

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0302a.10
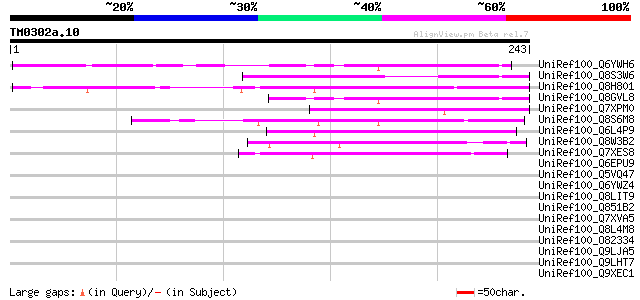
(243 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6YWH6 ATP synthase subunit-like protein [Oryza sativa] 70 6e-11
UniRef100_Q8S3W6 Hypothetical protein [Hordeum vulgare var. dist... 68 2e-10
UniRef100_Q8H801 Putative transposon protein [Oryza sativa] 66 7e-10
UniRef100_Q8GVL8 ATP synthase subunit-like protein [Oryza sativa] 59 8e-08
UniRef100_Q7XPM0 OSJNBa0085I10.6 protein [Oryza sativa] 59 1e-07
UniRef100_Q8S6M8 Putative TNP2-like transposon protein [Oryza sa... 58 2e-07
UniRef100_Q6L4P9 Hypothetical protein P0708D12.13 [Oryza sativa] 54 3e-06
UniRef100_Q8W3B2 Putative far-red impaired response protein [Ory... 47 6e-04
UniRef100_Q7XES8 Putative TNP-like transposable element [Oryza s... 46 7e-04
UniRef100_Q6EPU9 Hypothetical protein OSJNBa0016D04.28 [Oryza sa... 44 0.005
UniRef100_Q5VQ47 Hypothetical protein P0662B01.10 [Oryza sativa] 44 0.005
UniRef100_Q6YWZ4 Hypothetical protein OSJNBa0091D16.32 [Oryza sa... 43 0.006
UniRef100_Q8LIT9 Tnp1 protein-like [Oryza sativa] 43 0.006
UniRef100_Q851B2 Putative ATP synthase subunit [Oryza sativa] 43 0.006
UniRef100_Q7XVA5 OSJNBa0081G05.10 protein [Oryza sativa] 43 0.006
UniRef100_Q8L4M8 Hypothetical protein OSJNBa0079H13.19 [Oryza sa... 41 0.024
UniRef100_O82334 En/Spm-like transposon protein [Arabidopsis tha... 40 0.069
UniRef100_Q9LJA5 Arabidopsis thaliana genomic DNA, chromosome 3,... 40 0.069
UniRef100_Q9LHT7 En/Spm-like transposon protein [Arabidopsis tha... 39 0.090
UniRef100_Q9XEC1 Putative transposon protein [Arabidopsis thaliana] 39 0.12
>UniRef100_Q6YWH6 ATP synthase subunit-like protein [Oryza sativa]
Length = 364
Score = 69.7 bits (169), Expect = 6e-11
Identities = 55/235 (23%), Positives = 102/235 (43%), Gaps = 38/235 (16%)
Query: 2 KKAGRGKTTGTSVTKKKMKSATGKLPLCIPLDKMVAVGPGAPDFVTEISVLVKKKAPFNV 61
++ GRG T ++ K + + + K+ G F +E++V V+++ P V
Sbjct: 11 RRKGRGLTINGTLAKLRARGVPLDIQFAAQFGKVC--GRHTSVFKSEVTVCVRQEVPLKV 68
Query: 62 KGWKKVPDTAKDHILSQVFVNIII*FIFLFSFVLIQYLQYISCNCWVQDTFDIDQTEHNK 121
K WK + + A +S ++ + F I Y
Sbjct: 69 KKWKVI-EKAFPGTMSSIWN------LLKAKFPKISMADY-------------------- 101
Query: 122 EVILNTAKRLYGSHRGKLHEHFKKYETNEMALEHKPDEISEEDWEHLVDK-FSSPTYKEM 180
+ ++ +R Y R KL +K Y T + P ++ EDW+ L+D +S +++
Sbjct: 102 QCVMTQVERQYNVRRHKL---YKTYCTTKQC----PSHVAPEDWQWLIDNLWSDEQFQKR 154
Query: 181 SARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHTNNNGEW 235
S +N N+SKQ + G KS ++++ R+P T + P+ D+W+ T+ NG W
Sbjct: 155 SKQNSINRSKQEMKSHVGTKSIVQIAHELRNPVTGEWPSAIDVWKATYL-KNGTW 208
