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RIL+PE+AI+ ILPCVKELS+DSSQHVRSALASVIMGMAPVLGKDATIE LL
Sbjct: 301 AGKVTKFCRILNPEIAIQDILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLGVGFFDDKLGALCMQWL+DKV+SIRDAAANN+KRLAEEFGP+WAMQHI+PQV
Sbjct: 421 IPLLASQLGVGFFDDKLGALCMQWLQDKVHSIRDAAANNLKRLAEEFGPEWAMQHIVPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L+M+N+PHYLYRMTIL+A+SLLAPV+GSEIT SKLLP+V+ ASKDRVPNIKFNVAKVLQS
Sbjct: 481 LEMVNNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPVVMTASKDRVPNIKFNVAKVLQS 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
LIPIV++SVVE T+RP LVELSEDPDVDVRFFA+QALQS D V MSS
Sbjct: 541 LIPIVDQSVVEKTIRPGLVELSEDPDVDVRFFANQALQSIDNVMMSS 587
>UniRef100_Q6K4K9 Phosphatase 2A regulatory A subunit [Oryza sativa]
Length = 587
Score = 1041 bits (2693), Expect = 0.0
Identities = 519/587 (88%), Positives = 569/587 (96%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
MA +D+PLYPIA+LIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDD+
Sbjct: 1 MASMDEPLYPIAILIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDE 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIPYVGGVEHA+VLLPPLETLCTVEETCVRDK+VESLCRIGAQM+E
Sbjct: 61 DEVLLAMAEELGVFIPYVGGVEHAHVLLPPLETLCTVEETCVRDKAVESLCRIGAQMKES 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
D+V+ F+P+VKRLAAGEWFTARVSSCGLFHIAYPSAP+ KAELR +YGQLCQDDMPMVR
Sbjct: 121 DIVDWFVPVVKRLAAGEWFTARVSSCGLFHIAYPSAPDQLKAELRTIYGQLCQDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AA+NLGKFAATVE +LK+++MS+FDDLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RAAASNLGKFAATVEQNYLKTEVMSIFDDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPE +R +LVPAYVRLLRDNEAEVRIAA
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEHSREQLVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RILSP+LAI+HILPCVKELS+DSSQHVRSALASVIMGMAPVLGK+ATIEQLL
Sbjct: 301 AGKVTKFCRILSPQLAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKEATIEQLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLGVGFFDDKLGALCMQWL+DKV+SIRDAAANN+KRLAEEFGP+WAMQHIIPQV
Sbjct: 421 IPLLASQLGVGFFDDKLGALCMQWLEDKVFSIRDAAANNLKRLAEEFGPEWAMQHIIPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L+ IN+PHYLYRMTILQAISLLAPV+G+EIT +LLP+V+N+SKDRVPNIKFNVAKVLQ+
Sbjct: 481 LEKINNPHYLYRMTILQAISLLAPVMGAEITCQQLLPVVINSSKDRVPNIKFNVAKVLQA 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
LIPI+++SVVE ++PCLVELSEDPDVDVR++A+QALQ+ DQ+ MSS
Sbjct: 541 LIPILDQSVVEKNVKPCLVELSEDPDVDVRYYANQALQACDQIMMSS 587
>UniRef100_Q8LPH9 Protein phosphatase 2A regulatory subunit, putative [Arabidopsis
thaliana]
Length = 587
Score = 1034 bits (2673), Expect = 0.0
Identities = 518/587 (88%), Positives = 564/587 (95%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+MVD+PLYPIAVLIDELKN+DIQ RLNSI+RLS IARALGEERTRKELIPFLSENNDDD
Sbjct: 1 MSMVDEPLYPIAVLIDELKNDDIQRRLNSIKRLSIIARALGEERTRKELIPFLSENNDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELG FI YVGGVE+A VLLPPLETL TVEETCVR+K+V+SLCRIGAQMRE
Sbjct: 61 DEVLLAMAEELGGFILYVGGVEYAYVLLPPLETLSTVEETCVREKAVDSLCRIGAQMRES 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVEHF PL KRL+AGEWFTARVS+CG+FHIAYPSAP+ K ELR++YGQLCQDDMPMVR
Sbjct: 121 DLVEHFTPLAKRLSAGEWFTARVSACGIFHIAYPSAPDVLKTELRSIYGQLCQDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AATNLGKFAAT+E+AHLK+DIMS+F+DLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RAAATNLGKFAATIESAHLKTDIMSMFEDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRT+LVPAY RLL DNEAEVRIAA
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTDLVPAYARLLCDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RIL+PELAI+HILPCVKELS+DSSQHVRSALASVIMGMAPVLGKDATIE LL
Sbjct: 301 AGKVTKFCRILNPELAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLGVGFFD+KLGALCMQWL+DKV+SIR+AAANN+KRLAEEFGP+WAMQHI+PQV
Sbjct: 421 IPLLASQLGVGFFDEKLGALCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIVPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L+MIN+PHYLYRMTIL+A+SLLAPV+GSEIT SKLLP V+ ASKDRVPNIKFNVAK++QS
Sbjct: 481 LEMINNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPAVITASKDRVPNIKFNVAKMMQS 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
LIPIV+++VVEN +RPCLVELSEDPDVDVR+FA+QALQS D V MSS
Sbjct: 541 LIPIVDQAVVENMIRPCLVELSEDPDVDVRYFANQALQSIDNVMMSS 587
>UniRef100_Q40556 Protein phosphatase 2A [Nicotiana tabacum]
Length = 586
Score = 1025 bits (2649), Expect = 0.0
Identities = 521/586 (88%), Positives = 554/586 (93%), Gaps = 1/586 (0%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M MVD PLYPI VLID+LK ++ QLRLNSIRRLSTIARALGEE+TRKELIPFLSENNDDD
Sbjct: 1 MPMVDDPLYPIPVLIDDLKTDEKQLRLNSIRRLSTIARALGEEKTRKELIPFLSENNDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
EVLLAMAEELGVFIPYVGGVEHA+VLLPPLETLCTVEETCVRDK VESLCRIG+QMRE
Sbjct: 61 -EVLLAMAEELGVFIPYVGGVEHAHVLLPPLETLCTVEETCVRDKPVESLCRIGSQMRES 119
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
LV+ F+PLVKRLAAGEWFTARVS+CGLFHIAY SAPE KAELR++Y QLCQDDMPMVR
Sbjct: 120 GLVDWFVPLVKRLAAGEWFTARVSACGLFHIAYSSAPEMLKAELRSIYSQLCQDDMPMVR 179
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
RSA TNLGKFAATVE+ + KSDI S+FDDLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 180 RSATTNLGKFAATVESTYFKSDITSIFDDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVA 239
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRT+LVPAYVRLLRDNEAEVRIAA
Sbjct: 240 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTDLVPAYVRLLRDNEAEVRIAA 299
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RILSPELAI+HILPCVKELS+DSSQ+VRSALASVIMGMAPVLGKDATIE LL
Sbjct: 300 AGKVTKFCRILSPELAIQHILPCVKELSSDSSQYVRSALASVIMGMAPVLGKDATIEHLL 359
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 360 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 419
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLG+GFFDDKLGALCMQWL+DKVYSIRDAAANN+KRLAEEFGP+WAMQHIIPQV
Sbjct: 420 IPLLASQLGIGFFDDKLGALCMQWLQDKVYSIRDAAANNLKRLAEEFGPEWAMQHIIPQV 479
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
LDM PHYLYRMTIL+AISLLAPV+GSEIT SKLLP+V+ A+KDRVPNIKFNVAKVLQS
Sbjct: 480 LDMTTSPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVITATKDRVPNIKFNVAKVLQS 539
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMS 586
LIPIV+ SVVE T+RP LVEL+EDPDVDVRF+A+QALQS D MS
Sbjct: 540 LIPIVDHSVVEKTIRPSLVELAEDPDVDVRFYANQALQSIDNAMMS 585
>UniRef100_Q9FX65 Hypothetical protein T6J4.8 [Arabidopsis thaliana]
Length = 591
Score = 1019 bits (2635), Expect = 0.0
Identities = 514/591 (86%), Positives = 561/591 (93%), Gaps = 4/591 (0%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+MVD+PLYPIAVLIDELKN+DIQ RLNSI+RLS IARALGEERTRKELIPFLSENNDDD
Sbjct: 1 MSMVDEPLYPIAVLIDELKNDDIQRRLNSIKRLSIIARALGEERTRKELIPFLSENNDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELG FI YVGGVE+A VLLPPLETL TVEETCVR+K+V+SLCRIGAQMRE
Sbjct: 61 DEVLLAMAEELGGFILYVGGVEYAYVLLPPLETLSTVEETCVREKAVDSLCRIGAQMRES 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVEHF PL KRL+AGEWFTARVS+CG+FHIAYPSAP+ K ELR++YGQLCQDDMPMVR
Sbjct: 121 DLVEHFTPLAKRLSAGEWFTARVSACGIFHIAYPSAPDVLKTELRSIYGQLCQDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AATNLGKFAAT+E+AHLK+DIMS+F+DLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RAAATNLGKFAATIESAHLKTDIMSMFEDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRT+LVPAY RLL DNEAEVRIAA
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTDLVPAYARLLCDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RIL+PELAI+HILPCVKELS+DSSQHVRSALASVIMGMAPVLGKDATIE LL
