

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0301c.1
(229 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
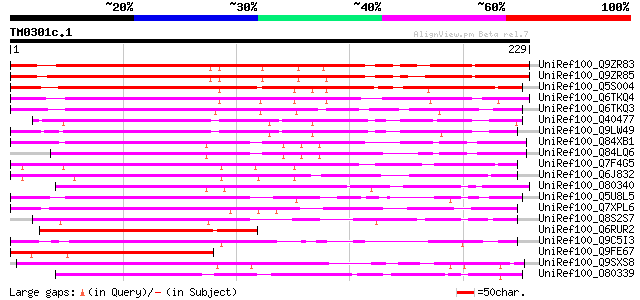
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZR83 EREBP-3 homolog [Stylosanthes hamata] 261 1e-68
UniRef100_Q9ZR85 Ethylene-responsive element binding protein hom... 254 9e-67
UniRef100_Q5S004 Ethylene response factor 3 [Cucumis sativus] 223 4e-57
UniRef100_Q6TKQ4 Putative ethylene response factor ERF3a [Vitis ... 214 2e-54
UniRef100_Q6TKQ3 Putative ethylene response factor ERF3b [Vitis ... 192 7e-48
UniRef100_Q40477 Ethylene-responsive transcription factor 4 [Nic... 181 1e-44
UniRef100_Q9LW49 Ethylene-responsive transcription factor 4 [Nic... 174 2e-42
UniRef100_Q84XB1 Ethylene response factor 3 [Lycopersicon escule... 172 8e-42
UniRef100_Q84LQ6 Ethylene response factor 2 [Lycopersicon escule... 166 4e-40
UniRef100_Q7F4G5 Ethylene responsive element binding factor3 [Or... 165 7e-40
UniRef100_Q6J832 Ethylene-responsive element binding factor [Ory... 163 4e-39
UniRef100_O80340 Ethylene-responsive transcription factor 4 [Ara... 161 1e-38
UniRef100_Q5U8L5 AP2/EREBP transcription factor ERF-1 [Gossypium... 158 1e-37
UniRef100_Q7XPL6 OSJNBa0085I10.10 protein [Oryza sativa] 145 6e-34
UniRef100_Q8S2S7 Ethylene responsive element binding factor 4-li... 144 1e-33
UniRef100_Q6RUR2 Ethylene-responsive element binding factor [Cap... 137 2e-31
UniRef100_Q9C5I3 Ethylene-responsive transcription factor 11 [Ar... 136 5e-31
UniRef100_Q9FE67 Ethylene-responsive transcription factor 9 [Ara... 133 3e-30
UniRef100_Q9SXS8 Ethylene-responsive transcription factor 3 [Nic... 128 1e-28
UniRef100_O80339 Ethylene-responsive transcription factor 3 [Ara... 124 2e-27
>UniRef100_Q9ZR83 EREBP-3 homolog [Stylosanthes hamata]
Length = 239
Score = 261 bits (666), Expect = 1e-68
Identities = 160/260 (61%), Positives = 175/260 (66%), Gaps = 52/260 (20%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAPR+KT AVKV N GVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT
Sbjct: 1 MAPREKTPAVKV--------NAGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 52
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFP-SDNV------------KNQSPSQSSTVESSSR 107
AE+AARAYD AAREFRGPKAKTNFPFP SD++ N+SPSQSSTVESSSR
Sbjct: 53 AEDAARAYDAAAREFRGPKAKTNFPFPDSDDINSNNNNIVVVKNNNRSPSQSSTVESSSR 112
Query: 108 DRD-------VAAAADSSSLDLNLAP-------SVRFPFRPQFP----QMPAANQVLFFD 149
DRD A ADSS LDLNLAP S+RFPF+ F MPAA Q L+ D
Sbjct: 113 DRDSYSAAAAATAVADSSPLDLNLAPAGAGFAGSIRFPFQQPFAVFPGGMPAAKQALYLD 172
Query: 150 AILRAGMVGGSPASQRYGFDFHTYNHPVAASEIQAVAGAQSDSDSSSVIDLNHYESGVVI 209
A+LRA M + ++GF YN P AA+ AGAQSDSDSSSVIDLN E G V
Sbjct: 173 AVLRASMA----SHGQFGFG---YNRPAAAA-----AGAQSDSDSSSVIDLNQNE-GDVA 219