>UniRef100_Q8S3W6 Hypothetical protein [Hordeum vulgare var. distichum]
Length = 427
Score = 67.8 bits (164), Expect = 2e-10
Identities = 38/134 (28%), Positives = 61/134 (45%), Gaps = 25/134 (18%)
Query: 110 DTFDIDQTEHNKEVILNTAKRLYGSHRGKLHEHFKKYETNEMALEHKPDEISEEDWEHLV 169
D +D++ T +E +L K Y R L +K Y+T+ L + P+++ E+WE ++
Sbjct: 104 DRWDLEVTPDTEEKVLKITKERYRGWRSTLSSTYKAYKTDTARLANLPEDLQPEEWEWMI 163
Query: 170 DKFSSPTYKEMSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHT 229
F + FQ +S R+PET +EP+ LW +THT
Sbjct: 164 QYFGKDS------------------------KFQVISKTKRNPETGEEPDCITLWELTHT 199
Query: 230 NNNGEWVNEASREV 243
NG W N S++V
Sbjct: 200 -KNGIWSNTESQDV 212
>UniRef100_Q8H801 Putative transposon protein [Oryza sativa]
Length = 1620
Score = 66.2 bits (160), Expect = 7e-10
Identities = 61/246 (24%), Positives = 103/246 (41%), Gaps = 43/246 (17%)
Query: 2 KKAGRGKTTGTSVTKKKMKSATGKLPLCIPLDKM-VAVGPGAPDFVTEISVLVKKKAPFN 60
KK GRG T K + S G+ + I L++ VG + +F T + V+KK P
Sbjct: 1085 KKEGRGITQ-----KLNIISRVGEAKIKITLNEFGQPVGLDSEEFATTVGTFVRKKIPVA 1139
Query: 61 VKGWKKVPDTAKDHILSQVFVNIII*FIFLFSFVLIQYLQYISCNCW--VQDTFDIDQTE 118
W+ V KD + W VQ ++I+ E
Sbjct: 1140 CGDWRDVD--VKDKL-----------------------------KVWEDVQKHYEIN--E 1166
Query: 119 HNKEVILNTAKRLYGSHRGKLHE-HFKKYETNEMALEHKPDEISEEDWEHLVDKFSSPTY 177
+ +L T+ ++ ++ L + HF T+E ++ + ++E W+ L++ + SP
Sbjct: 1167 YGLHFVLETSHMIWKDYKADLKKKHFDANLTDEELMDRRDLRVNEAQWKWLINHWRSPEA 1226
Query: 178 KEMSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHTNNNGEWVN 237
S R KAN+ + H G KS V +D +T + P +++ TH NGE +
Sbjct: 1227 VARSIRGKANRGMLRMLHTAGSKSHARVGHD-MGVKTGRPPRRDEVFVETHKRKNGEIIP 1285
Query: 238 EASREV 243
EA+ V
Sbjct: 1286 EAAETV 1291
>UniRef100_Q8GVL8 ATP synthase subunit-like protein [Oryza sativa]
Length = 267
Score = 59.3 bits (142), Expect = 8e-08
Identities = 34/123 (27%), Positives = 62/123 (49%), Gaps = 9/123 (7%)
Query: 122 EVILNTAKRLYGSHRGKLHEHFKKYETNEMALEHKPDEISEEDWEHLVDK-FSSPTYKEM 180
+ ++ +R Y R KL +K Y T + P ++ EDW+ L+D +S +++
Sbjct: 5 QCVMTQVERQYNVRRHKL---YKTYCTTKQC----PSHVAPEDWQWLIDNLWSDEQFQKR 57
Query: 181 SARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHTNNNGEWVNEAS 240
S +N N+SKQ + G KS ++++ R+P T + + D+W+ T+ NG W
Sbjct: 58 SKQNSINRSKQEMKSHVGTKSIVQIAHELRNPVTGEWTSAIDVWKATYL-KNGTWSVSNG 116
Query: 241 REV 243
E+
Sbjct: 117 EEI 119
>UniRef100_Q7XPM0 OSJNBa0085I10.6 protein [Oryza sativa]
Length = 969
Score = 58.9 bits (141), Expect = 1e-07
Identities = 29/104 (27%), Positives = 56/104 (52%), Gaps = 1/104 (0%)
Query: 141 EHFKKYETNEMALEHKPDEISEEDWEHLVDKFSSPTYKEMSARNKANKSKQVINHRCGRK 200