Sbjct: 301 AGKVTKFCRILNPELAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLGVGFFD+KLGALCMQWL+DKV+SIR+AAANN+KRLAEEFGP+WAMQHI+PQV
Sbjct: 421 IPLLASQLGVGFFDEKLGALCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIVPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVN----ASKDRVPNIKFNVAK 536
L+MIN+PHYLYRMTIL+A+SLLAPV+GSEIT SKLLP + +S RVPNIKFNVAK
Sbjct: 481 LEMINNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPALTKRTTLSSSFRVPNIKFNVAK 540
Query: 537 VLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
++QSLIPIV+++VVEN +RPCLVELSEDPDVDVR+FA+QALQS D V MSS
Sbjct: 541 MMQSLIPIVDQAVVENMIRPCLVELSEDPDVDVRYFANQALQSIDNVMMSS 591
>UniRef100_Q9ZRU5 Protein phosphatase [Cicer arietinum]
Length = 538
Score = 1004 bits (2595), Expect = 0.0
Identities = 510/536 (95%), Positives = 526/536 (97%)
Query: 52 FLSENNDDDDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLC 111
FLSENNDDDDEVLLA AEELGVF+PYVGGVE+A+VLLPPLETLCTVEETCVRDKSVESLC
Sbjct: 3 FLSENNDDDDEVLLATAEELGVFVPYVGGVEYASVLLPPLETLCTVEETCVRDKSVESLC 62
Query: 112 RIGAQMREQDLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQL 171
RIGAQMREQDLVEHFIPLVKRLA+GEWFTARVSSCGLFHIAYPSA E K ELRA+YGQL
Sbjct: 63 RIGAQMREQDLVEHFIPLVKRLASGEWFTARVSSCGLFHIAYPSASEGLKTELRAIYGQL 122
Query: 172 CQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGK 231
CQDDMPMVRRSAATNLGKFA+TVEAAHLK+DIMSVFDDLTQDDQDSVRLLAVEGCAALGK
Sbjct: 123 CQDDMPMVRRSAATNLGKFASTVEAAHLKTDIMSVFDDLTQDDQDSVRLLAVEGCAALGK 182
Query: 232 LLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRD 291
LLEPQ+CVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGP+ TR ELVPAYVRLLRD
Sbjct: 183 LLEPQDCVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPDSTRAELVPAYVRLLRD 242
Query: 292 NEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLG 351
NEAEVRIAAAGKVTKFSRIL+PELAI+HILPCVKELS DSSQHVRSALASVIMGMAPVLG
Sbjct: 243 NEAEVRIAAAGKVTKFSRILNPELAIQHILPCVKELSTDSSQHVRSALASVIMGMAPVLG 302
Query: 352 KDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHW 411
KDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHW
Sbjct: 303 KDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHW 362
Query: 412 RVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDW 471
RVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGP+W
Sbjct: 363 RVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPEW 422
Query: 472 AMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIK 531
AMQHIIPQVLDMINDPHYLYRMTIL AISLLAPVLGSEITSS LLPLVVNA+KDRVPNIK
Sbjct: 423 AMQHIIPQVLDMINDPHYLYRMTILHAISLLAPVLGSEITSSNLLPLVVNAAKDRVPNIK 482
Query: 532 FNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
FNVAKVLQSLIPIV+ESVVE+T+RPCLVELSEDPDVDVRFFASQALQSSDQVKMSS
Sbjct: 483 FNVAKVLQSLIPIVDESVVESTIRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 538
Score = 78.6 bits (192), Expect = 5e-13
Identities = 74/380 (19%), Positives = 154/380 (40%), Gaps = 41/380 (10%)
Query: 11 IAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIP-FLSENNDDDDEVLLAMAE 69
+ V+++ +++ ++R +L + A+G + TR EL+P ++ D++ EV +A A
Sbjct: 194 LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPDSTRAELVPAYVRLLRDNEAEVRIAAAG 253
Query: 70 ELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVEHFIPL 129
++ F + +L P L ++H +P
Sbjct: 254 KVTKF---------SRILNPEL------------------------------AIQHILPC 274
Query: 130 VKRLAAGEWFTARVSSCG-LFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLG 188
VK L+ R + + +A +A +L ++ L +D+ P VR + + L
Sbjct: 275 VKELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLD 334
Query: 189 KFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVN 248
+ + L ++ +L +D VRL +E L L + + +
Sbjct: 335 QVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQ 394
Query: 249 FSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFS 308
+ +DK + +R AN + L E GPE ++P + ++ D R+ ++ +
Sbjct: 395 WLKDKVYSIRDAAANNVKRLAEEFGPEWAMQHIIPQVLDMINDPHYLYRMTILHAISLLA 454
Query: 309 RILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLK 368
+L E+ ++LP V + D +++ +A V+ + P++ + + P + L +
Sbjct: 455 PVLGSEITSSNLLPLVVNAAKDRVPNIKFNVAKVLQSLIPIVDESVVESTIRPCLVELSE 514
Query: 369 DEFPDVRLNIISKLDQVNQV 388
D DVR L +QV
Sbjct: 515 DPDVDVRFFASQALQSSDQV 534
>UniRef100_Q38845 Phosphoprotein phosphatase 2A, regulatory subunit A [Arabidopsis
thaliana]
Length = 588
Score = 994 bits (2571), Expect = 0.0
Identities = 492/581 (84%), Positives = 551/581 (94%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
MAMVD+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFLSEN+DDD
Sbjct: 1 MAMVDEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENSDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIP+VGG+E A+VLLPPLE+LCTVEETCVR+K+VESLC+IG+QM+E
Sbjct: 61 DEVLLAMAEELGVFIPFVGGIEFAHVLLPPLESLCTVEETCVREKAVESLCKIGSQMKEN 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVE F+PLVKRLA GEWF ARVS+CG+FH+AY + K ELRA Y QLC+DDMPMVR
Sbjct: 121 DLVESFVPLVKRLAGGEWFAARVSACGIFHVAYQGCTDVLKTELRATYSQLCKDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AA+NLGKFA TVE+ L ++IM++FDDLT+DDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RAAASNLGKFATTVESTFLIAEIMTMFDDLTKDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
ILPVIVNFSQDKSWRVRYMVANQLYELCEAVGP+ TRT+LVPAYVRLLRDNEAEVRIAA
Sbjct: 241 RILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPDCTRTDLVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF R+L+PELAI+HILPCVKELS+DSSQHVRSALASVIMGMAP+LGKD+TIE LL
Sbjct: 301 AGKVTKFCRLLNPELAIQHILPCVKELSSDSSQHVRSALASVIMGMAPILGKDSTIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
+PLLASQLG+GFFDDKLGALCMQWL+DKVYSIR+AAANN+KRLAEEFGP+WAMQH++PQV
Sbjct: 421 VPLLASQLGIGFFDDKLGALCMQWLQDKVYSIREAAANNLKRLAEEFGPEWAMQHLVPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
LDM+N+PHYL+RM +L+AISL+APV+GSEIT SK LP+VV ASKDRVPNIKFNVAK+LQS
Sbjct: 481 LDMVNNPHYLHRMMVLRAISLMAPVMGSEITCSKFLPVVVEASKDRVPNIKFNVAKLLQS 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSD 581
LIPIV++SVV+ T+R CLV+LSEDPDVDVR+FA+QAL S D
Sbjct: 541 LIPIVDQSVVDKTIRQCLVDLSEDPDVDVRYFANQALNSID 581
>UniRef100_Q38855 Serine/threonine protein phosphatase type 2A regulatory subunit A
[Arabidopsis thaliana]
Length = 590
Score = 989 bits (2558), Expect = 0.0
Identities = 492/583 (84%), Positives = 551/583 (94%), Gaps = 2/583 (0%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
MAMVD+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFLSEN+DDD
Sbjct: 1 MAMVDEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENSDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIP+VGG+E A+VLLPPLE+LCTVEETCVR+K+VESLC+IG+QM+E
Sbjct: 61 DEVLLAMAEELGVFIPFVGGIEFAHVLLPPLESLCTVEETCVREKAVESLCKIGSQMKEN 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVE F+PLVKRLA GEWF ARVS+CG+FH+AY + K ELRA Y QLC+DDMPMVR
Sbjct: 121 DLVESFVPLVKRLAGGEWFAARVSACGIFHVAYQGCTDVLKTELRATYSQLCKDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AA+NLGKFA TVE+ L ++IM++FDDLT+DDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RAAASNLGKFATTVESTFLIAEIMTMFDDLTKDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
ILPVIVNFSQDKSWRVRYMVANQLYELCEAVGP+ TRT+LVPAYVRLLRDNEAEVRIAA
Sbjct: 241 RILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPDCTRTDLVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF R+L+PELAI+HILPCVKELS+DSSQHVRSALASVIMGMAP+LGKD+TIE LL
Sbjct: 301 AGKVTKFCRLLNPELAIQHILPCVKELSSDSSQHVRSALASVIMGMAPILGKDSTIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRD--AAANNVKRLAEEFGPDWAMQHIIP 478
+PLLASQLG+GFFDDKLGALCMQWL+DKVYSIR+ AAANN+KRLAEEFGP+WAMQH++P
