

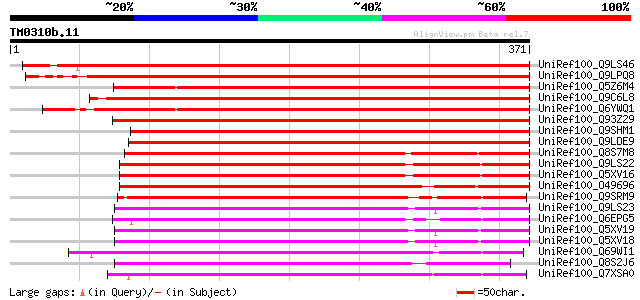
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.11
(371 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LS46 Gb|AAF25997.1 [Arabidopsis thaliana] 481 e-134
UniRef100_Q9LPQ8 F15H18.9 [Arabidopsis thaliana] 453 e-126
UniRef100_Q5Z6M4 Aluminum-activated malate transporter-like [Ory... 433 e-120
UniRef100_Q9C6L8 Hypothetical protein F2J7.18 [Arabidopsis thali... 419 e-116
UniRef100_Q6YWQ1 Putative aluminum-activated malate transporter ... 406 e-112
UniRef100_Q93Z29 At1g68600/F24J5_14 [Arabidopsis thaliana] 394 e-108
UniRef100_Q9SHM1 Hypothetical protein At2g17470 [Arabidopsis tha... 381 e-104
UniRef100_Q9LDE9 Similar to Arabidopsis thaliana chromosome 1 [O... 249 9e-65
UniRef100_Q8S7M8 Hypothetical protein OSJNBa0095C07.34 [Oryza sa... 233 6e-60
UniRef100_Q9LS22 Arabidopsis thaliana genomic DNA, chromosome 5,... 231 2e-59
UniRef100_Q5XV16 Hypothetical protein [Arabidopsis thaliana] 229 7e-59
UniRef100_O49696 Hypothetical protein AT4g17970 [Arabidopsis tha... 228 3e-58
UniRef100_Q9SRM9 T19F11.8 protein [Arabidopsis thaliana] 219 1e-55
UniRef100_Q9LS23 Arabidopsis thaliana genomic DNA, chromosome 5,... 214 2e-54
UniRef100_Q6EPG5 Aluminum-activated malate transporter-like [Ory... 214 3e-54
UniRef100_Q5XV19 Hypothetical protein [Arabidopsis thaliana] 213 9e-54
UniRef100_Q5XV18 Hypothetical protein [Arabidopsis thaliana] 213 9e-54
UniRef100_Q69WI1 Putative aluminum-activated malate transporter ... 212 1e-53
UniRef100_Q8S2J6 OSJNBb0021A09.9 protein [Oryza sativa] 210 6e-53
UniRef100_Q7XSA0 OSJNBa0005N02.7 protein [Oryza sativa] 209 8e-53
>UniRef100_Q9LS46 Gb|AAF25997.1 [Arabidopsis thaliana]
Length = 598
Score = 481 bits (1237), Expect = e-134
Identities = 236/364 (64%), Positives = 286/364 (77%), Gaps = 6/364 (1%)
Query: 10 KLGSFRYSFKERKEKLLSMKSSLIGYSQIGIPLPESDD--DNDGPPGSKNCCRFCSYRAL 67
K GSFR+ E++E+LLS G+S ES+D +N+ C CS L
Sbjct: 4 KQGSFRHGILEKRERLLSNN----GFSDFRFTDIESNDLLENENCGRRTRLCCCCSCGNL 59
Query: 68 SDRLVGAWKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSV 127
S+++ G + K VA +AWEMG+SDPRKIVF+AK+GLAL +++LLIF + P DL R+SV
Sbjct: 60 SEKISGVYDDAKDVARKAWEMGVSDPRKIVFSAKIGLALTIVALLIFYQEPNPDLSRYSV 119
Query: 128 WAILTVVTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIV 187
WAILTVV VFEF+IGATLSKG NR LGTL+AGGLA+GM LS L G WEEI +SIF +
Sbjct: 120 WAILTVVVVFEFTIGATLSKGFNRALGTLSAGGLALGMAELSTLFGDWEEIFCTLSIFCI 179
Query: 188 GFCATYAKLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLG 247
GF AT+ KLYP+MKAYEYGFRVFL+TYCYI++SG++TG+FI+ A+SRFLLIALGA VSLG
Sbjct: 180 GFLATFMKLYPSMKAYEYGFRVFLLTYCYILISGFRTGQFIEVAISRFLLIALGAGVSLG 239
Query: 248 VNVCLYPIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVY 307
VN+ +YPIWAGEDLH+LV KNFM VA SLEG VN YL C+EYER+PSKILTYQAS+D VY
Sbjct: 240 VNMFIYPIWAGEDLHNLVVKNFMNVATSLEGCVNGYLRCLEYERIPSKILTYQASEDPVY 299
Query: 308 SGYRSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCI 367
GYRSA++STS E++LM FA+WEPPHG YK YPW NYVK+SGAL+HCAF VMA+HGCI
Sbjct: 300 KGYRSAVESTSQEESLMSFAIWEPPHGPYKSFNYPWKNYVKLSGALKHCAFTVMALHGCI 359
Query: 368 LSEI 371
LSEI
Sbjct: 360 LSEI 363
>UniRef100_Q9LPQ8 F15H18.9 [Arabidopsis thaliana]
Length = 581
Score = 453 bits (1165), Expect = e-126
Identities = 228/361 (63%), Positives = 273/361 (75%), Gaps = 20/361 (5%)
Query: 12 GSFRYSFKERKEKLLSMKSSLIGYSQIGIPLPESDDDNDGPPGSKNCCRFCSYRALSDRL 71
GSFR S ++R L ++S GYS + +DDD K R CSY SD++
Sbjct: 27 GSFRQSMRDR----LILQSR--GYSNV------NDDD-------KTSVRCCSYSYFSDKI 67