+H+ +ET E L + + +E W++LV + + K +S+RNKA ++ H G K
Sbjct: 721 KHYDTHETVEECLADQNPRVLKEQWQYLVAYWGTEKAKAVSSRNKACRANVTATHTAGTK 780
Query: 201 SF-QAVSYDARDPETQKEPNYQDLWRMTHTNNNGEWVNEASREV 243
SF + + + + +EP DL+ +THT+ NG+ + + ++
Sbjct: 781 SFARIIEEEKQKRPNNEEPTAADLFLLTHTHRNGKPMKKEKADI 824
>UniRef100_Q8S6M8 Putative TNP2-like transposon protein [Oryza sativa]
Length = 1571
Score = 57.8 bits (138), Expect = 2e-07
Identities = 52/187 (27%), Positives = 83/187 (43%), Gaps = 30/187 (16%)
Query: 58 PFNVKGWKKVPDTAKDHILSQVFVNIII*FIFLFSFVLIQYLQYISCNCWVQDTFDID-Q 116
P VK W + +T ++HI FV+I F + D D +
Sbjct: 1283 PVKVKKWSDISNTEQEHI----FVSITDKF----------------------ENEDPDIK 1316
Query: 117 TEHNKEVILNTAKRLYGSHRGKLHEHF-KKYETNEMALEHKPDEISEEDWEHLVDK-FSS 174
+ K+ I+ AK L+ + RG L HF K +T + A+++KP + DW+ LV + F S
Sbjct: 1317 IDVYKDEIMEHAKSLWINWRGDLDRHFVKPAKTMQQAIKNKPKDFDPSDWQWLVQEHFYS 1376
Query: 175 PTYKEMSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHTNNNGE 234
+ MS RN N++ + HR G K F + Y+ E + P +L+ T +
Sbjct: 1377 KDFIAMSLRNSQNRAGLKMPHRTGSKPFIEIIYNMGGKE-NRPPTLDELFFETRKKGDTL 1435
Query: 235 WVNEASR 241
E+SR
Sbjct: 1436 VDVESSR 1442
>UniRef100_Q6L4P9 Hypothetical protein P0708D12.13 [Oryza sativa]
Length = 196
Score = 54.3 bits (129), Expect = 3e-06
Identities = 31/118 (26%), Positives = 56/118 (47%), Gaps = 1/118 (0%)
Query: 121 KEVILNTAKRLYGSHRGKLHE-HFKKYETNEMALEHKPDEISEEDWEHLVDKFSSPTYKE 179
+E IL T + + H+ KL E +F ++ E + P ++ + W L++ + S K+
Sbjct: 18 EEFILKTIRERWRGHKAKLKEKYFDVNKSKEANCNNVPADVLPDQWIALINHWMSEKSKK 77
Query: 180 MSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHTNNNGEWVN 237
+S +NK N K+ H G SF + R + K P+ ++ TH G+ +N
Sbjct: 78 ISKQNKENCRKKKALHTAGTNSFARTREEMRQKDPAKNPHRAIVYIQTHKRKTGKNLN 135
>UniRef100_Q8W3B2 Putative far-red impaired response protein [Oryza sativa]
Length = 515
Score = 46.6 bits (109), Expect = 6e-04
Identities = 38/134 (28%), Positives = 60/134 (44%), Gaps = 11/134 (8%)
Query: 112 FDIDQTEHN-KEVILNTAKRLYGSHRGKLHEHFKKYETNEMAL--EHKPDEISEEDWEHL 168
FD+D E N K + Y S R + +EH+ K+ E A HK E DW L
Sbjct: 17 FDLDLNEENVKGCVELMFSSSYKSFRHRFYEHYVKHGGGENARMNPHKALEDRLADWFWL 76
Query: 169 VDKFSSPTYKEMSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTH 228
D F + +++ S K N+S H G KSF A+ ++ + L++ +
Sbjct: 77 CDHFETEEFQKRSTIGKDNRSNLPYMHWKGTKSFVALQHELGCGDI-------TLYKECY 129
Query: 229 TNNNGEWVNEASRE 242
+N+ G W + +RE
Sbjct: 130 SNDKG-WASSDARE 142
>UniRef100_Q7XES8 Putative TNP-like transposable element [Oryza sativa]
Length = 1522
Score = 46.2 bits (108), Expect = 7e-04