Sbjct: 421 VPLLASQLGIGFFDDKLGALCMQWLQDKVYSIREAAAAANNLKRLAEEFGPEWAMQHLVP 480
Query: 479 QVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVL 538
QVLDM+N+PHYL+RM +L+AISL+APV+GSEIT SK LP+VV ASKDRVPNIKFNVAK+L
Sbjct: 481 QVLDMVNNPHYLHRMMVLRAISLMAPVMGSEITCSKFLPVVVEASKDRVPNIKFNVAKLL 540
Query: 539 QSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSD 581
QSLIPIV++SVV+ T+R CLV+LSEDPDVDVR+FA+QAL S D
Sbjct: 541 QSLIPIVDQSVVDKTIRQCLVDLSEDPDVDVRYFANQALNSID 583
>UniRef100_Q38952 65 kDa regulatory subunit of protein phosphatase 2A [Arabidopsis
thaliana]
Length = 590
Score = 983 bits (2542), Expect = 0.0
Identities = 490/583 (84%), Positives = 549/583 (94%), Gaps = 2/583 (0%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
MAMVD+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFLSEN+DDD
Sbjct: 1 MAMVDEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENSDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIP+VGG+E A+VLLPPLE+LCTVEETCVR K+VESLC+IG+QM+E
Sbjct: 61 DEVLLAMAEELGVFIPFVGGIEFAHVLLPPLESLCTVEETCVRGKAVESLCKIGSQMKEN 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVE F+PLVKRLA GEWF ARVS+CG+FH+AY + K ELRA Y QLC+DDMPMVR
Sbjct: 121 DLVESFVPLVKRLAGGEWFAARVSACGIFHVAYQGCTDVLKTELRATYSQLCKDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AA+NLGKFA TVE+ L ++IM++FDDLT+DDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RAAASNLGKFATTVESTFLIAEIMTMFDDLTKDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
ILPVIVNFSQDKSWRVRYMVANQLYELC+AVGP+ TRT+LVPAYVRLLRDNEAEVRIAA
Sbjct: 241 RILPVIVNFSQDKSWRVRYMVANQLYELCKAVGPDCTRTDLVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF R+L+PELAI+HILPCVKELS+DSSQHVRSALASVIMGMAP+LGKD+TIE LL
Sbjct: 301 AGKVTKFCRLLNPELAIQHILPCVKELSSDSSQHVRSALASVIMGMAPILGKDSTIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRD--AAANNVKRLAEEFGPDWAMQHIIP 478
+PLLASQLG+GFFD KLGALCMQWL+DKVYSIR+ AAANN+KRLAEEFGP+WAMQH++P
Sbjct: 421 VPLLASQLGIGFFDYKLGALCMQWLQDKVYSIREAAAAANNLKRLAEEFGPEWAMQHLVP 480
Query: 479 QVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVL 538
QVLDM+N+PHYL+RM +L+AISL+APV+GSEIT SK LP+VV ASKDRVPNIKFNVAK+L
Sbjct: 481 QVLDMVNNPHYLHRMMVLRAISLMAPVMGSEITCSKFLPVVVEASKDRVPNIKFNVAKLL 540
Query: 539 QSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSD 581
QSLIPIV++SVV+ T+R CLV+LSEDPDVDVR+FA+QAL S D
Sbjct: 541 QSLIPIVDQSVVDKTIRQCLVDLSEDPDVDVRYFANQALNSID 583
>UniRef100_Q38951 Protein phosphatase 2A 65 kDa regulatory subunit [Arabidopsis
thaliana]
Length = 508
Score = 902 bits (2332), Expect = 0.0
Identities = 448/508 (88%), Positives = 490/508 (96%)
Query: 80 GVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVEHFIPLVKRLAAGEWF 139
GVE+A VLLPPLETL TVEETCVR+K+V+SLCRIGAQMRE DLVEHF PL KRL+AGEWF
Sbjct: 1 GVEYAYVLLPPLETLSTVEETCVREKAVDSLCRIGAQMRESDLVEHFTPLAKRLSAGEWF 60
Query: 140 TARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLGKFAATVEAAHL 199
TARVS+CG+FHIAYPSAP+ K ELR++YGQLCQDDMPMVRR+AATNLGKFAAT+E+AHL
Sbjct: 61 TARVSACGIFHIAYPSAPDVLKTELRSIYGQLCQDDMPMVRRAAATNLGKFAATIESAHL 120
Query: 200 KSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRY 259
K+DIMS+F+DLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVAHILPVIVNFSQDKSWRVRY
Sbjct: 121 KTDIMSMFEDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVAHILPVIVNFSQDKSWRVRY 180
Query: 260 MVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAIEH 319
MVANQLYELCEAVGPEPTRT+LVPAY RLLRDNEAEVRIAAAGKVTKF RIL+PELAI+H
Sbjct: 181 MVANQLYELCEAVGPEPTRTDLVPAYARLLRDNEAEVRIAAAGKVTKFCRILNPELAIQH 240
Query: 320 ILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNII 379
ILPCVKELS+DSSQHVRSALASVIMGMAPVLGKDATIE LLPIFLSLLKDEFPDVRLNII
Sbjct: 241 ILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLNII 300
Query: 380 SKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGA 439
SKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFD+KLGA
Sbjct: 301 SKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDEKLGA 360
Query: 440 LCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAI 499
LCMQWL+DKV+SIR+AAANN+KRLAEEFGP+WAMQHI+PQVL+MIN+PHYLYRMTIL+A+
Sbjct: 361 LCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIVPQVLEMINNPHYLYRMTILRAV 420
Query: 500 SLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVENTLRPCLV 559
SLLAPV+GSEIT SKLLP V+ ASKDRVPNIKFNVAK++QSLIPIV+++VVEN +RPCLV
Sbjct: 421 SLLAPVMGSEITCSKLLPAVITASKDRVPNIKFNVAKMMQSLIPIVDQAVVENMIRPCLV 480
Query: 560 ELSEDPDVDVRFFASQALQSSDQVKMSS 587
ELSEDPDVDVR+FA+QALQS D V MSS
Sbjct: 481 ELSEDPDVDVRYFANQALQSIDNVMMSS 508
Score = 126 bits (317), Expect = 2e-27
Identities = 103/452 (22%), Positives = 192/452 (41%), Gaps = 40/452 (8%)
Query: 58 DDDDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQM 117
DD V A A LG F + ++ E L ++ VR +VE +G +
Sbjct: 95 DDMPMVRRAAATNLGKFAATIESAHLKTDIMSMFEDLTQDDQDSVRLLAVEGCAALGKLL 154
Query: 118 REQDLVEHFIPLVKRLAAGE-WFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDM 176
QD V H +P++ + + W + + L+ + PE + +L Y +L +D+
Sbjct: 155 EPQDCVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTDLVPAYARLLRDNE 214
Query: 177 PMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQ 236
VR +AA + KF ++L P+
Sbjct: 215 AEVRIAAAGKVTKFC---------------------------------------RILNPE 235
Query: 237 ECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEV 296
+ HILP + S D S VR +A+ + + +G + T L+P ++ LL+D +V
Sbjct: 236 LAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDV 295
Query: 297 RIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATI 356
R+ K+ + ++++ +L + +LP + EL+ D VR A+ I +A LG
Sbjct: 296 RLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFD 355
Query: 357 EQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLA 416
E+L + + L+D+ +R + L ++ + G + Q ++P ++E+ + H+ R+
Sbjct: 356 EKLGALCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIVPQVLEMINNPHYLYRMT 415
Query: 417 IIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHI 476
I+ + LLA +G KL + KD+V +I+ A ++ L I
Sbjct: 416 ILRAVSLLAPVMGSEITCSKLLPAVITASKDRVPNIKFNVAKMMQSLIPIVDQAVVENMI 475
Query: 477 IPQVLDMINDPHYLYRMTILQAISLLAPVLGS 508
P ++++ DP R QA+ + V+ S
Sbjct: 476 RPCLVELSEDPDVDVRYFANQALQSIDNVMMS 507
Score = 76.6 bits (187), Expect = 2e-12
Identities = 75/381 (19%), Positives = 151/381 (38%), Gaps = 41/381 (10%)
Query: 11 IAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIP-FLSENNDDDDEVLLAMAE 69
+ V+++ +++ ++R +L + A+G E TR +L+P + D++ EV +A A
Sbjct: 164 LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTDLVPAYARLLRDNEAEVRIAAAG 223
Query: 70 ELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVEHFIPL 129
+ V CRI + + ++H +P
Sbjct: 224 K------------------------------------VTKFCRI---LNPELAIQHILPC 244
Query: 130 VKRLAAGEWFTARVSSCG-LFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLG 188
VK L++ R + + +A +A L ++ L +D+ P VR + + L
Sbjct: 245 VKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLNIISKLD 304
Query: 189 KFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVN 248
+ + L ++ +L +D VRL +E L L + + +
Sbjct: 305 QVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDEKLGALCMQ 364
Query: 249 FSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFS 308
+ QDK +R AN L L E GPE +VP + ++ + R+ V+ +
Sbjct: 365 WLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIVPQVLEMINNPHYLYRMTILRAVSLLA 424
Query: 309 RILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLK 368
++ E+ +LP V S D +++ +A ++ + P++ + + P + L +
Sbjct: 425 PVMGSEITCSKLLPAVITASKDRVPNIKFNVAKMMQSLIPIVDQAVVENMIRPCLVELSE 484
Query: 369 DEFPDVRLNIISKLDQVNQVI 389