Query: 210 KAGGRNFDLDLNYPPMEDMA 229
K GR LDLN PP ++MA
Sbjct: 220 KNNGRGLVLDLNEPPPQEMA 239
>UniRef100_Q9ZR85 Ethylene-responsive element binding protein homolog [Stylosanthes
hamata]
Length = 238
Score = 254 bits (650), Expect = 9e-67
Identities = 158/257 (61%), Positives = 170/257 (65%), Gaps = 47/257 (18%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAPR+KT AVKV NG N GVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT
Sbjct: 1 MAPREKTPAVKV----NGKSNAGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 56
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFP-SDNV----------KNQSPSQSSTVESSSRDR 109
AE+AARAYD AAREFRGPKAKTNFPFP SD++ N+SPSQSSTVESSSRDR
Sbjct: 57 AEDAARAYDAAAREFRGPKAKTNFPFPDSDDINNNNIVVVKNNNRSPSQSSTVESSSRDR 116
Query: 110 D------VAAAADSSSLDLNLAP---SVRFPFRPQFPQ--------MPAANQVLFFDAIL 152
D A ADSS LDLNLAP PF Q MPAA Q L+ DA+L
Sbjct: 117 DSYSAAAATAVADSSPLDLNLAPQELDSPDPFGSHSTQPFAVFPGGMPAAKQALYLDAVL 176
Query: 153 RAGMVGGSPASQRYGFDFHTYNHPVAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAG 212
RA M + ++GF YN P A AGAQSDSDSSSVIDLN E G V K
Sbjct: 177 RASMA----SHGQFGFG---YNRP-------AAAGAQSDSDSSSVIDLNQNE-GDVAKNN 221
Query: 213 GRNFDLDLNYPPMEDMA 229
GR LDLN PP ++MA
Sbjct: 222 GRGLVLDLNEPPPQEMA 238
>UniRef100_Q5S004 Ethylene response factor 3 [Cucumis sativus]
Length = 246
Score = 223 bits (567), Expect = 4e-57
Identities = 144/259 (55%), Positives = 165/259 (63%), Gaps = 48/259 (18%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAPR+K AVK S G VKEVHFRGVRKRPWGRYAAEIRDP KKSRVWLGTFDT
Sbjct: 1 MAPREKAVAVKPSV-------GNVKEVHFRGVRKRPWGRYAAEIRDPSKKSRVWLGTFDT 53
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFPSDNV------------KNQSPSQSSTVESSSRD 108
AEEAARAYD+AAR+FRG KAKTNFP PSD+ NQSPSQSSTVESSSR+
Sbjct: 54 AEEAARAYDSAARDFRGVKAKTNFPLPSDDQLLNLNNKINNINNNQSPSQSSTVESSSRE 113
Query: 109 RDVAAAADSSSLDLNL---------APSVRFPF-RPQFPQ--------MPAANQVLFFDA 150
+ A DSS L+LNL A + FPF R Q P +P +N VL+FDA
Sbjct: 114 Q--ALMVDSSPLNLNLGHGIGGLTNAGPISFPFQRYQIPMIGEVFTRGIPPSNHVLYFDA 171
Query: 151 ILRAGMVGGSPASQRYGFDFHTYNHPVAASEIQ---AVAGAQSDSDSSSVIDLNHYESGV 207
LRAGM+ P +QR FD A S+ + A +G QSDSDSSSV+D+N +
Sbjct: 172 ALRAGMINSHP-NQRLHFD----RIREAVSDFRREFAGSGVQSDSDSSSVVDMNGQDLKP 226
Query: 208 VIKAGGRNFDLDLNYPPME 226
+GGR DLDLN+PP E
Sbjct: 227 RGGSGGR-LDLDLNFPPPE 244
>UniRef100_Q6TKQ4 Putative ethylene response factor ERF3a [Vitis aestivalis]
Length = 249
Score = 214 bits (544), Expect = 2e-54
Identities = 139/270 (51%), Positives = 160/270 (58%), Gaps = 62/270 (22%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAP++K VK S N KEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT
Sbjct: 1 MAPKEKVAGVKPSANA--------KEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 52
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFPSDNV--KNQSPSQSSTVESSSRDR-DVAAAADS 117