Identities = 34/127 (26%), Positives = 65/127 (50%), Gaps = 4/127 (3%)
Query: 108 VQDTFDIDQTEHNKEVILNTAKRLYGSHRGKLH-EHFKKYETNEMALEHKPDEISEEDWE 166
V+ FD+D E KE ++ +A + + + +L ++F +TN+ + IS + W+
Sbjct: 1095 VKRYFDLD--ECLKEYVMRSAHKKWKDFKNRLKAKYFDPEKTNKELWKKHDSRISLKVWK 1152
Query: 167 HLVDKFSSPTYKEMSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRM 226
LV + S K S R KAN++K V H G +S V +D + +K + +++++
Sbjct: 1153 WLVCFWRSEKGKARSNRGKANRAKMVAVHTIGTRSLANVRHDMEKRKGRK-VSRAEVFKV 1211
Query: 227 THTNNNG 233
+T +G
Sbjct: 1212 AYTRKDG 1218
>UniRef100_Q6EPU9 Hypothetical protein OSJNBa0016D04.28 [Oryza sativa]
Length = 644
Score = 43.5 bits (101), Expect = 0.005
Identities = 20/69 (28%), Positives = 34/69 (48%)
Query: 160 ISEEDWEHLVDKFSSPTYKEMSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPN 219
+S E W L++ + SP EM +NK N+ +H G +++ + D +EPN
Sbjct: 294 MSNEQWTQLLESWKSPQKMEMCQKNKENRGNVKYHHTTGSRAYMVHVENLDDKYNDEEPN 353
Query: 220 YQDLWRMTH 228
DL++ H
Sbjct: 354 AFDLFKEFH 362
>UniRef100_Q5VQ47 Hypothetical protein P0662B01.10 [Oryza sativa]
Length = 613
Score = 43.5 bits (101), Expect = 0.005
Identities = 53/226 (23%), Positives = 86/226 (37%), Gaps = 32/226 (14%)
Query: 6 RGKTTGTSVTKKKMKSATGKLPLCIPLDKMVAVGPG-APDFVTEISVLVKKKAPFNVKGW 64
RG G + ++ +S GKLP+ I KM A F TE ++ V+ P K W
Sbjct: 273 RGPNVGRGL-ERMSRSKKGKLPVVIEPGKMRPNSVKIAAKFATECNIAVRNHVPIFPK-W 330
Query: 65 KKVPDTAKDHILSQVFVNIII*FIFLFSFVLIQYLQYISCNCWVQDTFDIDQTEHN-KEV 123
K D K L + ++N V FD+D T ++
Sbjct: 331 KDYKDNKK---LLRFYINK------------------------VGSKFDMDTTAGPVRKA 363
Query: 124 ILNTAKRLYGSHRGKLHEHFKKYETNEMALEHKPDE-ISEEDWEHLVDKFSSPTYKEMSA 182
+ KR R KL + + + L+ P +S+ W LV+ + +
Sbjct: 364 CVAMMKRAVCQQRHKLKKKYFDPYPLHLVLKTSPVTCMSDLQWLQLVEHWKDEKKMLICE 423
Query: 183 RNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTH 228
+NKAN++K + G +S+ + D PN DL++ H
Sbjct: 424 KNKANRAKVQFHQTTGSRSYDMHIIELGDKYKDAPPNGLDLFKELH 469
>UniRef100_Q6YWZ4 Hypothetical protein OSJNBa0091D16.32 [Oryza sativa]
Length = 274
Score = 43.1 bits (100), Expect = 0.006
Identities = 23/102 (22%), Positives = 50/102 (48%), Gaps = 1/102 (0%)
Query: 141 EHFKKYETNEMALEHKPDEISEEDWEHLVDKFSSPTYKEMSARNKANKSKQVINHRCGRK 200
++F NE+ +++ +W+ LVD +S+P +KE +NK ++ + G +
Sbjct: 44 KYFNGIPANEVRTTSPLSSMTDNEWKQLVDMWSTPKHKEKCIKNKDSRELVQYHQMTGSR 103
Query: 201 SFQAVSYDARDPETQKEPNYQDLWRMTHTNNNGEWVNEASRE 242
S+ A Y + P D+++ TH ++ + NE +++
Sbjct: 104 SYVAQCYVMQTKFKDVPPTAIDIFKDTHCSSKSGF-NENAKD 144
>UniRef100_Q8LIT9 Tnp1 protein-like [Oryza sativa]
Length = 436
Score = 43.1 bits (100), Expect = 0.006
Identities = 35/138 (25%), Positives = 61/138 (43%), Gaps = 9/138 (6%)