D DVR L ++ V+
Sbjct: 485 DPDVDVRYFANQALQSIDNVM 505
>UniRef100_P36875 Protein phosphatase PP2A regulatory subunit A [Pisum sativum]
Length = 395
Score = 720 bits (1859), Expect = 0.0
Identities = 372/395 (94%), Positives = 385/395 (97%), Gaps = 1/395 (0%)
Query: 194 VEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQDK 253
VEAAHLK+DIMSVFDDLTQDDQDS R LAVEGCAALGKLLEPQ+C+AHILPVIVNFSQDK
Sbjct: 1 VEAAHLKTDIMSVFDDLTQDDQDSFRFLAVEGCAALGKLLEPQDCLAHILPVIVNFSQDK 60
Query: 254 SWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSP 313
SWRVRYMVANQLYELCEAVGP+ T+TELVPAYVRLLRDN AEVRIAAAGKV+KFSRILSP
Sbjct: 61 SWRVRYMVANQLYELCEAVGPDSTKTELVPAYVRLLRDNVAEVRIAAAGKVSKFSRILSP 120
Query: 314 ELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPD 373
ELAI+HILPCVKELS DSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPD
Sbjct: 121 ELAIQHILPCVKELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPD 180
Query: 374 VRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFF 433
VRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFF
Sbjct: 181 VRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFF 240
Query: 434 DDKLGALCMQWLKDKVYSIRDAAANNVKRL-AEEFGPDWAMQHIIPQVLDMINDPHYLYR 492
DDKLGAL MQWLKDK YSIR+AAANNVKRL AEEFGP+WAMQHIIPQVLDMINDPHYLYR
Sbjct: 241 DDKLGALIMQWLKDKEYSIRNAAANNVKRLAAEEFGPEWAMQHIIPQVLDMINDPHYLYR 300
Query: 493 MTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVEN 552
MTIL AISLLAPVLGSEITS+ LLPLVVNASKDRVPNIKFNVAKVLQSLIPIV+ESVVE+
Sbjct: 301 MTILHAISLLAPVLGSEITSTNLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVDESVVES 360
Query: 553 TLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
T+RPCLVELSEDPDVDVRFFASQALQSSDQVKMSS
Sbjct: 361 TIRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 395
Score = 112 bits (281), Expect = 2e-23
Identities = 82/369 (22%), Positives = 159/369 (42%), Gaps = 41/369 (11%)
Query: 87 LLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVEHFIPLVKRLAAGE-WFTARVSS 145
++ + L ++ R +VE +G + QD + H +P++ + + W + +
Sbjct: 10 IMSVFDDLTQDDQDSFRFLAVEGCAALGKLLEPQDCLAHILPVIVNFSQDKSWRVRYMVA 69
Query: 146 CGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMS 205
L+ + P++ K EL Y +L +D++ VR +AA + KF+ + I+
Sbjct: 70 NQLYELCEAVGPDSTKTELVPAYVRLLRDNVAEVRIAAAGKVSKFSRILSPELAIQHILP 129
Query: 206 VFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQL 265
+L+ D VR + +L + +LP+ ++ +D+ VR + ++L
Sbjct: 130 CVKELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKL 189
Query: 266 YELCEAVGPEPTRTELVPAYVRL------------------------------------- 288
++ + +G + L+PA V L
Sbjct: 190 DQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALIM 249
Query: 289 --LRDNEAEVRIAAAGKVTKF-SRILSPELAIEHILPCVKELSADSSQHVRSALASVIMG 345
L+D E +R AAA V + + PE A++HI+P V ++ D R + I
Sbjct: 250 QWLKDKEYSIRNAAANNVKRLAAEEFGPEWAMQHIIPQVLDMINDPHYLYRMTILHAISL 309
Query: 346 MAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVEL 405
+APVLG + T LLP+ ++ KD P+++ N+ L + ++ ++ ++ P +VEL
Sbjct: 310 LAPVLGSEITSTNLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVDESVVESTIRPCLVEL 369
Query: 406 AEDRHWRVR 414
+ED VR
Sbjct: 370 SEDPDVDVR 378
Score = 75.9 bits (185), Expect = 3e-12
Identities = 75/381 (19%), Positives = 154/381 (39%), Gaps = 42/381 (11%)
Query: 11 IAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIP-FLSENNDDDDEVLLAMAE 69
+ V+++ +++ ++R +L + A+G + T+ EL+P ++ D+ EV +A A
Sbjct: 50 LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPDSTKTELVPAYVRLLRDNVAEVRIAAAG 109
Query: 70 ELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVEHFIPL 129
++ F + +L P L ++H +P
Sbjct: 110 KVSKF---------SRILSPEL------------------------------AIQHILPC 130
Query: 130 VKRLAAGEWFTARVSSCG-LFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLG 188
VK L+ R + + +A +A +L ++ L +D+ P VR + + L
Sbjct: 131 VKELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLD 190
Query: 189 KFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVN 248
+ + L ++ +L +D VRL +E L L + +I+
Sbjct: 191 QVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALIMQ 250
Query: 249 FSQDKSWRVRYMVANQLYEL-CEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKF 307
+ +DK + +R AN + L E GPE ++P + ++ D R+ ++
Sbjct: 251 WLKDKEYSIRNAAANNVKRLAAEEFGPEWAMQHIIPQVLDMINDPHYLYRMTILHAISLL 310
Query: 308 SRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLL 367
+ +L E+ ++LP V S D +++ +A V+ + P++ + + P + L
Sbjct: 311 APVLGSEITSTNLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVDESVVESTIRPCLVELS 370
Query: 368 KDEFPDVRLNIISKLDQVNQV 388
+D DVR L +QV
Sbjct: 371 EDPDVDVRFFASQALQSSDQV 391
>UniRef100_Q7ZWS7 Ppp2r1a-prov protein [Xenopus laevis]
Length = 589
Score = 667 bits (1720), Expect = 0.0
Identities = 341/577 (59%), Positives = 441/577 (76%), Gaps = 4/577 (0%)
Query: 5 DQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDDEVL 64
D LYPIAVLIDEL+NED+QLRLNSI++LSTIA ALG ERTR EL+PFL++ D+DEVL
Sbjct: 7 DDSLYPIAVLIDELRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDEVL 66
Query: 65 LAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVE 124
LA+AE+LG F VGG E A+ LLPPLE+L TVEET VRD++VESL I + DL
Sbjct: 67 LALAEQLGTFTTLVGGPEFAHCLLPPLESLATVEETVVRDRAVESLRAISHEHSPSDLEA 126
Query: 125 HFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAA 184
HF+PLVKRLA+G+WFT+R S+CGLF + YP KAELR + LC DD PMVRR+AA
Sbjct: 127 HFVPLVKRLASGDWFTSRTSACGLFSVCYPRVSSTVKAELRQHFRNLCSDDTPMVRRAAA 186
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILP 244
+ LG+FA +E ++KS+I+ +F +L D+QDSVRLLAVE C + +LL +E ++P
Sbjct: 187 SKLGEFAKVLELENVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEELEPLVMP 246
Query: 245 VIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKV 304
+ ++DKSWRVRYMVA++ EL AVGPE T+T+LVPA+ L++D EAEVR A++ KV
Sbjct: 247 TLRQAAEDKSWRVRYMVADKFTELQNAVGPEITKTDLVPAFQNLMKDCEAEVRAASSHKV 306
Query: 305 TKFSRILSPE----LAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
+F LS + + + ILPCVKEL +D++QHV+SALASVIMG++P+LGKD TIE LL
Sbjct: 307 KEFCENLSADCRENVIMTQILPCVKELVSDANQHVKSALASVIMGLSPILGKDNTIEHLL 366
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
P+FL+ LKDE P+VRLNIIS LD VN+VIGI LSQSLLPAIVELAED WRVRLAIIEY
Sbjct: 367 PLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAEDAKWRVRLAIIEY 426
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
+PLLA QLGV FFD+KL +LCM WL D VY+IR+AA +N+K+L E+FG DWA IIP+V
Sbjct: 427 MPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFGKDWAQATIIPKV 486
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L M NDP+YL+RMT L I++L+ V G +IT+ +LP VV ++D V N++FNVAK LQ
Sbjct: 487 LAMSNDPNYLHRMTTLFCINVLSEVCGQDITTKHMLPTVVRMAEDAVANVRFNVAKSLQK 546
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQAL 577
+ P ++ S ++N ++P L +L++D DVDV++FA +AL
Sbjct: 547 IGPTLDNSTLQNEVKPVLEKLTQDQDVDVKYFAQEAL 583
Score = 90.1 bits (222), Expect = 2e-16
Identities = 89/455 (19%), Positives = 182/455 (39%), Gaps = 48/455 (10%)
Query: 94 LCTVEETCVRDKSVESLCRIGAQMREQDLVEHFIPLVKRLAAGEWFTAR---VSSCGLFH 150
LC+ + VR + L + +++ IP+ LA+ E + R V +C +
Sbjct: 173 LCSDDTPMVRRAAASKLGEFAKVLELENVKSEIIPMFSNLASDEQDSVRLLAVEAC--VN 230
Query: 151 IAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDL 210
IA E + + Q +D VR A + V K+D++ F +L
Sbjct: 231 IAQLLPQEELEPLVMPTLRQAAEDKSWRVRYMVADKFTELQNAVGPEITKTDLVPAFQNL 290
Query: 211 TQDDQDSVRLLAV----EGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLY 266
+D + VR + E C L + ILP + D + V+ +A+ +
Sbjct: 291 MKDCEAEVRAASSHKVKEFCENLSADCRENVIMTQILPCVKELVSDANQHVKSALASVIM 350