Query: 72 VGAWKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAIL 131
G K K V AWEMG +DPRK++F+AKMGLAL L S+LIF K P +L H +WAIL
Sbjct: 68 TGVVKKLKDVLVTAWEMGTADPRKMIFSAKMGLALTLTSILIFFKIPGLELSGHYLWAIL 127
Query: 132 TVVTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCA 191
TVV +FEFSIGAT SKG NRGLGTL+AGGLA+GM +S++ G W ++ SIF+V F A
Sbjct: 128 TVVVIFEFSIGATFSKGCNRGLGTLSAGGLALGMSWISEMTGNWADVFNAASIFVVAFFA 187
Query: 192 TYAKLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVC 251
TYAKLYPTMK YEYGFRVFL+TYCY+IVSGY+TG+F++TAVSRFLLIALGA+V L VN C
Sbjct: 188 TYAKLYPTMKPYEYGFRVFLLTYCYVIVSGYKTGEFMETAVSRFLLIALGASVGLIVNTC 247
Query: 252 LYPIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQA-SDDVVYSGY 310
+YPIWAGEDLH+LVAKNF+ VA SLEG VN YL C+ Y+ +PS+IL Y+A ++D VYSGY
Sbjct: 248 IYPIWAGEDLHNLVAKNFVNVATSLEGCVNGYLECVAYDTIPSRILVYEAVAEDPVYSGY 307
Query: 311 RSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSE 370
RSA+QSTS ED LM FA WEPPHG YK +YPW+ YVKV GALRHCA MVMA+HGCILSE
Sbjct: 308 RSAVQSTSQEDTLMSFASWEPPHGPYKSFRYPWALYVKVGGALRHCAIMVMALHGCILSE 367
Query: 371 I 371
I
Sbjct: 368 I 368
>UniRef100_Q5Z6M4 Aluminum-activated malate transporter-like [Oryza sativa]
Length = 388
Score = 433 bits (1113), Expect = e-120
Identities = 205/297 (69%), Positives = 247/297 (83%), Gaps = 1/297 (0%)
Query: 75 WKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVV 134
W + A W +DPRK VFAAK+GLAL LISLL+FL+ P D+ HSVWAILTVV
Sbjct: 78 WGAVRGAAEELWAFARADPRKAVFAAKVGLALALISLLVFLREP-RDIVSHSVWAILTVV 136
Query: 135 TVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCATYA 194
VFEFSIGAT SKG NRGLGTLTAGGLA+ + LSK G+ EE+I+++SIFIV F T
Sbjct: 137 VVFEFSIGATFSKGFNRGLGTLTAGGLALAVAELSKHLGKLEEVILIISIFIVAFFTTLT 196
Query: 195 KLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYP 254
KL+P MKAYEYGFRVFL+T+CY++VSGY TGKF DTAVSRF+LIA+GAAVSLG+NV +YP
Sbjct: 197 KLHPKMKAYEYGFRVFLLTFCYVMVSGYNTGKFTDTAVSRFILIAIGAAVSLGINVGIYP 256
Query: 255 IWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSAL 314
IWAG+DLH+LVAKNF+GVA SLEG V+ YL C+EYER+PSKIL YQASDD +YSGYR+A+
Sbjct: 257 IWAGQDLHNLVAKNFIGVAKSLEGCVDGYLKCMEYERIPSKILVYQASDDPLYSGYRAAV 316
Query: 315 QSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
++++ E+ L+GFA+WEPPHG YKM+KYPW N+ KV GALRHC+F VMA+HGCILSEI
Sbjct: 317 EASAQEETLLGFAIWEPPHGAYKMMKYPWRNFTKVGGALRHCSFAVMALHGCILSEI 373
>UniRef100_Q9C6L8 Hypothetical protein F2J7.18 [Arabidopsis thaliana]
Length = 548
Score = 419 bits (1077), Expect = e-116
Identities = 200/314 (63%), Positives = 244/314 (77%), Gaps = 5/314 (1%)
Query: 58 CCRFCSYRALSDRLVGAWKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKT 117
C RFCS D + +WK + A+ +EMG SD RK+ F+ KMG+AL L S +I+LK
Sbjct: 35 CFRFCS-----DGITASWKALYDIGAKLYEMGRSDRRKVYFSVKMGMALALCSFVIYLKE 89
Query: 118 PFEDLGRHSVWAILTVVTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEE 177
P D +++VWAILTVV VFE+SIGATL KG NR +GTL+AGGLA+G+ LS AG++EE
Sbjct: 90 PLRDASKYAVWAILTVVVVFEYSIGATLVKGFNRAIGTLSAGGLALGIARLSVSAGEFEE 149
Query: 178 IIVMVSIFIVGFCATYAKLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLL 237
+I+++SIFI GF A+Y KLYP MK+YEY FRVFL+TYC ++VSG + F TA RFLL
Sbjct: 150 LIIIISIFIAGFSASYLKLYPAMKSYEYAFRVFLLTYCIVLVSGNNSRDFFSTAYYRFLL 209
Query: 238 IALGAAVSLGVNVCLYPIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKIL 297
I +GA + LGVN+ + PIWAGEDLH LV KNF VA SLEG VN YL C+EYER+PSKIL
Sbjct: 210 ILVGAGICLGVNIFILPIWAGEDLHKLVVKNFKSVANSLEGCVNGYLQCVEYERIPSKIL 269
Query: 298 TYQASDDVVYSGYRSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCA 357
TYQASDD +YSGYRS +QSTS ED+L+ FAVWEPPHG YK +PW+NYVK+SGA+RHCA
Sbjct: 270 TYQASDDPLYSGYRSVVQSTSQEDSLLDFAVWEPPHGPYKTFHHPWANYVKLSGAVRHCA 329
Query: 358 FMVMAMHGCILSEI 371
FMVMAMHGCILSEI
Sbjct: 330 FMVMAMHGCILSEI 343
>UniRef100_Q6YWQ1 Putative aluminum-activated malate transporter [Oryza sativa]
Length = 584
Score = 406 bits (1044), Expect = e-112
Identities = 210/349 (60%), Positives = 256/349 (73%), Gaps = 10/349 (2%)
Query: 24 KLLSMKSSLIGYSQIGIPLPESDDDNDGPPGSKNCCRFCSYRALSDRLVGAWKTTKRVAA 83
+L S++S+L ++ PL D G PG + RL A + A
Sbjct: 15 ELRSLRSTLDQRGELRAPLLSFDW---GFPGG-----IARWEGEEGRLRRAAGAARAAAT 66
Query: 84 RAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIGA 143
W DPRK VFAAK+ AL LI+LL+FL+ P DL H+VWAILTVV VFEFSIGA
Sbjct: 67 EMWAFARKDPRKPVFAAKVATALALITLLVFLREP-TDLANHAVWAILTVVVVFEFSIGA 125
Query: 144 TLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCATYAKLYPTMKAY 203
TLSKGLNRGLGTLTAGG A+ + LS G + +I+++ F+V F AT KL+P MK Y
Sbjct: 126 TLSKGLNRGLGTLTAGGFALAVSELSSSMGNFGNVILIICTFVVAFGATLTKLHPKMKPY 185
Query: 204 EYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLHD 263
EYGFRVFL+T+CY+ VSGY TGKFI TA+SRFLLIA+GAAVSL +N+ ++PIWAGEDLH+
Sbjct: 186 EYGFRVFLLTFCYVTVSGYNTGKFIATAISRFLLIAIGAAVSLALNIGIHPIWAGEDLHN 245
Query: 264 LVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQAS-DDVVYSGYRSALQSTSTEDA 322
LVAKNF GVA SLEG V+ YL C+EYERVPS ILTYQAS DD +YSG R+A++S++ E+A
Sbjct: 246 LVAKNFDGVAKSLEGCVDGYLKCMEYERVPSTILTYQASDDDHLYSGCRAAVESSAQEEA 305
Query: 323 LMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
L+GFA+WEPPHG YKM+KYPW NY KV GALRHC+F VMA+HGCILSEI
Sbjct: 306 LLGFAIWEPPHGPYKMMKYPWMNYTKVGGALRHCSFSVMALHGCILSEI 354
>UniRef100_Q93Z29 At1g68600/F24J5_14 [Arabidopsis thaliana]
Length = 537
Score = 394 bits (1012), Expect = e-108
Identities = 187/298 (62%), Positives = 231/298 (76%)
Query: 74 AWKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTV 133
+W+ A+ + +G SD RK+ F+ KMG+AL L S +IFLK P +D + +VWAILTV
Sbjct: 35 SWRALYEAPAKLYALGHSDRRKLYFSIKMGIALALCSFVIFLKEPLQDASKFAVWAILTV 94
Query: 134 VTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCATY 193
V +FE+ +GATL KG NR LGT+ AGGLA+G+ LS LAG++EE+I+++ IF+ GF A+Y
Sbjct: 95 VLIFEYYVGATLVKGFNRALGTMLAGGLALGVAQLSVLAGEFEEVIIVICIFLAGFGASY 154
Query: 194 AKLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLY 253
KLY +MK YEY FRVF +TYC ++VSG + F+ TA R LLI LGA + L VNV L+
Sbjct: 155 LKLYASMKPYEYAFRVFKLTYCIVLVSGNNSRDFLSTAYYRILLIGLGATICLLVNVFLF 214
Query: 254 PIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSA 313
PIWAGEDLH LVAKNF VA SLEG VN YL C+EYER+PSKILTYQASDD +YSGYRSA
Sbjct: 215 PIWAGEDLHKLVAKNFKNVANSLEGCVNGYLQCVEYERIPSKILTYQASDDPLYSGYRSA 274
Query: 314 LQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
+QSTS ED+L+ FA+WEPPHG YK +PW NYVK+SGA+RHCAF VMAMHGCILSEI
Sbjct: 275 VQSTSQEDSLLDFAIWEPPHGPYKTFNHPWKNYVKLSGAVRHCAFTVMAMHGCILSEI 332
>UniRef100_Q9SHM1 Hypothetical protein At2g17470 [Arabidopsis thaliana]
Length = 538
Score = 381 bits (979), Expect = e-104
Identities = 179/285 (62%), Positives = 223/285 (77%)
Query: 87 EMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIGATLS 146
E+G SD R+I FA KMG+AL L S++IFLK P D ++SVW ILTVV VFE+S+GATL
Sbjct: 31 ELGHSDRRRIFFAVKMGMALALCSVVIFLKEPLHDASKYSVWGILTVVVVFEYSVGATLV 90
Query: 147 KGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCATYAKLYPTMKAYEYG 206
KG NR +GT++AGGLA+G+ LS L+ +E+ I++ IF+ GF A+Y+KL+P MK YEY