AEEAA+AYD+AAREFRG KAKTNFP S+N+ NQSPSQSSTVESSSR+ A DS
Sbjct: 53 AEEAAKAYDSAAREFRGAKAKTNFPLVSENLNNNNQSPSQSSTVESSSREGFSPALMVDS 112
Query: 118 SSLDLNL--------------------APSVRFPFRPQFPQ---------------MPAA 142
S LDLNL A ++RFPF+ Q +PAA
Sbjct: 113 SPLDLNLLHGGGVGVGVGVGVGAAAGYATAMRFPFQHHQFQVSSPSPAAGIVPTGGLPAA 172
Query: 143 NQVLFFDAILRAGMVGGSPASQRYGFDFHTYNHPVAASEIQA--VAGAQSDSDSSSVIDL 200
N + +FDA+LR G R DF AAS+ +A G QSDSDSSSV+DL
Sbjct: 173 NHLFYFDAMLRTG---------RVNQDFQRLRFDRAASDFRAALTGGVQSDSDSSSVVDL 223
Query: 201 NHYESGVVIKAGGR-NFDLDLNYPPMEDMA 229
NH + +K R DLDLN PP ++A
Sbjct: 224 NHND----LKPRARVLIDLDLNRPPPPEIA 249
>UniRef100_Q6TKQ3 Putative ethylene response factor ERF3b [Vitis aestivalis]
Length = 220
Score = 192 bits (487), Expect = 7e-48
Identities = 123/242 (50%), Positives = 143/242 (58%), Gaps = 40/242 (16%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAPRDK V GN KE+ +RGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT
Sbjct: 1 MAPRDKPTGVTAGATGN-------KEIRYRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 53
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFPSD------NVKNQSPSQSSTVESSSRDR-DVAA 113
AEEAARAYD AAREFRG KAKTNFP P+D N+SPSQSSTVESSSR+ A
Sbjct: 54 AEEAARAYDAAAREFRGAKAKTNFPSPTDLAAAAATTANRSPSQSSTVESSSREALSPGA 113
Query: 114 AADSSSLDLNLA--------PSVRFPFRPQFPQMPAANQVLFFDAILRAGMVGGSPASQR 165
A +LDLNL+ +VR+P F P A + FF+ R P + R
Sbjct: 114 IAGPPALDLNLSHPAAAGQFSAVRYPAVGVF---PIAQPLFFFEPFSRP----EKPKTHR 166
Query: 166 YGFDFHTYNHPVAASEIQAVAGA-QSDSDSSSVIDLNHYESGVVIKAGGRNFDLDLNYPP 224
FD A A+AG+ SDSDSSSV+D N+++ R +LDLN+PP
Sbjct: 167 DMFDLDR----AVADFHPAIAGSVHSDSDSSSVVDFNYHDRST------RLLNLDLNHPP 216
Query: 225 ME 226
E
Sbjct: 217 AE 218
>UniRef100_Q40477 Ethylene-responsive transcription factor 4 [Nicotiana tabacum]
Length = 225
Score = 181 bits (459), Expect = 1e-44
Identities = 122/241 (50%), Positives = 143/241 (58%), Gaps = 47/241 (19%)
Query: 11 KVSGNGNGNGNG----GVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAAR 66
KVS NGN G GVKEVH+RGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAA+
Sbjct: 6 KVS-NGNLKGGNVKTDGVKEVHYRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAAK 64
Query: 67 AYDTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVESSSRDRDVAA------------- 113
AYDTAAREFRGPKAKTNFP P++ NQSPS SSTVESSS + V A
Sbjct: 65 AYDTAAREFRGPKAKTNFPSPTE---NQSPSHSSTVESSSGENGVHAPPHAPLELDLTRR 121
Query: 114 ----AADSSSLDLNLAPSVRFPF---RPQFPQMPAANQVLFFDAILRAGMVGGSPASQRY 166
AAD + + V +P +P +P VL FD++ RAG+V Q Y
Sbjct: 122 LGSVAADGGD-NCRRSGEVGYPIFHQQPTVAVLPNGQPVLLFDSLWRAGVVN---RPQPY 177
Query: 167 GFDFHTYNHPVAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRNFDLDLNY-PPM 225
+ P+ + + A G + SDSSS ++ N Y+ G R DLDLN PPM
Sbjct: 178 ------HVTPMGFNGVNAGVG-PTVSDSSSAVEENQYD-------GKRGIDLDLNLAPPM 223