Query: 108 VQDTFDIDQTEHNKEVILNTAKRLYGSHRGKLHE-HFKKYETNEMALEHKPDEISEEDWE 166
V+ F ID +N L+ + ++ KL + ++K + E L+ P ++E W
Sbjct: 159 VERKFAIDGRANNWN--LHQLDGKWRQYKSKLKKGYYKPNLSMERVLQTMPKTVAESQWA 216
Query: 167 HLVDKFSSPTYKEMSARNKANKSKQVINHRCGRKSF--QAVSYDARDPETQKEPNYQDLW 224
LV + S K++S +NK N H GRKSF + + E + + +
Sbjct: 217 TLVSYWYSEDSKKISDKNKENAQNIKHPHTLGRKSFARKRKELEVNGVEVDRATFFDE-- 274
Query: 225 RMTHTNNNGEWVNEASRE 242
H +G +VN+A+ E
Sbjct: 275 --CHKTKDGRYVNDATEE 290
>UniRef100_Q851B2 Putative ATP synthase subunit [Oryza sativa]
Length = 357
Score = 43.1 bits (100), Expect = 0.006
Identities = 17/58 (29%), Positives = 34/58 (58%)
Query: 178 KEMSARNKANKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHTNNNGEW 235
++ S +N AN++KQ + + G KS ++++ R+ ET + P +W+ T+ +G W
Sbjct: 135 QKRSNQNAANRAKQEMGSKVGTKSIAQIAHELRNKETGEWPTTMQVWKATYQKADGTW 192
>UniRef100_Q7XVA5 OSJNBa0081G05.10 protein [Oryza sativa]
Length = 1490
Score = 43.1 bits (100), Expect = 0.006
Identities = 29/119 (24%), Positives = 50/119 (41%), Gaps = 34/119 (28%)
Query: 58 PFNVKGWKKVPDTAKDHILSQVFVNIII*FIFLFSFVLIQYLQYISCNCWVQDTFDIDQT 117
P VK W + +T ++HI + + D F+ +
Sbjct: 1233 PIKVKKWSDISNTEQEHIFASI-----------------------------TDKFENEDP 1263
Query: 118 EHN----KEVILNTAKRLYGSHRGKLHEHF-KKYETNEMALEHKPDEISEEDWEHLVDK 171
+ K+ I+ AK L+ + RG LH HF K +T + A+++KP + DW+ LV +
Sbjct: 1264 DIKIDVYKDEIMEHAKSLWINWRGDLHRHFVKPAKTMQQAIKNKPKDFDPSDWQWLVQE 1322
>UniRef100_Q8L4M8 Hypothetical protein OSJNBa0079H13.19 [Oryza sativa]
Length = 275
Score = 41.2 bits (95), Expect = 0.024
Identities = 23/103 (22%), Positives = 52/103 (50%), Gaps = 2/103 (1%)
Query: 141 EHFKKYETNEMALEHKPDEISEEDWEHLVDKFSSPTYKEMSARNKANKSKQVINHRCGRK 200
++F NE+ +++ +W+ LVD +S+P +KE +NK ++ + G +
Sbjct: 44 KYFNGIPANEVRTTSPLSSMTDNEWKQLVDMWSTPKHKEKCIKNKDSRELVQYHQMTGSR 103
Query: 201 SFQAVSYDARDPETQK-EPNYQDLWRMTHTNNNGEWVNEASRE 242
S+ A Y + + + P D+++ TH ++ + NE +++
Sbjct: 104 SYVAQCYVMKQTKFKDVPPTAIDIFKDTHCSSKSGF-NENAKD 145
>UniRef100_O82334 En/Spm-like transposon protein [Arabidopsis thaliana]
Length = 771
Score = 39.7 bits (91), Expect = 0.069
Identities = 26/145 (17%), Positives = 57/145 (38%), Gaps = 30/145 (20%)
Query: 38 VGPGAPDFVTEISVLVKKKAPFNVKGWKKVPDTAKDHILSQVFVNIII*FIFLFSFVLIQ 97
+G G+ + LV++ P + W K+ + + + V
Sbjct: 212 IGKGSVKLASYAGALVREHVPITINRWTKIGEEIRTLLWKSV------------------ 253
Query: 98 YLQYISCNCWVQDTFDIDQTEHNKEVILNTAKRLYGSHRGKLHEHFKKYETNEMALEHKP 157
Q F++D+ E+ K +L L+ S + + F++ +TN+ + +P
Sbjct: 254 -----------QAKFELDE-EYQKVAVLKQMGCLWRSWKSRQVTKFREAKTNQQRMNLRP 301