Sbjct: 91 KGFNRAIGTVSAGGLALGIARLSVLSRDFEQTIIITCIFLAGFIASYSKLHPAMKPYEYA 150
Query: 207 FRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLHDLVA 266
FRVFL+T+C ++VSG TG F TA RFL I +GA L VN+ ++PIWAGEDLH LVA
Sbjct: 151 FRVFLLTFCIVLVSGNNTGDFFSTAYYRFLFIVVGATTCLVVNIFIFPIWAGEDLHKLVA 210
Query: 267 KNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQSTSTEDALMGF 326
NF VA SLEG VN YL C+EYERVPSKILTYQ SDD +YSGYRSA+QST+ E++L+ F
Sbjct: 211 NNFKSVANSLEGCVNGYLQCVEYERVPSKILTYQTSDDPLYSGYRSAIQSTNQEESLLDF 270
Query: 327 AVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
A+WEPPHG Y+ +PW NYVK+SGA+RHCAF VMA+HGCILSEI
Sbjct: 271 AIWEPPHGPYRTFNHPWKNYVKLSGAVRHCAFTVMAIHGCILSEI 315
>UniRef100_Q9LDE9 Similar to Arabidopsis thaliana chromosome 1 [Oryza sativa]
Length = 524
Score = 249 bits (636), Expect = 9e-65
Identities = 127/288 (44%), Positives = 183/288 (63%), Gaps = 2/288 (0%)
Query: 86 WEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIGATL 145
W+ D ++ FA K+GLA +L+SLLI + P++ G + +W+ILTV +FE+++GAT
Sbjct: 43 WDFARQDTNRVTFALKVGLACLLVSLLILFRAPYDIFGANIIWSILTVAIMFEYTVGATF 102
Query: 146 SKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEE-IIVMVSIFIVGFCATYAKLYPTMKAYE 204
++G NR +G++ AG AV + ++ +G E I+ SIF++G ++ KL+P++ YE
Sbjct: 103 NRGFNRAVGSVFAGVFAVVVIQVAMSSGHIAEPYIIGFSIFLIGAVTSFMKLWPSLVPYE 162
Query: 205 YGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLHDL 264
YGFRV L TYC IIVSGY+ G I TA+ R IA+GA +++ VNV + PIWAGE LH
Sbjct: 163 YGFRVILFTYCLIIVSGYRMGNPIRTAMDRLYSIAIGALIAVLVNVFICPIWAGEQLHRE 222
Query: 265 VAKNFMGVAASLEGVVNNYLNCIEYERVP-SKILTYQASDDVVYSGYRSALQSTSTEDAL 323
+ +F +A SLE V YL+ E SK + D+ + R+ L S++ D+L
Sbjct: 223 LVNSFNSLADSLEECVKKYLSDDGSEHPEFSKTVMDNFPDEPAFRKCRATLNSSAKFDSL 282
Query: 324 MGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
A WEPPHGR+K YPW+ YVKV LRHCA+ VMA+HGC+ SEI
Sbjct: 283 ANSAKWEPPHGRFKHFFYPWAEYVKVGNVLRHCAYEVMALHGCVHSEI 330
>UniRef100_Q8S7M8 Hypothetical protein OSJNBa0095C07.34 [Oryza sativa]
Length = 529
Score = 233 bits (594), Expect = 6e-60
Identities = 120/291 (41%), Positives = 183/291 (62%), Gaps = 6/291 (2%)
Query: 83 ARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIG 142
A AW +G DPR+ + A K+G AL L+SLL L+ F+ +G++++WA++TVV V EF+ G
Sbjct: 33 AHAWGIGREDPRRAIHALKVGTALTLVSLLYILEPLFKGVGKNAMWAVMTVVVVLEFTAG 92
Query: 143 ATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYAKLYPTMK 201
AT+ KGLNRGLGT+ AG LA + +++ +G+ + + V S+F++GF ATY + +P++K
Sbjct: 93 ATICKGLNRGLGTILAGSLAFIIELVAVRSGKVFRALFVGSSVFLIGFAATYLRFFPSIK 152
Query: 202 A-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIWAGED 260
Y+YG +FL+T+ I VS ++ + A R IA+G A+ L +++ + P W+GED
Sbjct: 153 KNYDYGVVIFLLTFNLITVSSFRQEDVVPLARDRLSTIAIGCAICLFMSLFVLPNWSGED 212
Query: 261 LHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQSTSTE 320
LH + F G+A S+E V Y + + KIL QAS ++ GYR+ L S S++
Sbjct: 213 LHSSTVRKFEGLARSIEACVTEY---FQDQDKDDKILDKQASRASIHIGYRAVLDSKSSD 269
Query: 321 DALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
+ L +A WEP H + YPW YVK+ LRH A+ V A+HGC+ SEI
Sbjct: 270 ETLAHYASWEPRHS-MQCYSYPWQKYVKIGSVLRHFAYTVAALHGCLESEI 319
>UniRef100_Q9LS22 Arabidopsis thaliana genomic DNA, chromosome 5, BAC clone:F10E10
[Arabidopsis thaliana]
Length = 543
Score = 231 bits (590), Expect = 2e-59
Identities = 117/295 (39%), Positives = 181/295 (60%), Gaps = 8/295 (2%)
Query: 79 KRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFE 138
K++ W++G DPR++ A K+G++L L+SLL ++ F+ +G ++WA++TVV V E