Query: 226 E 226
E
Sbjct: 224 E 224
>UniRef100_Q9LW49 Ethylene-responsive transcription factor 4 [Nicotiana sylvestris]
Length = 227
Score = 174 bits (440), Expect = 2e-42
Identities = 118/245 (48%), Positives = 142/245 (57%), Gaps = 43/245 (17%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MA ++K + + G GN NG VKEVH+RGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT
Sbjct: 1 MAVKNKVSNGDLKG-GNVKTNG-VKEVHYRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 58
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVESSSRDRDVAA------- 113
AEEAA+AYDTAAREFRGPKAKTNFP PS+ NQS S SST+ESSS + + A
Sbjct: 59 AEEAAKAYDTAAREFRGPKAKTNFPLPSE---NQSTSHSSTMESSSGETGIHAPPHAPLE 115
Query: 114 ----------AADSSSLDLNLAPSVRFPF---RPQFPQMPAANQVLFFDAILRAGMVGGS 160
AAD + + V +P +P +P VL FD++ R G+V
Sbjct: 116 LDLTRRLGSVAADGGD-NCRRSGEVGYPIFHQQPTVAVLPNGQPVLLFDSLWRPGVVN-- 172
Query: 161 PASQRYGFDFHTYNHPVAASEIQAVAGAQ-SDSDSSSVIDLNHYESGVVIKAGGRNFDLD 219
Q Y + P+A AG + SDSSSV++ N Y+ G R DLD
Sbjct: 173 -RPQPY------HVMPMAMGFNGVNAGVDPTVSDSSSVVEENQYD-------GKRGIDLD 218
Query: 220 LNYPP 224
LN P
Sbjct: 219 LNLAP 223
>UniRef100_Q84XB1 Ethylene response factor 3 [Lycopersicon esculentum]
Length = 222
Score = 172 bits (435), Expect = 8e-42
Identities = 117/250 (46%), Positives = 141/250 (55%), Gaps = 50/250 (20%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAP++K AV N NG KEVH+RGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT
Sbjct: 1 MAPKEKIGAVTAMAMVNLNGIS--KEVHYRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 58
Query: 61 AEEAARAYDTAAREFRGPKAKTNFP-----------FPSDNVKNQSPSQSSTVESSSRDR 109
AEEAARAYD AAREFRG KAKTNFP F N N+SP Q+STVESSS
Sbjct: 59 AEEAARAYDNAAREFRGAKAKTNFPKLEMEKEEDLKFAVKNEINRSPGQTSTVESSS--- 115
Query: 110 DVAAAADSSS-LDLNLAPS--------VRFPFRPQ--FPQMPAANQVLFFDAILRAGMVG 158
DSSS LDL+L S V+FP + A N++ + +A+ RAG++
Sbjct: 116 --PVMVDSSSPLDLSLCGSIGGFNHHTVKFPSSGGGFTGSVQAVNRMYYIEALARAGVI- 172
Query: 159 GSPASQRYGFDFHTYNHPVAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRNFDL 218
+ + + G DSDSS+VID V +K+ +L
Sbjct: 173 --------------KLEQIGRKRLDYLGG--GDSDSSTVIDFMR----VDVKSTTAGLNL 212
Query: 219 DLNYPPMEDM 228
DLN+PP E+M
Sbjct: 213 DLNFPPPENM 222
>UniRef100_Q84LQ6 Ethylene response factor 2 [Lycopersicon esculentum]
Length = 210
Score = 166 bits (420), Expect = 4e-40
Identities = 112/232 (48%), Positives = 134/232 (57%), Gaps = 48/232 (20%)
Query: 19 NGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGP 78
N NG KEVH+RGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYD AAREFRG
Sbjct: 5 NLNGISKEVHYRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDNAAREFRGA 64
Query: 79 KAKTNFP-----------FPSDNVKNQSPSQSSTVESSSRDRDVAAAADSSS-LDLNLAP 126
KAKTNFP F N N+SPSQ+STVESSS DSSS LDL+L