Query: 158 DEISEEDWEHLVDKFSSPTYKEMSA 182
+S +W V +SP +K++ +
Sbjct: 302 KNVSPFEWRKFVKSKTSPEFKKIGS 326
>UniRef100_Q9LJA5 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MTO24
[Arabidopsis thaliana]
Length = 317
Score = 39.7 bits (91), Expect = 0.069
Identities = 42/166 (25%), Positives = 71/166 (42%), Gaps = 23/166 (13%)
Query: 74 HILSQVFVNIII*FIFLFSFVLIQYLQYISCNCWVQDTFDIDQTEHNKEVILNTAKRLYG 133
HI++++ +N II F C QD F +N++ ++
Sbjct: 5 HIINKININKIISFF---------------CGQVWQDGFGRTLLSNNEKKKSRRRRKYVL 49
Query: 134 SHRG----KLHEH-FKKYETN--EMALEHKPDEISEEDWEHLVDKFSSPTYKEMSARNKA 186
S G L +H + KY N E A E + I E+ W V+ + K++ RN
Sbjct: 50 SALGGRCKDLKQHIWMKYRRNTIEKAFEARRGLIPEDQWREFVEIQFTDKAKKVRERNIN 109
Query: 187 NKSKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHTNNN 232
+++ I H RKS A + + ET KEPN +L+ ++ T ++
Sbjct: 110 SRNNHKIPHTLERKSL-ARKPNEMEEETSKEPNRAELFIVSRTKSD 154
>UniRef100_Q9LHT7 En/Spm-like transposon protein [Arabidopsis thaliana]
Length = 433
Score = 39.3 bits (90), Expect = 0.090
Identities = 27/115 (23%), Positives = 54/115 (46%), Gaps = 7/115 (6%)
Query: 136 RGKLHEHFKKYETN--EMALEHKPDEISEEDWEHLVDKFSSPTYKEMS-----ARNKANK 188
+ ++ + +++ N E E KP I + W+ LV + P ++ S AR
Sbjct: 155 KSRMTDQVSRWKGNWKEKGDEAKPKWIDPDVWKGLVLFWQDPKSEKKSNNSRNARYHDPD 214
Query: 189 SKQVINHRCGRKSFQAVSYDARDPETQKEPNYQDLWRMTHTNNNGEWVNEASREV 243
K + HR G+ S++A + + +K P++ +L TH +G +++ S E+
Sbjct: 215 GKGIYKHRSGQTSYKARARKRCEKTGEKTPDFLELLDETHRKADGSFIDGKSEEI 269
>UniRef100_Q9XEC1 Putative transposon protein [Arabidopsis thaliana]
Length = 350
Score = 38.9 bits (89), Expect = 0.12
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 5/95 (5%)
Query: 154 EHKPDEISEEDWEHLVDKFSSPTYKEMS-----ARNKANKSKQVINHRCGRKSFQAVSYD 208
E KP I + W+ LV + P ++ S AR K + HR G+ S++A +
Sbjct: 129 EAKPKWIDPDVWKGLVQFWQDPKSEKKSNNSRNARYHDLDGKDIYKHRSGQTSYKARARK 188
Query: 209 ARDPETQKEPNYQDLWRMTHTNNNGEWVNEASREV 243
+ + P++ +L TH +G +++ S+E+
Sbjct: 189 RCEKTGETTPDFLELLDETHRKADGTFIDGKSKEI 223
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.133 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 420,271,489
Number of Sequences: 2790947
Number of extensions: 16946378
Number of successful extensions: 48915
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 48864
Number of HSP's gapped (non-prelim): 77
length of query: 243
length of database: 848,049,833
effective HSP length: 124
effective length of query: 119
effective length of database: 501,972,405
effective search space: 59734716195
effective search space used: 59734716195
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0302a.10