Sbjct: 33 KKILKNIWKVGKDDPRRVKHALKVGVSLTLVSLLYLMEPLFKGIGNSAIWAVMTVVVVLE 92
Query: 139 FSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYAKLY 197
FS GATL KGLNRGLGTL AG LA + ++ +G+ + I + ++FI+G TY +
Sbjct: 93 FSAGATLCKGLNRGLGTLIAGSLAFFIEFVANDSGKIFRAIFIGAAVFIIGALITYLRFI 152
Query: 198 PTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIW 256
P +K Y+YG +FL+T+ I VS Y+ I A RF IA+G + L +++ ++PIW
Sbjct: 153 PYIKKNYDYGMLIFLLTFNLITVSSYRVDTVIKIAHERFYTIAMGVGICLLMSLLVFPIW 212
Query: 257 AGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQS 316
+GEDLH G++ S+E VN Y +E T S+D +Y+GY++ L S
Sbjct: 213 SGEDLHKSTVAKLQGLSYSIEACVNEY-----FEEEEKDEETSDLSEDTIYNGYKTVLDS 267
Query: 317 TSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
S ++AL +A WEP H R+ ++PW +YVKV LR + V+A+HGC+ +EI
Sbjct: 268 KSADEALAMYASWEPRHTRH-CHRFPWKHYVKVGSVLRQFGYTVVALHGCLKTEI 321
>UniRef100_Q5XV16 Hypothetical protein [Arabidopsis thaliana]
Length = 543
Score = 229 bits (585), Expect = 7e-59
Identities = 116/295 (39%), Positives = 180/295 (60%), Gaps = 8/295 (2%)
Query: 79 KRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFE 138
K++ W++G DPR++ A K+G++L L+SLL ++ F+ +G ++WA++TVV V E
Sbjct: 33 KKILKNIWKVGKDDPRRVXHALKVGVSLTLVSLLYLMEPLFKGIGNSAIWAVMTVVVVLE 92
Query: 139 FSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYAKLY 197
FS GATL KGLN GLGTL AG LA + ++ +G+ + I + ++FI+G TY +
Sbjct: 93 FSXGATLCKGLNXGLGTLIAGSLAFFIEFVANDSGKIFRAIFIGAAVFIIGALITYLRFI 152
Query: 198 PTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIW 256
P +K Y+YG +FL+T+ I VS Y+ I A RF IA+G + L +++ ++PIW
Sbjct: 153 PYIKKNYDYGMLIFLLTFNLITVSSYRVDTVIKIAHERFYTIAMGVGICLLMSLLVFPIW 212
Query: 257 AGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQS 316
+GEDLH G++ S+E VN Y +E T S+D +Y+GY++ L S
Sbjct: 213 SGEDLHKSTVAKLQGLSYSIEACVNEY-----FEEEEKDEETSDLSEDTIYNGYKTVLDS 267
Query: 317 TSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
S ++AL +A WEP H R+ ++PW +YVKV LR + V+A+HGC+ +EI
Sbjct: 268 KSADEALAMYASWEPRHTRH-CHRFPWKHYVKVGSVLRQFGYTVVALHGCLKTEI 321
>UniRef100_O49696 Hypothetical protein AT4g17970 [Arabidopsis thaliana]
Length = 560
Score = 228 bits (580), Expect = 3e-58
Identities = 118/295 (40%), Positives = 180/295 (61%), Gaps = 11/295 (3%)
Query: 79 KRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFE 138
K++ R W +G DPR+++ A K+GL+L L+SLL ++ F+ +G +++WA++TVV V E
Sbjct: 31 KKIPKRLWNVGKEDPRRVIHALKVGLSLTLVSLLYLMEPLFKGIGSNAIWAVMTVVVVLE 90
Query: 139 FSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYAKLY 197
FS GATL KGLNRGLGTL AG LA + ++ +G+ I + ++FI+G ATY +
Sbjct: 91 FSAGATLCKGLNRGLGTLIAGSLAFFIEFVANDSGKVLRAIFIGTAVFIIGAAATYIRFI 150
Query: 198 PTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIW 256
P +K Y+YG +FL+T+ I VS Y+ I+ A RF IA+G + L +++ ++PIW
Sbjct: 151 PYIKKNYDYGVVIFLLTFNLITVSSYRVDSVINIAHDRFYTIAVGCGICLFMSLLVFPIW 210
Query: 257 AGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQS 316
+GEDLH G++ S+E V+ Y E E+ SK D +Y GY++ L S
Sbjct: 211 SGEDLHKTTVGKLQGLSRSIEACVDEYFEEKEKEKTDSK--------DRIYEGYQAVLDS 262
Query: 317 TSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
ST++ L +A WEP H + ++P YVKV LR + V+A+HGC+ +EI
Sbjct: 263 KSTDETLALYANWEPRH-TLRCHRFPCQQYVKVGAVLRQFGYTVVALHGCLQTEI 316
>UniRef100_Q9SRM9 T19F11.8 protein [Arabidopsis thaliana]
Length = 488
Score = 219 bits (557), Expect = 1e-55
Identities = 117/294 (39%), Positives = 179/294 (60%), Gaps = 14/294 (4%)
Query: 78 TKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVF 137