Query: 267 ELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKE 326
L +G + T L+P ++ L+D EVR+ + + ++ + +LP + E
Sbjct: 351 GLSPILGKDNTIEHLLPLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVE 410
Query: 327 LSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVN 386
L+ D+ VR A+ + +A LG + E+L + ++ L D +R S L ++
Sbjct: 411 LAEDAKWRVRLAIIEYMPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLV 470
Query: 387 QVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLK 446
+ G D +++P ++ ++ D ++ R+ + C
Sbjct: 471 EKFGKDWAQATIIPKVLAMSNDPNYLHRMTTL---------------------FC----- 504
Query: 447 DKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVL 506
+ L+E G D +H++P V+ M D R + +++ + P L
Sbjct: 505 -------------INVLSEVCGQDITTKHMLPTVVRMAEDAVANVRFNVAKSLQKIGPTL 551
Query: 507 GSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSL 541
+ +++ P++ ++D+ ++K+ + L L
Sbjct: 552 DNSTLQNEVKPVLEKLTQDQDVDVKYFAQEALTVL 586
Score = 88.6 bits (218), Expect = 5e-16
Identities = 87/413 (21%), Positives = 165/413 (39%), Gaps = 44/413 (10%)
Query: 57 NDDDDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQ 116
+DD V A A +LG F + + ++P L + E+ VR +VE+ I
Sbjct: 175 SDDTPMVRRAAASKLGEFAKVLELENVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQL 234
Query: 117 MREQDLVEHFIPLVKRLAAGEWFTARVSSCGLF-HIAYPSAPEAQKAELRAMYGQLCQDD 175
+ +++L +P +++ A + + R F + PE K +L + L +D
Sbjct: 235 LPQEELEPLVMPTLRQAAEDKSWRVRYMVADKFTELQNAVGPEITKTDLVPAFQNLMKDC 294
Query: 176 MPMVRRSAATNLGKFAATVEA-----------------------AHLKSDIMSVFDDLT- 211
VR +++ + +F + A H+KS + SV L+
Sbjct: 295 EAEVRAASSHKVKEFCENLSADCRENVIMTQILPCVKELVSDANQHVKSALASVIMGLSP 354
Query: 212 -------------------QDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQD 252
+D+ VRL + + +++ ++ +LP IV ++D
Sbjct: 355 ILGKDNTIEHLLPLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAED 414
Query: 253 KSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILS 312
WRVR + + L +G E +L + L D+ +R AA + K
Sbjct: 415 AKWRVRLAIIEYMPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFG 474
Query: 313 PELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFP 372
+ A I+P V +S D + R I ++ V G+D T + +LP + + +D
Sbjct: 475 KDWAQATIIPKVLAMSNDPNYLHRMTTLFCINVLSEVCGQDITTKHMLPTVVRMAEDAVA 534
Query: 373 DVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLA 425
+VR N+ L ++ + L + P + +L +D+ V+ E + +LA
Sbjct: 535 NVRFNVAKSLQKIGPTLDNSTLQNEVKPVLEKLTQDQDVDVKYFAQEALTVLA 587
Score = 51.2 bits (121), Expect = 8e-05
Identities = 58/290 (20%), Positives = 115/290 (39%), Gaps = 2/290 (0%)
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAP 348
LR+ + ++R+ + K++ + L E +LP + + D + V ALA +
Sbjct: 20 LRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDE-VLLALAEQLGTFTT 78
Query: 349 VLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAED 408
++G LLP SL E VR + L ++ L +P + LA
Sbjct: 79 LVGGPEFAHCLLPPLESLATVEETVVRDRAVESLRAISHEHSPSDLEAHFVPLVKRLASG 138
Query: 409 RHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFG 468
+ R + + ++ +L D +R AAA+ + A+
Sbjct: 139 DWFTSRTSACGLFSVCYPRVS-STVKAELRQHFRNLCSDDTPMVRRAAASKLGEFAKVLE 197
Query: 469 PDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVP 528
+ IIP ++ +D R+ ++A +A +L E ++P + A++D+
Sbjct: 198 LENVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEELEPLVMPTLRQAAEDKSW 257
Query: 529 NIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQ 578
+++ VA L V + + L P L +D + +VR +S ++
Sbjct: 258 RVRYMVADKFTELQNAVGPEITKTDLVPAFQNLMKDCEAEVRAASSHKVK 307
>UniRef100_Q6P1U8 Hypothetical protein MGC76072 [Xenopus tropicalis]
Length = 589
Score = 666 bits (1719), Expect = 0.0
Identities = 342/576 (59%), Positives = 439/576 (75%), Gaps = 4/576 (0%)
Query: 5 DQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDDEVL 64
D LYPIAVLIDEL+NED+QLRLNSI++LSTIA ALG ERTR EL+PFL++ D+DEVL
Sbjct: 7 DDSLYPIAVLIDELRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDEVL 66
Query: 65 LAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVE 124
LA+AE+LG F VGG E + LLPPLE+L TVEET VRDK+VESL I + DL
Sbjct: 67 LALAEQLGTFTSLVGGPEFVHCLLPPLESLATVEETVVRDKAVESLRAISHEHSPSDLEA 126
Query: 125 HFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAA 184
HF+PLVKRLA+G+WFT+R S+CGLF + YP KAELR + LC DD PMVRR+AA
Sbjct: 127 HFVPLVKRLASGDWFTSRTSACGLFSVCYPRVSSTVKAELRQHFRNLCSDDTPMVRRAAA 186
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILP 244
+ LG+FA +E ++KS+I+ +F +L D+QDSVRLLAVE C + +LL +E ++P
Sbjct: 187 SKLGEFAKVLELENVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEELEPLVMP 246
Query: 245 VIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKV 304
+ ++DKSWRVRYMVA++ EL +AVGPE T+T+LVPA+ L++D EAEVR AA+ KV
Sbjct: 247 TLRQAAEDKSWRVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKV 306
Query: 305 TKFSRILSPE----LAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
+F LS + + + ILPCVKEL +D++QHV+SALASVIMG++P+LGKD TIE LL
Sbjct: 307 KEFCENLSADCRENVIMTQILPCVKELVSDANQHVKSALASVIMGLSPILGKDNTIEHLL 366
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
P+FL+ LKDE P+VRLNIIS LD VN+VIGI LSQSLLPAIVELAED WRVRLAIIEY
Sbjct: 367 PLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAEDAKWRVRLAIIEY 426
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
+PLLA QLGV FFD+KL +LCM WL D VY+IR+AA +N+K+L E+FG DWA IIP+V
Sbjct: 427 MPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFGKDWAQATIIPKV 486
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L M NDP+YL+RMT L I++L+ V G EIT+ +LP VV + D V N++FNVAK LQ
Sbjct: 487 LAMSNDPNYLHRMTTLFCINVLSEVCGQEITTKHMLPTVVRMAGDAVANVRFNVAKSLQK 546
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQA 576
+ P ++ S ++N ++P L +L++D DVDV++FA +A
Sbjct: 547 IGPTLDNSTLQNEVKPVLEKLTQDQDVDVKYFAQEA 582
Score = 85.1 bits (209), Expect = 5e-15
Identities = 87/413 (21%), Positives = 163/413 (39%), Gaps = 44/413 (10%)
Query: 57 NDDDDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQ 116
+DD V A A +LG F + + ++P L + E+ VR +VE+ I
Sbjct: 175 SDDTPMVRRAAASKLGEFAKVLELENVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQL 234
Query: 117 MREQDLVEHFIPLVKRLAAGEWFTARVSSCGLF-HIAYPSAPEAQKAELRAMYGQLCQDD 175
+ +++L +P +++ A + + R F + PE K +L + L +D
Sbjct: 235 LPQEELEPLVMPTLRQAAEDKSWRVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDC 294
Query: 176 MPMVRRSAATNLGKFAATVEA-----------------------AHLKSDIMSVFDDLT- 211
VR +A+ + +F + A H+KS + SV L+
Sbjct: 295 EAEVRAAASHKVKEFCENLSADCRENVIMTQILPCVKELVSDANQHVKSALASVIMGLSP 354
Query: 212 -------------------QDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQD 252
+D+ VRL + + +++ ++ +LP IV ++D
Sbjct: 355 ILGKDNTIEHLLPLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAED 414
Query: 253 KSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILS 312
WRVR + + L +G E +L + L D+ +R AA + K
Sbjct: 415 AKWRVRLAIIEYMPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFG 474
Query: 313 PELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFP 372
+ A I+P V +S D + R I ++ V G++ T + +LP + + D
Sbjct: 475 KDWAQATIIPKVLAMSNDPNYLHRMTTLFCINVLSEVCGQEITTKHMLPTVVRMAGDAVA 534
Query: 373 DVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLA 425
+VR N+ L ++ + L + P + +L +D+ V+ E +LA
Sbjct: 535 NVRFNVAKSLQKIGPTLDNSTLQNEVKPVLEKLTQDQDVDVKYFAQEAHTVLA 587
Score = 54.3 bits (129), Expect = 1e-05
Identities = 59/290 (20%), Positives = 116/290 (39%), Gaps = 2/290 (0%)
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAP 348
LR+ + ++R+ + K++ + L E +LP + + D + V ALA +