Sbjct: 65 KAKTNFPKLEMEKEEDLKFAVKNEINRSPSQTSTVESSS-----PVMVDSSSPLDLSLCG 119
Query: 127 S--------VRFPFRPQ--FPQMPAANQVLFFDAILRAGMVGGSPASQRYGFDFHTYNHP 176
S V+FP + A N + + +A+ RAG++ ++
Sbjct: 120 SIGGFNHHTVKFPSSGGGFTGSVQAVNHMYYIEALARAGVIKLETNRKK----------- 168
Query: 177 VAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRNFDLDLNYPPMEDM 228
+ + G DSDSS+VID V +K+ +LDLN+PP E+M
Sbjct: 169 ----TVDYLGG--GDSDSSTVIDFMR----VDVKSTTAGLNLDLNFPPPENM 210
>UniRef100_Q7F4G5 Ethylene responsive element binding factor3 [Oryza sativa]
Length = 235
Score = 165 bits (418), Expect = 7e-40
Identities = 112/244 (45%), Positives = 132/244 (53%), Gaps = 33/244 (13%)
Query: 1 MAPR----DKTNAVKVSGNGNGNGNG-GVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWL 55
MAPR +K +G G G G G G H+RGVRKRPWGRYAAEIRDP KKSRVWL
Sbjct: 1 MAPRAATVEKVAVAPPTGLGLGVGGGVGAGGPHYRGVRKRPWGRYAAEIRDPAKKSRVWL 60
Query: 56 GTFDTAEEAARAYDTAAREFRGPKAKTNFPFPSDNVK--NQSPSQSSTVESSSR--DRDV 111
GT+DTAEEAARAYD AAREFRG KAKTNFPF S ++ SPS +STV++ +
Sbjct: 61 GTYDTAEEAARAYDAAAREFRGAKAKTNFPFASQSMVGCGGSPSSNSTVDTGGGGVQTPM 120
Query: 112 AAAADSSSLDLNL-----------APSVRFPFRPQFPQMPAANQVLFFDAILRAGMVGGS 160
A +LDL+L VRFPFR PA + F++ A
Sbjct: 121 RAMPLPPTLDLDLFHRAAAVTAVAGTGVRFPFRGYPVARPATHPYFFYEQAAAA------ 174
Query: 161 PASQRYGFDFHTYNHPVAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRNFDLDL 220
A+ G+ PV + + AQSDSDSSSV+DL V FDLDL
Sbjct: 175 -AAAEAGYRMMKLAPPVTVAAV-----AQSDSDSSSVVDLAPSPPAVTANKAAA-FDLDL 227
Query: 221 NYPP 224
N PP
Sbjct: 228 NRPP 231
>UniRef100_Q6J832 Ethylene-responsive element binding factor [Oryza sativa]
Length = 236
Score = 163 bits (412), Expect = 4e-39
Identities = 113/249 (45%), Positives = 131/249 (52%), Gaps = 42/249 (16%)
Query: 1 MAPR--DKTNAVKVSGNGNGNGNGGVKEV-------HFRGVRKRPWGRYAAEIRDPGKKS 51
MAPR DKT + + G G GGV HFRGVRKRPWGRYAAEIRDP KKS
Sbjct: 1 MAPRTSDKTMSPAAAATGLALGVGGVAGAAAVGTGQHFRGVRKRPWGRYAAEIRDPAKKS 60
Query: 52 RVWLGTFDTAEEAARAYDTAAREFRGPKAKTNFPFPSDNVK---NQSPSQSSTVESSSRD 108
RVWLGTFDTAEEAARAYD AARE+RG KAKTNFP+P+ N S SSTVES D
Sbjct: 61 RVWLGTFDTAEEAARAYDAAAREYRGAKAKTNFPYPNGAPAAGVNSGSSNSSTVESFGSD 120
Query: 109 --RDVAAAADSSSLDLNL-----------APSVRFPFRPQFPQMPAANQVLFFDAILRAG 155
+ A SL+L+L A +RFPF P ++ FF A
Sbjct: 121 VQAPMKAMPIPPSLELDLFHRAAAAAAAGAGGMRFPFE----GYPVSHPYYFFGQAAAAA 176
Query: 156 MVGGSPASQRYGFDFHTYNHPVAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRN 215
G + +A + + A AQSDSDSSS++DL + K
Sbjct: 177 AASGCRMLK------------IAPAPVTVAALAQSDSDSSSIVDLAPSPPAALAKK-AIA 223
Query: 216 FDLDLNYPP 224
FDLDLN PP
Sbjct: 224 FDLDLNCPP 232
>UniRef100_O80340 Ethylene-responsive transcription factor 4 [Arabidopsis thaliana]
Length = 222
Score = 161 bits (408), Expect = 1e-38
Identities = 103/217 (47%), Positives = 128/217 (58%), Gaps = 19/217 (8%)