TKRV + DPR+I+ + K+G+AL L+SLL +++ + G +WAILTVV VF
Sbjct: 28 TKRVK-NVQKFAKDDPRRIIHSMKVGVALTLVSLLYYVRPLYISFGVTGMWAILTVVVVF 86
Query: 138 EFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAG-QWEEIIVMVSIFIVGFCATYAKL 196
EF++G TLSKGLNRG TL AG L VG L++ G Q E I++ + +F +G AT+++
Sbjct: 87 EFTVGGTLSKGLNRGFATLIAGALGVGAVHLARFFGHQGEPIVLGILVFSLGAAATFSRF 146
Query: 197 YPTMK-AYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPI 255
+P +K Y+YG +F++T+ ++ +SGY+T + + A R I +G + + V++ + P+
Sbjct: 147 FPRIKQRYDYGALIFILTFSFVAISGYRTDEILIMAYQRLSTILIGGTICILVSIFICPV 206
Query: 256 WAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQ 315
WAGEDLH ++A N +A LEG Y P KI +S Y+S L
Sbjct: 207 WAGEDLHKMIANNINKLAKYLEGFEGEYFQ-------PEKISKETSS---CVREYKSILT 256
Query: 316 STSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILS 369
S STED+L A WEP HGR++ L++PW Y+K++G +R CA + ++G +LS
Sbjct: 257 SKSTEDSLANLARWEPGHGRFR-LRHPWKKYLKIAGLVRQCAVHLEILNGYVLS 309
>UniRef100_Q9LS23 Arabidopsis thaliana genomic DNA, chromosome 5, BAC clone:F10E10
[Arabidopsis thaliana]
Length = 539
Score = 214 bits (546), Expect = 2e-54
Identities = 115/300 (38%), Positives = 177/300 (58%), Gaps = 9/300 (3%)
Query: 76 KTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVT 135
K K++ W +G DPR+++ A K+G+AL L+SLL ++ FE +G++++WA++TVV
Sbjct: 31 KKMKKILRNLWNVGKEDPRRVIHALKVGVALTLVSLLYLMEPFFEGVGKNALWAVMTVVV 90
Query: 136 VFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYA 194
V EFS GATL KGLNRGLGTL AG LA + ++ +G+ I + S+F +G TY
Sbjct: 91 VLEFSAGATLRKGLNRGLGTLIAGSLAFFIEWVAIHSGKILGGIFIGTSVFTIGSMITYM 150
Query: 195 KLYPTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLY 253
+ P +K Y+YG VFL+T+ I VS Y+ I A R I +G + L +++ +
Sbjct: 151 RFIPYIKKNYDYGMLVFLLTFNLITVSSYRVDTVIKIAHERLYTIGMGIGICLFMSLLFF 210
Query: 254 PIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASD--DVVYSGYR 311
PIW+G+DLH G++ +E V+ Y E++ + SD D++Y+GY
Sbjct: 211 PIWSGDDLHKSTITKLQGLSRCIEACVSEYFE----EKLKDNETSDSESDDEDLIYNGYN 266
Query: 312 SALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
+ L S S ++AL +A WEP H R + K+P Y+KV LR + V+A+HGC+ +EI
Sbjct: 267 TVLDSKSADEALAMYAKWEPRHTR-RCNKFPSQQYIKVGSVLRKFGYTVVALHGCLQTEI 325
>UniRef100_Q6EPG5 Aluminum-activated malate transporter-like [Oryza sativa]
Length = 488
Score = 214 bits (545), Expect = 3e-54
Identities = 108/309 (34%), Positives = 184/309 (58%), Gaps = 26/309 (8%)
Query: 74 AWKTTKRVAARA---------WEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGR 124
AW VA RA W++G DPR+ V + K+GLAL L+S++ + + ++ +G
Sbjct: 40 AWVVRMLVAVRAAVAGFARKVWKIGADDPRRAVHSLKVGLALTLVSIVYYTRPVYDGVGG 99
Query: 125 HSVWAILTVVTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMG-VLSKLAGQWEEIIVMVS 183
+++WA++TVV VFE+++G + KG NR + T +AG LA+G+ V K + E I+ S
Sbjct: 100 NAMWAVMTVVVVFEYTVGGCMYKGFNRAVATASAGLLALGVNWVADKSGDKLEPFILSGS 159
Query: 184 IFIVGFCATYAKLYPTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGA 242
+F++ AT+++ PT+KA ++YG +F++T+ + VSGY+ + +D A R I +G
Sbjct: 160 LFLLAAAATFSRFIPTVKARFDYGVTIFILTFSLVAVSGYRVDQLLDLAQQRMSTIGIGI 219
Query: 243 AVSLGVNVCLYPIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQAS 302
+ L V V ++P+WAG++LH L +N +A ++EG V +Y + P+ +
Sbjct: 220 VICLAVCVVIWPVWAGQELHLLTVRNMEKLAGAVEGCVEDY-----FAAKPAAAKS---- 270
Query: 303 DDVVYSGYRSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMA 362
GY+ L S ++ED+ A WEPPHGR+ ++P++ Y KV A+RHCA+ V A
Sbjct: 271 -----EGYKCVLNSKASEDSQANLARWEPPHGRFG-FRHPYAQYTKVGAAMRHCAYCVEA 324