Sbjct: 20 LRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDE-VLLALAEQLGTFTS 78
Query: 349 VLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAED 408
++G + LLP SL E VR + L ++ L +P + LA
Sbjct: 79 LVGGPEFVHCLLPPLESLATVEETVVRDKAVESLRAISHEHSPSDLEAHFVPLVKRLASG 138
Query: 409 RHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFG 468
+ R + + ++ +L D +R AAA+ + A+
Sbjct: 139 DWFTSRTSACGLFSVCYPRVS-STVKAELRQHFRNLCSDDTPMVRRAAASKLGEFAKVLE 197
Query: 469 PDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVP 528
+ IIP ++ +D R+ ++A +A +L E ++P + A++D+
Sbjct: 198 LENVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEELEPLVMPTLRQAAEDKSW 257
Query: 529 NIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQ 578
+++ VA L V + + L P L +D + +VR AS ++
Sbjct: 258 RVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKVK 307
>UniRef100_Q6AZF1 Ppp2r1a-B-prov protein [Xenopus laevis]
Length = 589
Score = 664 bits (1713), Expect = 0.0
Identities = 340/577 (58%), Positives = 439/577 (75%), Gaps = 4/577 (0%)
Query: 5 DQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDDEVL 64
D LYPIAVLIDEL+NED+QLRLNSI++LSTIA ALG ERTR EL+PFL++ D+DEVL
Sbjct: 7 DDSLYPIAVLIDELRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDEVL 66
Query: 65 LAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVE 124
LA+AE+LG F VGG E + LLPPLE+L TVEET VRDK+V+SL I + DL
Sbjct: 67 LALAEQLGTFTSLVGGPEFVHCLLPPLESLATVEETVVRDKAVDSLRAISHEHSPSDLEA 126
Query: 125 HFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAA 184
HF+PLVKRLA+G+WFT+R S+CGLF + YP KAELR + LC DD PMVRR+AA
Sbjct: 127 HFVPLVKRLASGDWFTSRTSACGLFSVCYPRVSSTVKAELRQHFRNLCSDDTPMVRRAAA 186
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILP 244
+ LG+FA +E ++KS+++ +F +L D+QDSVRLLAVE C + +LL +E ++P
Sbjct: 187 SKLGEFAKVLELENVKSELIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEELEPLVMP 246
Query: 245 VIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKV 304
++ ++DKSWRVRYMVA++ EL AVGPE T+T+LVPA+ L++D EAEVR AA+ KV
Sbjct: 247 ILRQAAEDKSWRVRYMVADKFIELQNAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKV 306
Query: 305 TKFSRILSPE----LAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
+F LS E + + ILPCVKEL +D++QHV+SALASVIMG++P+LGKD TIE LL
Sbjct: 307 KEFCENLSAECRENVIMTQILPCVKELVSDANQHVKSALASVIMGLSPILGKDNTIEHLL 366
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
P+FL+ LKDE P+VRLNIIS LD VN+VIGI LSQSLLPAIVELAED WRVRLAIIEY
Sbjct: 367 PLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAEDAKWRVRLAIIEY 426
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
+PLLA QLGV FFD+KL +LCM WL D VY+IR+AA +N+K+L E+FG DWA IIP+V
Sbjct: 427 MPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFGKDWAQATIIPKV 486
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L M NDP+YL+RMT L I++L+ V +IT+ +LP VV + D V N++FNVAK LQ
Sbjct: 487 LAMSNDPNYLHRMTTLFCINVLSEVCEQDITTKHMLPTVVRMAGDAVANVRFNVAKSLQK 546
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQAL 577
+ P ++ S ++N ++P L +L++D DVDV++FA +AL
Sbjct: 547 IGPTLDNSTLQNEVKPVLEKLTQDQDVDVKYFAQEAL 583
Score = 87.4 bits (215), Expect = 1e-15
Identities = 88/413 (21%), Positives = 164/413 (39%), Gaps = 44/413 (10%)
Query: 57 NDDDDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQ 116
+DD V A A +LG F + + L+P L + E+ VR +VE+ I
Sbjct: 175 SDDTPMVRRAAASKLGEFAKVLELENVKSELIPMFSNLASDEQDSVRLLAVEACVNIAQL 234
Query: 117 MREQDLVEHFIPLVKRLAAGEWFTARVSSCGLF-HIAYPSAPEAQKAELRAMYGQLCQDD 175
+ +++L +P++++ A + + R F + PE K +L + L +D
Sbjct: 235 LPQEELEPLVMPILRQAAEDKSWRVRYMVADKFIELQNAVGPEITKTDLVPAFQNLMKDC 294
Query: 176 MPMVRRSAATNLGKFAATVEAA-----------------------HLKSDIMSVFDDLT- 211
VR +A+ + +F + A H+KS + SV L+
Sbjct: 295 EAEVRAAASHKVKEFCENLSAECRENVIMTQILPCVKELVSDANQHVKSALASVIMGLSP 354
Query: 212 -------------------QDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQD 252
+D+ VRL + + +++ ++ +LP IV ++D
Sbjct: 355 ILGKDNTIEHLLPLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAED 414
Query: 253 KSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILS 312
WRVR + + L +G E +L + L D+ +R AA + K
Sbjct: 415 AKWRVRLAIIEYMPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFG 474
Query: 313 PELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFP 372
+ A I+P V +S D + R I ++ V +D T + +LP + + D
Sbjct: 475 KDWAQATIIPKVLAMSNDPNYLHRMTTLFCINVLSEVCEQDITTKHMLPTVVRMAGDAVA 534
Query: 373 DVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLA 425
+VR N+ L ++ + L + P + +L +D+ V+ E + +LA
Sbjct: 535 NVRFNVAKSLQKIGPTLDNSTLQNEVKPVLEKLTQDQDVDVKYFAQEALTVLA 587
Score = 53.5 bits (127), Expect = 2e-05
Identities = 58/290 (20%), Positives = 117/290 (40%), Gaps = 2/290 (0%)
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAP 348
LR+ + ++R+ + K++ + L E +LP + + D + V ALA +
Sbjct: 20 LRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDE-VLLALAEQLGTFTS 78
Query: 349 VLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAED 408
++G + LLP SL E VR + L ++ L +P + LA
Sbjct: 79 LVGGPEFVHCLLPPLESLATVEETVVRDKAVDSLRAISHEHSPSDLEAHFVPLVKRLASG 138
Query: 409 RHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFG 468
+ R + + ++ +L D +R AAA+ + A+
Sbjct: 139 DWFTSRTSACGLFSVCYPRVS-STVKAELRQHFRNLCSDDTPMVRRAAASKLGEFAKVLE 197
Query: 469 PDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVP 528
+ +IP ++ +D R+ ++A +A +L E ++P++ A++D+
Sbjct: 198 LENVKSELIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEELEPLVMPILRQAAEDKSW 257
Query: 529 NIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQ 578
+++ VA L V + + L P L +D + +VR AS ++
Sbjct: 258 RVRYMVADKFIELQNAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKVK 307
>UniRef100_Q96DH3 Alpha isoform of regulatory subunit A, protein phosphatase 2 [Homo
sapiens]
Length = 589
Score = 664 bits (1712), Expect = 0.0
Identities = 339/577 (58%), Positives = 442/577 (75%), Gaps = 4/577 (0%)
Query: 5 DQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDDEVL 64
D LYPIAVLIDEL+NED+QLRLNSI++LSTIA ALG ERTR EL+PFL++ D+DEVL
Sbjct: 7 DDSLYPIAVLIDELRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDEVL 66
Query: 65 LAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVE 124
LA+AE+LG F VGG E+ + LLPPLE+L TVEET VRDK+VESL I + DL
Sbjct: 67 LALAEQLGTFTTLVGGPEYVHCLLPPLESLATVEETVVRDKAVESLRAISHEHSPSDLEA 126
Query: 125 HFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAA 184
HF+PLVKRLA G+WFT+R S+CGLF + YP A KAELR + LC DD PMVRR+AA
Sbjct: 127 HFVPLVKRLAGGDWFTSRTSACGLFSVCYPRVSSAVKAELRQYFRNLCSDDTPMVRRAAA 186
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILP 244
+ LG+FA +E ++KS+I+ +F +L D+QDSVRLLAVE C + +LL ++ A ++P
Sbjct: 187 SKLGEFAKVLELDNVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEDLEALVMP 246
Query: 245 VIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKV 304
+ ++DKSWRVRYMVA++ EL +AVGPE T+T+LVPA+ L++D EAEVR AA+ KV
Sbjct: 247 TLRQAAEDKSWRVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKV 306
Query: 305 TKFSRILSPE----LAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
+F LS + + + ILPC+KEL +D++QHV+SALASVIMG++P+LGKD TIE LL
Sbjct: 307 KEFCENLSADCRENVIMSQILPCIKELVSDANQHVKSALASVIMGLSPILGKDNTIEHLL 366
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
P+FL+ LKDE P+VRLNIIS LD VN+VIGI LSQSLLPAIVELAED WRVRLAIIEY
Sbjct: 367 PLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAEDAKWRVRLAIIEY 426
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
+PLLA QLGV FFD+KL +LCM WL D VY+IR+AA +N+K+L E+FG +WA IIP+V