Query: 21 NGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKA 80
+ KE+ +RGVRKRPWGRYAAEIRDPGKK+RVWLGTFDTAEEAARAYDTAAR+FRG KA
Sbjct: 17 HNNAKEIRYRGVRKRPWGRYAAEIRDPGKKTRVWLGTFDTAEEAARAYDTAARDFRGAKA 76
Query: 81 KTNFP----FPSDNVKN---QSPSQSSTVESSSRDRDVAAAADSSSLDLNLAPSVRFPFR 133
KTNFP V +SPSQSST++ +S V +A + ++ L S+
Sbjct: 77 KTNFPTFLELSDQKVPTGFARSPSQSSTLDCASPPTLVVPSATAGNVPPQLELSLGGGGG 136
Query: 134 PQFPQMPAANQVLFFDAILRAGMVG-GSPASQRYGFDFHTYNHPVAASEIQAVAGAQSDS 192
Q+P + V F D ++ G VG G P F PV + GAQSDS
Sbjct: 137 GSCYQIPMSRPVYFLD-LMGIGNVGRGQPPPVTSAF-----RSPVVHVATKMACGAQSDS 190
Query: 193 DSSSVIDLNHYESGVVIKAGGRNFDLDLNYPPMEDMA 229
DSSSV+D +E G ++ + DLDLN PP + A
Sbjct: 191 DSSSVVD---FEGG--MEKRSQLLDLDLNLPPPSEQA 222
>UniRef100_Q5U8L5 AP2/EREBP transcription factor ERF-1 [Gossypium hirsutum]
Length = 176
Score = 158 bits (399), Expect = 1e-37
Identities = 115/228 (50%), Positives = 126/228 (54%), Gaps = 56/228 (24%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAPR K + + + + N KE+ +RGVRKRPWGRYAAEIRDP KK+RVWLGTFDT
Sbjct: 1 MAPRSKPSPISPNPDPNS------KEIRYRGVRKRPWGRYAAEIRDPRKKTRVWLGTFDT 54
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVESSSRDRDVAAAADSSSL 120
AEEAARAYD AREFRG KAKTNF + N +SPSQSSTVESSS L
Sbjct: 55 AEEAARAYDAKAREFRGAKAKTNFADNNANDFTRSPSQSSTVESSS----------PPPL 104
Query: 121 DLNLA-PSVRFPFRPQFPQMPAANQVLFFDAILRAGMVGGSPASQRYGFDFHTYNHPVAA 179
DL LA P P Q P V FFDA G G PAS GF
Sbjct: 105 DLTLASPCSSLPVTAQRP-------VYFFDAFATGG--SGCPAS---GF----------- 141
Query: 180 SEIQAVAGAQSDSD-SSSVIDLNHYESGVVIKAGGRNFDLDLNYPPME 226
AQSDSD SSSV+D +E GV R FDLDLN P E
Sbjct: 142 --------AQSDSDSSSSVVD---FEGGV----RRRVFDLDLNQLPAE 174
>UniRef100_Q7XPL6 OSJNBa0085I10.10 protein [Oryza sativa]
Length = 222
Score = 145 bits (367), Expect = 6e-34
Identities = 107/247 (43%), Positives = 123/247 (49%), Gaps = 53/247 (21%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAPR+ AV V+ G G E FRGVRKRPWGRYAAEIRDP +K+RVWLGTFDT
Sbjct: 1 MAPRNAAEAVAVAV---AEGGGAGMEPRFRGVRKRPWGRYAAEIRDPARKARVWLGTFDT 57
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFPSDNVKNQSP--------------SQSSTVESSS 106
AE AARAYD+AA FRGPKAKTNFP + + +P SSTVESSS
Sbjct: 58 AEAAARAYDSAALHFRGPKAKTNFPVAFAHAHHHAPPPPLPKAAALAVVSPTSSTVESSS 117
Query: 107 RD----RDVAAAAD-----SSSLDLNLAPSVRFPFRPQFPQMPAANQVLFFDAILRAGMV 157
RD VAAAA S SLDL+L S M AA LF D + +
Sbjct: 118 RDTPAAAPVAAAAKAQVPASPSLDLSLGMSA----------MVAAQPFLFLDPRVAVTVA 167
Query: 158 GGSPASQRYGFDFHTYNHPVAASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRNFD 217
+P +R P S + VA SD+ S S VV +
Sbjct: 168 VAAPVPRR----------PAVVSVKKEVARLDEQSDTGS-------SSSVVDASPAVGVG 210
Query: 218 LDLNYPP 224
LDLN PP
Sbjct: 211 LDLNLPP 217
>UniRef100_Q8S2S7 Ethylene responsive element binding factor 4-like protein
[Thellungiella halophila]
Length = 195