Query: 363 MHGCILSEI 371
++ C+ +E+
Sbjct: 325 LNSCVRAEV 333
>UniRef100_Q5XV19 Hypothetical protein [Arabidopsis thaliana]
Length = 335
Score = 213 bits (541), Expect = 9e-54
Identities = 114/300 (38%), Positives = 176/300 (58%), Gaps = 9/300 (3%)
Query: 76 KTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVT 135
K K++ W +G DPR+++ A K+G+AL L+SLL ++ FE +G++++WA++TVV
Sbjct: 31 KKMKKILRNLWNVGKEDPRRVIHALKVGVALTLVSLLYLMEPFFEGVGKNALWAVMTVVV 90
Query: 136 VFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYA 194
V EFS GATL KGLNRGLGTL AG LA + ++ +G+ + S+F +G TY
Sbjct: 91 VLEFSAGATLRKGLNRGLGTLIAGSLAFFIEWVAIHSGKILGGXFIGTSVFTIGSMITYM 150
Query: 195 KLYPTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLY 253
+ P +K Y+YG VFL+T+ I VS Y+ I A R I +G + L +++ +
Sbjct: 151 RFIPYIKKNYDYGMLVFLLTFNLITVSSYRVDTVIKIAHERLYTIGMGIGICLFMSLLFF 210
Query: 254 PIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASD--DVVYSGYR 311
PIW+G+DLH G++ +E V+ Y E++ + SD D++Y+GY
Sbjct: 211 PIWSGDDLHKSTITKLQGLSRCIEACVSEYFE----EKLKDNETSDSESDDEDLIYNGYN 266
Query: 312 SALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
+ L S S ++AL +A WEP H R + K+P Y+KV LR + V+A+HGC+ +EI
Sbjct: 267 TVLDSKSADEALAMYAKWEPRHTR-RCNKFPSQQYIKVGSVLRKFGYTVVALHGCLQTEI 325
>UniRef100_Q5XV18 Hypothetical protein [Arabidopsis thaliana]
Length = 539
Score = 213 bits (541), Expect = 9e-54
Identities = 114/300 (38%), Positives = 176/300 (58%), Gaps = 9/300 (3%)
Query: 76 KTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVT 135
K K++ W +G DPR+++ A K+G+AL L+SLL ++ FE +G++++WA++TVV
Sbjct: 31 KKMKKILRNLWNVGKEDPRRVIHALKVGVALTLVSLLYLMEPFFEGVGKNALWAVMTVVV 90
Query: 136 VFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYA 194
V EFS GATL KGLNRGLGTL AG LA + ++ +G+ + S+F +G TY
Sbjct: 91 VLEFSAGATLRKGLNRGLGTLIAGSLAFFIEWVAIHSGKILGGXFIGTSVFTIGSMITYM 150
Query: 195 KLYPTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLY 253
+ P +K Y+YG VFL+T+ I VS Y+ I A R I +G + L +++ +
Sbjct: 151 RFIPYIKKNYDYGMLVFLLTFNLITVSSYRVDTVIKIAHERLYTIGMGIGICLFMSLLFF 210
Query: 254 PIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASD--DVVYSGYR 311
PIW+G+DLH G++ +E V+ Y E++ + SD D++Y+GY
Sbjct: 211 PIWSGDDLHKSTITKLQGLSRCIEACVSEYFE----EKLKDNETSDSESDDEDLIYNGYN 266
Query: 312 SALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
+ L S S ++AL +A WEP H R + K+P Y+KV LR + V+A+HGC+ +EI
Sbjct: 267 TVLDSKSADEALAMYAKWEPRHTR-RCNKFPSQQYIKVGSVLRKFGYTVVALHGCLQTEI 325
>UniRef100_Q69WI1 Putative aluminum-activated malate transporter [Oryza sativa]
Length = 668
Score = 212 bits (540), Expect = 1e-53
Identities = 108/329 (32%), Positives = 188/329 (56%), Gaps = 8/329 (2%)
Query: 43 PESDDDNDGPPGSKN--CCRFCSYRALSDRLVGAWKTTKRVAARAWEMGLSDPRKIVFAA 100
PE++ +++ G++ CC + + L WK R AW++G DPR++V
Sbjct: 26 PEAEVEHENAKGARRGCCCAPAAAAWVLWWLAAPWKWVARFGRTAWKVGADDPRRVVHGF 85
Query: 101 KMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIGATLSKGLNRGLGTLTAGG 160
K+ LAL L S +++ + G+ ++WA+LTVV VFE+++G + KGLNR + T+ G
Sbjct: 86 KVALALTLCSAFYYVRPLYVFTGQTAMWAVLTVVVVFEYTVGGCMYKGLNRAMATVAGGA 145
Query: 161 LAVGM-GVLSKLAGQWEEIIVMVSIFIVGFCATYAKLYPTMKA-YEYGFRVFLITYCYII 218
LA+G+ V K E ++ S+F++ A++++ PT+KA ++YG +F++TY +
Sbjct: 146 LALGVHWVADKSGDDAEPFVLTASLFVLAAAASFSRFIPTLKARFDYGVTIFILTYSLVA 205
Query: 219 VSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLHDLVAKNFMGVAASLEG 278
VSGY+ + A R + IA+GA + V ++P+WAG++LH LVA+N +AA++E
Sbjct: 206 VSGYRVDTLVTMAQQRLITIAIGAFICFAVCTLVFPVWAGQELHVLVARNMDKLAAAIEA 265