Sbjct: 427 MPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFGKEWAHATIIPKV 486
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L M DP+YL+RMT L I++L+ V G +IT+ +LP V+ + D V N++FNVAK LQ
Sbjct: 487 LAMSGDPNYLHRMTTLFCINVLSEVCGQDITTKHMLPTVLRMAGDPVANVRFNVAKSLQK 546
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQAL 577
+ PI++ S +++ ++P L +L++D DVDV++FA +AL
Sbjct: 547 IGPILDNSTLQSEVKPILEKLTQDQDVDVKYFAQEAL 583
Score = 97.4 bits (241), Expect = 1e-18
Identities = 94/455 (20%), Positives = 185/455 (40%), Gaps = 48/455 (10%)
Query: 94 LCTVEETCVRDKSVESLCRIGAQMREQDLVEHFIPLVKRLAAGEWFTAR---VSSCGLFH 150
LC+ + VR + L + ++ IP+ LA+ E + R V +C +
Sbjct: 173 LCSDDTPMVRRAAASKLGEFAKVLELDNVKSEIIPMFSNLASDEQDSVRLLAVEAC--VN 230
Query: 151 IAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDL 210
IA E +A + Q +D VR A + V K+D++ F +L
Sbjct: 231 IAQLLPQEDLEALVMPTLRQAAEDKSWRVRYMVADKFTELQKAVGPEITKTDLVPAFQNL 290
Query: 211 TQDDQDSVRLLAV----EGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLY 266
+D + VR A E C L ++ ILP I D + V+ +A+ +
Sbjct: 291 MKDCEAEVRAAASHKVKEFCENLSADCRENVIMSQILPCIKELVSDANQHVKSALASVIM 350
Query: 267 ELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKE 326
L +G + T L+P ++ L+D EVR+ + + ++ + +LP + E
Sbjct: 351 GLSPILGKDNTIEHLLPLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVE 410
Query: 327 LSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVN 386
L+ D+ VR A+ + +A LG + E+L + ++ L D +R S L ++
Sbjct: 411 LAEDAKWRVRLAIIEYMPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLV 470
Query: 387 QVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLK 446
+ G + +++P ++ ++ D ++ R+ + C
Sbjct: 471 EKFGKEWAHATIIPKVLAMSGDPNYLHRMTTL---------------------FC----- 504
Query: 447 DKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVL 506
+ L+E G D +H++P VL M DP R + +++ + P+L
Sbjct: 505 -------------INVLSEVCGQDITTKHMLPTVLRMAGDPVANVRFNVAKSLQKIGPIL 551
Query: 507 GSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSL 541
+ S++ P++ ++D+ ++K+ + L L
Sbjct: 552 DNSTLQSEVKPILEKLTQDQDVDVKYFAQEALTVL 586
Score = 92.4 bits (228), Expect = 3e-17
Identities = 90/413 (21%), Positives = 165/413 (39%), Gaps = 44/413 (10%)
Query: 57 NDDDDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQ 116
+DD V A A +LG F + + ++P L + E+ VR +VE+ I
Sbjct: 175 SDDTPMVRRAAASKLGEFAKVLELDNVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQL 234
Query: 117 MREQDLVEHFIPLVKRLAAGEWFTARVSSCGLF-HIAYPSAPEAQKAELRAMYGQLCQDD 175
+ ++DL +P +++ A + + R F + PE K +L + L +D
Sbjct: 235 LPQEDLEALVMPTLRQAAEDKSWRVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDC 294
Query: 176 MPMVRRSAATNLGKFAATVEA-----------------------AHLKSDIMSVFDDLT- 211
VR +A+ + +F + A H+KS + SV L+
Sbjct: 295 EAEVRAAASHKVKEFCENLSADCRENVIMSQILPCIKELVSDANQHVKSALASVIMGLSP 354
Query: 212 -------------------QDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQD 252
+D+ VRL + + +++ ++ +LP IV ++D
Sbjct: 355 ILGKDNTIEHLLPLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAED 414
Query: 253 KSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILS 312
WRVR + + L +G E +L + L D+ +R AA + K
Sbjct: 415 AKWRVRLAIIEYMPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFG 474
Query: 313 PELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFP 372
E A I+P V +S D + R I ++ V G+D T + +LP L + D
Sbjct: 475 KEWAHATIIPKVLAMSGDPNYLHRMTTLFCINVLSEVCGQDITTKHMLPTVLRMAGDPVA 534
Query: 373 DVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLA 425
+VR N+ L ++ ++ L + P + +L +D+ V+ E + +L+
Sbjct: 535 NVRFNVAKSLQKIGPILDNSTLQSEVKPILEKLTQDQDVDVKYFAQEALTVLS 587
Score = 56.6 bits (135), Expect = 2e-06
Identities = 60/290 (20%), Positives = 117/290 (39%), Gaps = 2/290 (0%)
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAP 348
LR+ + ++R+ + K++ + L E +LP + + D + V ALA +
Sbjct: 20 LRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDE-VLLALAEQLGTFTT 78
Query: 349 VLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAED 408
++G + LLP SL E VR + L ++ L +P + LA
Sbjct: 79 LVGGPEYVHCLLPPLESLATVEETVVRDKAVESLRAISHEHSPSDLEAHFVPLVKRLAGG 138
Query: 409 RHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFG 468
+ R + + ++ +L D +R AAA+ + A+
Sbjct: 139 DWFTSRTSACGLFSVCYPRVSSAV-KAELRQYFRNLCSDDTPMVRRAAASKLGEFAKVLE 197
Query: 469 PDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVP 528
D IIP ++ +D R+ ++A +A +L E + ++P + A++D+
Sbjct: 198 LDNVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEDLEALVMPTLRQAAEDKSW 257
Query: 529 NIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQ 578
+++ VA L V + + L P L +D + +VR AS ++
Sbjct: 258 RVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKVK 307
>UniRef100_Q92138 Phosphorylase phosphatase [Xenopus laevis]
Length = 589
Score = 663 bits (1711), Expect = 0.0
Identities = 339/577 (58%), Positives = 439/577 (75%), Gaps = 4/577 (0%)
Query: 5 DQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDDEVL 64
D LYPIAVLIDEL+NED+QLRLNSI++LSTIA ALG ERTR EL+PFL++ D+DEVL
Sbjct: 7 DDSLYPIAVLIDELRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDEVL 66
Query: 65 LAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVE 124
LA+AE+LG F VGG E + LLPPLE+L TVEET VRDK+V+SL I + DL
Sbjct: 67 LALAEQLGTFTSLVGGPEFVHCLLPPLESLATVEETVVRDKAVDSLRAISHEHSPSDLEA 126
Query: 125 HFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAA 184
HF+PLVKRLA+G+WFT+R S+CGLF + YP KAELR + LC DD PMVRR+AA
Sbjct: 127 HFVPLVKRLASGDWFTSRTSACGLFSVCYPRVSSTVKAELRQHFRNLCSDDTPMVRRAAA 186
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILP 244
+ LG+FA +E ++KS+++ +F +L D+QDSVRLLAVE C + +LL +E ++P
Sbjct: 187 SKLGEFAKVLELENVKSELIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEELEPLVMP 246
Query: 245 VIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKV 304
++ ++DKSWRVRYMVA++ EL AVGPE T+T+LVPA+ L++D EAEVR AA+ KV
Sbjct: 247 ILRQAAEDKSWRVRYMVADKFIELQNAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKV 306
Query: 305 TKFSRILSPE----LAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
+F LS E + + ILPCVKEL +D++QHV+SALASVIMG++P+LGKD TIE L+
Sbjct: 307 KEFCENLSAECRENVIMTQILPCVKELVSDANQHVKSALASVIMGLSPILGKDNTIEHLI 366
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
P+FL+ LKDE P+VRLNIIS LD VN+VIGI LSQSLLPAIVELAED WRVRLAIIEY
Sbjct: 367 PLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAEDAKWRVRLAIIEY 426
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
+PLLA QLGV FFD+KL +LCM WL D VY+IR+AA +N+K+L E+FG DWA IIP+V
Sbjct: 427 MPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFGKDWAQATIIPKV 486
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L M NDP+YL+RMT L I++L+ V +IT+ +LP VV + D V N++FNVAK LQ
Sbjct: 487 LAMSNDPNYLHRMTTLFCINVLSEVCEQDITTKHMLPTVVRMAGDAVANVRFNVAKSLQK 546
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQAL 577
+ P ++ S ++N ++P L +L++D DVDV++FA +AL
Sbjct: 547 IGPTLDNSTLQNEVKPVLEKLTQDQDVDVKYFAQEAL 583
Score = 87.4 bits (215), Expect = 1e-15
Identities = 88/413 (21%), Positives = 164/413 (39%), Gaps = 44/413 (10%)
Query: 57 NDDDDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQ 116
+DD V A A +LG F + + L+P L + E+ VR +VE+ I
Sbjct: 175 SDDTPMVRRAAASKLGEFAKVLELENVKSELIPMFSNLASDEQDSVRLLAVEACVNIAQL 234
Query: 117 MREQDLVEHFIPLVKRLAAGEWFTARVSSCGLF-HIAYPSAPEAQKAELRAMYGQLCQDD 175
+ +++L +P++++ A + + R F + PE K +L + L +D
Sbjct: 235 LPQEELEPLVMPILRQAAEDKSWRVRYMVADKFIELQNAVGPEITKTDLVPAFQNLMKDC 294
Query: 176 MPMVRRSAATNLGKFAATVEAA-----------------------HLKSDIMSVFDDLT- 211
VR +A+ + +F + A H+KS + SV L+
