

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
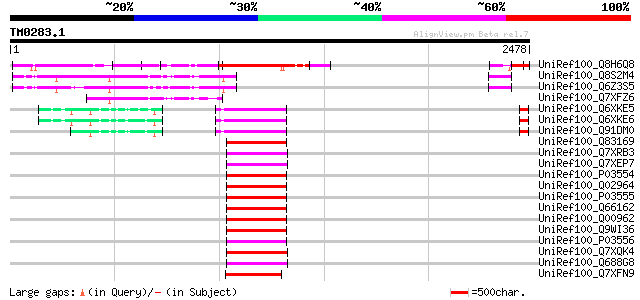
Query= TM0283.1
(2478 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8H6Q8 CTV.20 [Poncirus trifoliata] 533 e-149
UniRef100_Q8S2M4 Hypothetical protein OSJNBa0090K04.9 [Oryza sat... 400 e-109
UniRef100_Q6Z3S5 Hypothetical protein OSJNBa0025J22.29 [Oryza sa... 384 e-104
UniRef100_Q7XFZ6 Hypothetical protein [Oryza sativa] 290 5e-76
UniRef100_Q6XKE5 Polyprotein 1 [Petunia vein clearing virus] 250 4e-64
UniRef100_Q6XKE6 Polyprotein 1 [Petunia vein clearing virus] 250 4e-64
UniRef100_Q91DM0 ORF I polyprotein [Petunia vein clearing virus] 249 1e-63
UniRef100_Q83169 Reverse transcriptase [Cauliflower mosaic virus] 223 6e-56
UniRef100_Q7XRB3 OSJNBa0006B20.7 protein [Oryza sativa] 223 6e-56
UniRef100_Q7XEP7 Putative rice tungro bacilliform virus; reverse... 223 8e-56
UniRef100_P03554 Enzymatic polyprotein [Contains: Aspartic prote... 223 8e-56
UniRef100_Q02964 Enzymatic polyprotein [Contains: Aspartic prote... 223 8e-56
UniRef100_P03555 Enzymatic polyprotein [Contains: Aspartic prote... 223 8e-56
UniRef100_Q66162 ORF V [Cauliflower mosaic virus] 222 1e-55
UniRef100_Q00962 Enzymatic polyprotein [Contains: Aspartic prote... 222 1e-55
UniRef100_Q9WI36 Reverse transcriptase [Cauliflower mosaic virus] 221 2e-55
UniRef100_P03556 Enzymatic polyprotein [Contains: Aspartic prote... 220 4e-55
UniRef100_Q7XQK4 OSJNBa0017B10.4 protein [Oryza sativa] 219 8e-55
UniRef100_Q688G8 Putative polyprotein [Oryza sativa] 216 5e-54
UniRef100_Q7XFN9 Putative retroelement [Oryza sativa] 211 2e-52
>UniRef100_Q8H6Q8 CTV.20 [Poncirus trifoliata]
Length = 3148
Score = 533 bits (1374), Expect = e-149
Identities = 325/840 (38%), Positives = 487/840 (57%), Gaps = 56/840 (6%)
Query: 719 SSEYEYEGDED--QDINLVEYSSDSHSSSEEERECVKDSSGFCDCAGCLGQNINMLTQDQ 776
S+ + GD D Q L++ + +SS+ E E + IN+LT+DQ
Sbjct: 839 STSKDCNGDSDSLQIDELIDSDTSVSNSSDSESESYL-------------KKINILTKDQ 885
Query: 777 TMSLISIIDKMEDSPLRDEFLQQL--NLLVKKEETLKKDISPPIN-SMNEIFNRFRPKPT 833
+ + ++ + D L+ E+L++L + K ET K I + + EI ++ + K +
Sbjct: 886 E-TFLELVKHISDPNLQKEYLEKLLKTMDFNKAETSKVPIVKKNSYDLTEILDKKKTKKS 944
Query: 834 I-TLSDLQEEVKILKEEINQLKQNDLSLEFRLIELAGIKIIESQETLASASGIKQIPDKK 892
+ + DLQ+E+K +K EI LK+ S + I+L K ++ S I ++
Sbjct: 945 VPNIQDLQKEIKDIKSEIKDLKEKQKS-DSETIQLLLQKHLQDDSDNESTHSENHI--EQ 1001
Query: 893 NEDNNSQEPNENYLNLLNLVITHKWHSEITLVI-NDFRINIVALIDSGADINCIQEGLIP 951
N DN P++ +L +L V T K+ + TL+ NDF I+ +AL D+GAD+NCI++ ++P
Sbjct: 1002 NVDNIESVPHD-FLFVLKQVTTRKYLIKATLIFSNDFAIDAIALFDTGADLNCIRQDIVP 1060
Query: 952 TQFYEKTKEGVNSANGSKMNIQYKLSNAKICKNQICFKSSFVLVKNMTEKVILGTLFICL 1011
+F+EKTKE +++AN SK+ + K+ A I N FK+SF+L ++ VILGT FI L
Sbjct: 1061 KRFHEKTKERLSAANNSKLKVDSKVE-ASIHNNGFEFKTSFILTNDIHHAVILGTPFINL 1119
Query: 1012 LYPKVHTIALPYIDEFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSC 1071
+ PK H + LPY + F+EK IPTKARPIQMN ++C+EEI + + KGLI S+SPWSC
Sbjct: 1120 ITPKQHMVDLPYENSFDEKQIPTKARPIQMNMDLERHCKEEINDLVKKGLIVKSRSPWSC 1179