Query: 279 VVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQSTSTEDALMGFAVWEPPHGRYKM 338
V++Y + E+ T + GYR+ L + ++ED+L A WEP HG++
Sbjct: 266 CVDDYFSSAEHAGGGGDAATALSEK---ARGYRAVLNAKASEDSLANLARWEPGHGKFG- 321
Query: 339 LKYPWSNYVKVSGALRHCAFMVMAMHGCI 367
++P+ Y V A+R CA+ + A+ C+
Sbjct: 322 FRHPYGQYQNVGAAMRCCAYCIDALAACV 350
>UniRef100_Q8S2J6 OSJNBb0021A09.9 protein [Oryza sativa]
Length = 495
Score = 210 bits (534), Expect = 6e-53
Identities = 107/284 (37%), Positives = 166/284 (57%), Gaps = 10/284 (3%)
Query: 76 KTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVT 135
+ + AA AW D ++ FA K GLA++L SLL+ + PF G + +W+ILTV
Sbjct: 45 RCARAAAAAAWAFASEDAGRVAFALKAGLAMLLASLLVLVGEPFRLFGTNIIWSILTVGI 104
Query: 136 VFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEE-IIVMVSIFIVGFCATYA 194
+FE+++GA+ ++G NR +G++ AG +A+ + +S G E ++ +SIF+VG ++
Sbjct: 105 MFEYTVGASFNRGFNRAVGSMVAGVVAIAVIWISLRCGSVAEPYVIGLSIFLVGAVTSFV 164
Query: 195 KLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYP 254
K P + YEYGFRV L TYC I+VS Y+ G+ + + R IA+GA ++L VNV ++P
Sbjct: 165 KQLPALAPYEYGFRVILFTYCLIMVSVYRVGEPVAAGLDRLYAIAIGAVLALLVNVLIFP 224
Query: 255 IWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSAL 314
WAGE LH + +F VA SL V +YL+ E T + R+ L
Sbjct: 225 AWAGEQLHRELVASFAAVADSLHDCVRSYLSGDE---------TAVDGGEPAIEKCRAIL 275
Query: 315 QSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAF 358
+++ ++L A WEPPHGR++ +PWS+Y +V LRHCA+
Sbjct: 276 NASARIESLARSARWEPPHGRFRSFSFPWSHYARVGAVLRHCAY 319
>UniRef100_Q7XSA0 OSJNBa0005N02.7 protein [Oryza sativa]
Length = 513
Score = 209 bits (533), Expect = 8e-53
Identities = 111/311 (35%), Positives = 179/311 (56%), Gaps = 14/311 (4%)
Query: 71 LVGAWKTTKRVAA----------RAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFE 120
+V AW+ VAA R W++G DPR+ V K+GLAL L+S+ + + ++
Sbjct: 37 VVWAWQLVSCVAALGSRASGLAGRVWKIGADDPRRAVHGVKVGLALALVSVFYYTRPLYD 96
Query: 121 DLGRHSVWAILTVVTVFEFSIGATLSKGLNRGLGTLTAGGLAVGM-GVLSKLAGQWEEII 179
+G ++WA++TVV VFEF++G + KG NR T++AG +A+G+ + SK + E ++
Sbjct: 97 GVGGAAMWAVMTVVVVFEFTVGGCVYKGFNRATATVSAGAVALGVHWIASKSGDKLEPVV 156
Query: 180 VMVSIFIVGFCATYAKLYPTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLI 238
S+F++ AT+++ PT+KA ++YG +F++TY + VSGY+ + A R I
Sbjct: 157 RSGSVFLLAAAATFSRFIPTVKARFDYGVTIFILTYSLVAVSGYRVDALVAMAQQRVSTI 216
Query: 239 ALGAAVSLGVNVCLYPIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILT 298
A+G + L V + + P+WAG++LH L A+N +A ++E V Y E E +
Sbjct: 217 AIGIFICLAVCLLICPVWAGQELHRLTARNMDKLAGAVEACVEGYFVAGEEEAAGPEYKR 276
Query: 299 YQASDDVVYSGYRSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAF 358
A+ GY+ L S ++EDA A WEP HGR+ ++P++ Y V A+RHCA+
Sbjct: 277 RPAA-AAAAEGYKCVLNSKASEDAQANLARWEPAHGRFG-FRHPYAQYKAVGAAMRHCAY 334
Query: 359 MVMAMHGCILS 369
V A+ GCI S
Sbjct: 335 CVEALSGCIRS 345
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 614,653,117
Number of Sequences: 2790947
Number of extensions: 25159108
Number of successful extensions: 79036
Number of sequences better than 10.0: 144
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 109
Number of HSP's that attempted gapping in prelim test: 78883
Number of HSP's gapped (non-prelim): 157
length of query: 371
length of database: 848,049,833
effective HSP length: 129
effective length of query: 242
effective length of database: 488,017,670
effective search space: 118100276140
effective search space used: 118100276140
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0310b.11