Score = 144 bits (364), Expect = 1e-33
Identities = 102/226 (45%), Positives = 119/226 (52%), Gaps = 52/226 (23%)
Query: 11 KVSGNGNGNGN-GGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYD 69
K N + N N KE+ +RGVRKRPWGRYAAEIRDPGKK+RVWLGTFDTA++AARAYD
Sbjct: 7 KPDSNPSPNPNESNAKEIRYRGVRKRPWGRYAAEIRDPGKKTRVWLGTFDTAQQAARAYD 66
Query: 70 TAAREFRGPKAKTNFPF------PSDNVKNQSPSQSSTVESSSRDRDVAAAADSSSLDLN 123
AAREFRG KAKTNFP D +SPSQSSTV+S S
Sbjct: 67 AAAREFRGAKAKTNFPTFLELNAAKDGGFARSPSQSSTVDSVS----------------- 109
Query: 124 LAPSVRFPFRPQFPQMPAANQVLFFDAILRAGMVGGS-----PASQRYGFDFHTYNHPVA 178
S R PQ L G GG P ++R + ++ P A
Sbjct: 110 -PTSARLVTPPQLE--------------LSLGGGGGGACYQIPVARRPVYFYNMTTFPAA 154
Query: 179 ASEIQAVAGAQSDSDSSSVIDLNHYESGVVIKAGGRNFDLDLNYPP 224
A+ A G QS+SDSSSV+D +E G K R DLDLN P
Sbjct: 155 AA---ATCGVQSESDSSSVVD---FECGAEKKY--RPLDLDLNLAP 192
>UniRef100_Q6RUR2 Ethylene-responsive element binding factor [Capsicum annuum]
Length = 204
Score = 137 bits (346), Expect = 2e-31
Identities = 69/96 (71%), Positives = 75/96 (77%), Gaps = 1/96 (1%)
Query: 14 GNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAR 73
GN NG K+VH+RGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAA+AYD AAR
Sbjct: 4 GNSKCNGVNNNKDVHYRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAAKAYDAAAR 63
Query: 74 EFRGPKAKTNFPFPSDNVKNQSPSQSSTVESSSRDR 109
EFRG KAKTNFP PS+N +N S S S S +R
Sbjct: 64 EFRGCKAKTNFPLPSEN-QNGSDSGSPDESFSGENR 98
>UniRef100_Q9C5I3 Ethylene-responsive transcription factor 11 [Arabidopsis thaliana]
Length = 166
Score = 136 bits (342), Expect = 5e-31
Identities = 103/229 (44%), Positives = 122/229 (52%), Gaps = 72/229 (31%)
Query: 1 MAPRDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDT 60
MAP KT AVK + GNG V +RGVRKRPWGRYAAEIRDP KKSRVWLGTFDT
Sbjct: 1 MAPTVKTAAVKTN-----EGNG----VRYRGVRKRPWGRYAAEIRDPFKKSRVWLGTFDT 51
Query: 61 AEEAARAYDTAAREFRGPKAKTNFPFPSDNVK----NQSPSQSSTVESSSRDRDVAAAAD 116
EEAARAYD A EFRG KAKTNFP + N QS SQSSTVESS + ++ + +
Sbjct: 52 PEEAARAYDKRAIEFRGAKAKTNFPCYNINAHCLSLTQSLSQSSTVESSFPNLNLGSDSV 111
Query: 117 SSSLDLNLAPSVRFPFRPQFPQMPAANQVLFFDAILRAGMVGGSPASQRYGFDFHTYNHP 176
SS RFP FP++ ++AGM+
Sbjct: 112 SS----------RFP----FPKIQ-----------VKAGMM------------------- 127
Query: 177 VAASEIQAVAGAQSDSDSSSVI-DLNHYESGVVIKAGGRNFDLDLNYPP 224
V +S+SDSSSV+ D+ YE V+ DLDLN+PP
Sbjct: 128 --------VFDERSESDSSSVVMDVVRYEGRRVV------LDLDLNFPP 162
>UniRef100_Q9FE67 Ethylene-responsive transcription factor 9 [Arabidopsis thaliana]
Length = 200
Score = 133 bits (335), Expect = 3e-30
Identities = 68/94 (72%), Positives = 73/94 (77%), Gaps = 4/94 (4%)
Query: 1 MAPRDKTN-AVKVSGNGNGNGNGGV---KEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLG 56
MAPR ++ VS G G KEVHFRGVRKRPWGRYAAEIRDPGKK+RVWLG
Sbjct: 1 MAPRQANGRSIAVSEGGGGKTMTMTTMRKEVHFRGVRKRPWGRYAAEIRDPGKKTRVWLG 60