Sbjct: 295 EAEVRAAASHKVKEFCENLSAECRENVIMTQILPCVKELVSDANQHVKSALASVIMGLSP 354
Query: 212 -------------------QDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQD 252
+D+ VRL + + +++ ++ +LP IV ++D
Sbjct: 355 ILGKDNTIEHLIPLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAED 414
Query: 253 KSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILS 312
WRVR + + L +G E +L + L D+ +R AA + K
Sbjct: 415 AKWRVRLAIIEYMPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFG 474
Query: 313 PELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFP 372
+ A I+P V +S D + R I ++ V +D T + +LP + + D
Sbjct: 475 KDWAQATIIPKVLAMSNDPNYLHRMTTLFCINVLSEVCEQDITTKHMLPTVVRMAGDAVA 534
Query: 373 DVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLA 425
+VR N+ L ++ + L + P + +L +D+ V+ E + +LA
Sbjct: 535 NVRFNVAKSLQKIGPTLDNSTLQNEVKPVLEKLTQDQDVDVKYFAQEALTVLA 587
Score = 53.5 bits (127), Expect = 2e-05
Identities = 58/290 (20%), Positives = 117/290 (40%), Gaps = 2/290 (0%)
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAP 348
LR+ + ++R+ + K++ + L E +LP + + D + V ALA +
Sbjct: 20 LRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDE-VLLALAEQLGTFTS 78
Query: 349 VLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAED 408
++G + LLP SL E VR + L ++ L +P + LA
Sbjct: 79 LVGGPEFVHCLLPPLESLATVEETVVRDKAVDSLRAISHEHSPSDLEAHFVPLVKRLASG 138
Query: 409 RHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFG 468
+ R + + ++ +L D +R AAA+ + A+
Sbjct: 139 DWFTSRTSACGLFSVCYPRVS-STVKAELRQHFRNLCSDDTPMVRRAAASKLGEFAKVLE 197
Query: 469 PDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVP 528
+ +IP ++ +D R+ ++A +A +L E ++P++ A++D+
Sbjct: 198 LENVKSELIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEELEPLVMPILRQAAEDKSW 257
Query: 529 NIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQ 578
+++ VA L V + + L P L +D + +VR AS ++
Sbjct: 258 RVRYMVADKFIELQNAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKVK 307
>UniRef100_Q76MZ3 Serine/threonine protein phosphatase 2A, 65 kDa regulatory subunit
A, alpha isoform [Mus musculus]
Length = 588
Score = 663 bits (1711), Expect = 0.0
Identities = 339/577 (58%), Positives = 442/577 (75%), Gaps = 4/577 (0%)
Query: 5 DQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDDEVL 64
D LYPIAVLIDEL+NED+QLRLNSI++LSTIA ALG ERTR EL+PFL++ D+DEVL
Sbjct: 6 DDSLYPIAVLIDELRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDEVL 65
Query: 65 LAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQDLVE 124
LA+AE+LG F VGG E+ + LLPPLE+L TVEET VRDK+VESL I + DL
Sbjct: 66 LALAEQLGTFTTLVGGPEYVHCLLPPLESLATVEETVVRDKAVESLRAISHEHSPSDLEA 125
Query: 125 HFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAA 184
HF+PLVKRLA G+WFT+R S+CGLF + YP A KAELR + LC DD PMVRR+AA
Sbjct: 126 HFVPLVKRLAGGDWFTSRTSACGLFSVCYPRVSSAVKAELRQYFRNLCSDDTPMVRRAAA 185
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILP 244
+ LG+FA +E ++KS+I+ +F +L D+QDSVRLLAVE C + +LL ++ A ++P
Sbjct: 186 SKLGEFAKVLELDNVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEDLEALVMP 245
Query: 245 VIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKV 304
+ ++DKSWRVRYMVA++ EL +AVGPE T+T+LVPA+ L++D EAEVR AA+ KV
Sbjct: 246 TLRQAAEDKSWRVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKV 305
Query: 305 TKFSRILSPE----LAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
+F LS + + + ILPC+KEL +D++QHV+SALASVIMG++P+LGKD TIE LL
Sbjct: 306 KEFCENLSADCRENVIMTQILPCIKELVSDANQHVKSALASVIMGLSPILGKDNTIEHLL 365
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
P+FL+ LKDE P+VRLNIIS LD VN+VIGI LSQSLLPAIVELAED WRVRLAIIEY
Sbjct: 366 PLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAEDAKWRVRLAIIEY 425
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
+PLLA QLGV FFD+KL +LCM WL D VY+IR+AA +N+K+L E+FG +WA IIP+V
Sbjct: 426 MPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFGKEWAHATIIPKV 485
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L M DP+YL+RMT L I++L+ V G +IT+ +LP V+ + D V N++FNVAK LQ
Sbjct: 486 LAMSGDPNYLHRMTTLFCINVLSEVCGQDITTKHMLPTVLRMAGDPVANVRFNVAKSLQK 545
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQAL 577
+ PI++ S +++ ++P L +L++D DVDV++FA +AL
Sbjct: 546 IGPILDNSTLQSEVKPILEKLTQDQDVDVKYFAQEAL 582
Score = 92.4 bits (228), Expect = 3e-17
Identities = 90/413 (21%), Positives = 165/413 (39%), Gaps = 44/413 (10%)
Query: 57 NDDDDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQ 116
+DD V A A +LG F + + ++P L + E+ VR +VE+ I
Sbjct: 174 SDDTPMVRRAAASKLGEFAKVLELDNVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQL 233
Query: 117 MREQDLVEHFIPLVKRLAAGEWFTARVSSCGLF-HIAYPSAPEAQKAELRAMYGQLCQDD 175
+ ++DL +P +++ A + + R F + PE K +L + L +D
Sbjct: 234 LPQEDLEALVMPTLRQAAEDKSWRVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDC 293
Query: 176 MPMVRRSAATNLGKFAATVEA-----------------------AHLKSDIMSVFDDLT- 211
VR +A+ + +F + A H+KS + SV L+
Sbjct: 294 EAEVRAAASHKVKEFCENLSADCRENVIMTQILPCIKELVSDANQHVKSALASVIMGLSP 353
Query: 212 -------------------QDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQD 252
+D+ VRL + + +++ ++ +LP IV ++D
Sbjct: 354 ILGKDNTIEHLLPLFLAQLKDECPEVRLNIISNLDCVNEVIGIRQLSQSLLPAIVELAED 413
Query: 253 KSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILS 312
WRVR + + L +G E +L + L D+ +R AA + K
Sbjct: 414 AKWRVRLAIIEYMPLLAGQLGVEFFDEKLNSLCMAWLVDHVYAIREAATSNLKKLVEKFG 473
Query: 313 PELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFP 372
E A I+P V +S D + R I ++ V G+D T + +LP L + D
Sbjct: 474 KEWAHATIIPKVLAMSGDPNYLHRMTTLFCINVLSEVCGQDITTKHMLPTVLRMAGDPVA 533
Query: 373 DVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLA 425
+VR N+ L ++ ++ L + P + +L +D+ V+ E + +L+
Sbjct: 534 NVRFNVAKSLQKIGPILDNSTLQSEVKPILEKLTQDQDVDVKYFAQEALTVLS 586
Score = 56.6 bits (135), Expect = 2e-06
Identities = 60/290 (20%), Positives = 117/290 (39%), Gaps = 2/290 (0%)
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAP 348
LR+ + ++R+ + K++ + L E +LP + + D + V ALA +
Sbjct: 19 LRNEDVQLRLNSIKKLSTIALALGVERTRSELLPFLTDTIYDEDE-VLLALAEQLGTFTT 77
Query: 349 VLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAED 408
++G + LLP SL E VR + L ++ L +P + LA
Sbjct: 78 LVGGPEYVHCLLPPLESLATVEETVVRDKAVESLRAISHEHSPSDLEAHFVPLVKRLAGG 137
Query: 409 RHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFG 468
+ R + + ++ +L D +R AAA+ + A+
Sbjct: 138 DWFTSRTSACGLFSVCYPRVSSAV-KAELRQYFRNLCSDDTPMVRRAAASKLGEFAKVLE 196
Query: 469 PDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVP 528
D IIP ++ +D R+ ++A +A +L E + ++P + A++D+
Sbjct: 197 LDNVKSEIIPMFSNLASDEQDSVRLLAVEACVNIAQLLPQEDLEALVMPTLRQAAEDKSW 256
Query: 529 NIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQ 578
+++ VA L V + + L P L +D + +VR AS ++
Sbjct: 257 RVRYMVADKFTELQKAVGPEITKTDLVPAFQNLMKDCEAEVRAAASHKVK 306
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 878,552,501
Number of Sequences: 2790947
Number of extensions: 34586690
Number of successful extensions: 98421
Number of sequences better than 10.0: 388
Number of HSP's better than 10.0 without gapping: 173
Number of HSP's successfully gapped in prelim test: 218
Number of HSP's that attempted gapping in prelim test: 94496
Number of HSP's gapped (non-prelim): 1624
length of query: 587
length of database: 848,049,833
effective HSP length: 133
effective length of query: 454
effective length of database: 476,853,882
effective search space: 216491662428
effective search space used: 216491662428
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0310b.14