Query: 57 TFDTAEEAARAYDTAAREFRGPKAKTNFPFPSDN 90
TFDTAEEAARAYDTAAREFRG KAKTNFP P ++
Sbjct: 61 TFDTAEEAARAYDTAAREFRGSKAKTNFPLPGES 94
>UniRef100_Q9SXS8 Ethylene-responsive transcription factor 3 [Nicotiana tabacum]
Length = 225
Score = 128 bits (321), Expect = 1e-28
Identities = 92/240 (38%), Positives = 123/240 (50%), Gaps = 46/240 (19%)
Query: 4 RDKTNAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEE 63
R + A G NG+GG KE+ FRGVRKRPWGR+AAEIRDP KK+RVWLGTFD+AE+
Sbjct: 3 RGRAAAAPAPVTGEPNGSGGSKEIRFRGVRKRPWGRFAAEIRDPWKKTRVWLGTFDSAED 62
Query: 64 AARAYDTAAREFRGPKAKTNFPFPSDN----VKNQSPSQSSTVESS--SRDRDVAAAADS 117
AARAYD AAR RGPKAKTNFP P + + +P+ V+S +D + + +
Sbjct: 63 AARAYDAAARALRGPKAKTNFPLPYAHHHQFNQGHNPNNDPFVDSRFYPQDNPIISQRPT 122
Query: 118 SSLDLNLAPSVRFPFRPQFPQMPAANQVLFFDAILRAGMVGGSPASQRYGFDFHTYNHPV 177
SS + S P P P+ + +S++Y T + PV
Sbjct: 123 SSSMSSTVESFSGPRPPPAPRQQT------------------TASSRKY-----TRSPPV 159
Query: 178 AASEIQAVAGAQSDSD-SSSVID------LNHYESGVVIKAGGRN---FDLDLNYPPMED 227
+ SD D SSSV+D N ++ + + R FDL+L PPM+D
Sbjct: 160 VPDD------CHSDCDSSSSVVDHGDCEKENDNDNDNIASSSFRKPLLFDLNLP-PPMDD 212
>UniRef100_O80339 Ethylene-responsive transcription factor 3 [Arabidopsis thaliana]
Length = 225
Score = 124 bits (310), Expect = 2e-27
Identities = 82/208 (39%), Positives = 112/208 (53%), Gaps = 22/208 (10%)
Query: 21 NGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGTFDTAEEAARAYDTAAREFRGPKA 80
NG VKE+ FRGVRKRPWGR+AAEIRDP KK+RVWLGTFD+AEEAARAYD+AAR RGPKA
Sbjct: 20 NGSVKEIRFRGVRKRPWGRFAAEIRDPWKKARVWLGTFDSAEEAARAYDSAARNLRGPKA 79
Query: 81 KTNFPFPSDNVKNQSPSQSSTVESSSRDRDVAAAADSSSLDLNLAPSVRFPFRPQFPQMP 140
KTNFP + + SP + + R++ + + +D + + + QFP +
Sbjct: 80 KTNFP-----IDSSSPPPPNLRFNQIRNQ------NQNQVDPFMDHRLFTDHQQQFPIVN 128
Query: 141 AANQVLFFDAILRAGMVGGSPASQRYGFDFHTYNHPVAASEIQAVAGAQSDSDSSSVIDL 200
+ G P + + T +P + + DS SSSVID
Sbjct: 129 RPTSSSMSSTV--ESFSGPRPTTMK---PATTKRYPRTPPVVPEDCHSDCDS-SSSVID- 181
Query: 201 NHYESGVVIKAGGRN--FDLDLNYPPME 226
+ + + RN F DLN+PP++
Sbjct: 182 --DDDDIASSSRRRNPPFQFDLNFPPLD 207
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.131 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 396,195,831
Number of Sequences: 2790947
Number of extensions: 16137909
Number of successful extensions: 45005
Number of sequences better than 10.0: 613
Number of HSP's better than 10.0 without gapping: 571
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 44131
Number of HSP's gapped (non-prelim): 747
length of query: 229
length of database: 848,049,833
effective HSP length: 123
effective length of query: 106
effective length of database: 504,763,352
effective search space: 53504915312
effective search space used: 53504915312
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0301c.1