Query: 1072 IGFYVLNASELERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSG 1131
FYV SE+ERG RLVINYKPLNK LKWIRYP+PNK DL+++LH IFSKFDMKS
Sbjct: 1180 AAFYVNKNSEIERGTPRLVINYKPLNKALKWIRYPIPNKKDLLQKLHSAFIFSKFDMKSV 1239
Query: 1132 YYQISVREEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDD 1191
++QI + ++RYKTAF VPFG YEW VMP GLKNAPSE+Q IMNDI+ PY +F IVY+DD
Sbjct: 1240 FWQIQIHPKERYKTAFTVPFGQYEWTVMPFGLKNAPSEFQRIMNDIYNPYSDFCIVYIDD 1299
Query: 1192 VLVFSKGIDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSL 1251
VL+FS IDQH +HL+ F +K GL +S K+ +F+TK RFLG+ I +G ITPI+RSL
Sbjct: 1300 VLIFSNSIDQHFKHLKTFYFATRKAGLAISNSKVSLFQTKIRFLGHHISKGTITPIERSL 1359
Query: 1252 EFAKKFPDELKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKN---SPPW------ 1302
FA KFPD++ +KTQLQRFLG +NYV DF PNI + PL RL+K PP
Sbjct: 1360 AFADKFPDKILDKTQLQRFLGSLNYVLDFCPNISRLSKPLHDRLKKKPAAMPPKRRDKGK 1419
Query: 1303 -----------SEEMSHSVREVKKLVQSLPIVTPFQIF*QENFYRVNMSEKPKSKDQY*P 1351
S+E + + K L ++PI + ++ E+ + +S+ S Q
Sbjct: 1420 GIAKDTDSLKPSKESQSTPSKEKLLSSAMPIKSWIEMV--EDEEQKALSKSISSDQQVKE 1477
Query: 1352 PLGSSSTSKDKALAITPLKSAGSSAPATPSKSSQRLTTSQIVKTPPQNQKAPLVPLANKY 1411
+ S + S + A L+ S + ++ + +I K Q+Q + ++
Sbjct: 1478 WMESITKSPELMFA---LQGISKSKALSQIPEEEKPISKEITKLSSQSQNVVISGESSSS 1534
Query: 1412 TVLTQSSSKSPVKPESTEYFEKPEGLSSLIIEKEYFHLYPKEIATQLLPPNFQYVPGHPK 1471
++ S +P K +++++F+K + L +E ++H P + ++ P ++ + P
Sbjct: 1535 QIVL--SQPTPSK-KTSDWFDKSHFQNVLTMEHGFYHSDPFQAISKFFPQSWFFNPWDLT 1591
Query: 1472 KTRLFYEFILVDSDSIEVTH--NPDKQGEIAFSKIKIIKVLSAQDWNAPLHYFKNFSRQF 1529
K + +Y+ IL ++S++ H + E A+S I+KVLS W LH +K+F F
Sbjct: 1592 KPQSYYQSILEATESVKFKHFFLSETHSEPAYSTATILKVLSPNQWGDQLHKYKSFPPNF 1651
Score = 372 bits (956), Expect = e-101
Identities = 209/452 (46%), Positives = 275/452 (60%), Gaps = 24/452 (5%)
Query: 996 KNMTEKVILGTLFICLLYPKVHTIALPYIDEFNEKDIPTKARPIQMNEQYLKYCREEIGE 1055
K + EK I L K H + LPY + F+EK IPTKARPIQMN ++C+EEI +
Sbjct: 2569 KTLIEKEICADLPSAFWNRKQHMVDLPYENSFDEKQIPTKARPIQMNMDLERHCKEEIND 2628
Query: 1056 YLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYKPLNKVLKWIRYPLPNKTDLIK 1115
+ KGLI S+SPWSC FYV SE+ERG RLVINYKPLNK LKWIRYP+PNK DL++
Sbjct: 2629 LVKKGLIVKSRSPWSCAAFYVNKNSEIERGTPRLVINYKPLNKALKWIRYPIPNKKDLLQ 2688
Query: 1116 RLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMN 1175
+LH IFSKFDMKSG++QI + +DRYKTAF VPFG YEW VMP GLKNAPSE+Q IMN
Sbjct: 2689 KLHSAFIFSKFDMKSGFWQIQIHPKDRYKTAFTVPFGQYEWTVMPFGLKNAPSEFQRIMN 2748
Query: 1176 DIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFL 1235
DI+ PY +F IVY+DDVL+FS IDQH +HL+ F +K GL +S K+ +F+TK RFL
Sbjct: 2749 DIYNPYSDFCIVYIDDVLIFSNSIDQHFKHLKTFYFATRKAGLAISNSKVSLFQTKIRFL 2808
Query: 1236 GYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCVNYVADFIPNI------RIVCA 1289
G+ I +G ITPI+RSL FA KFPD++ +KTQLQRFLG +NYV DF PNI RI
Sbjct: 2809 GHHISKGTITPIERSLAFADKFPDKILDKTQLQRFLGSLNYVLDFCPNISRFSNSRISSK 2868
Query: 1290 PLFKRLRK----------NSPPWSEEMSHSVREVKKLVQSLPIVTPFQIF*QENFYRVNM 1339
+ R+ +S S+E + + K L ++PI + ++ E+ + +
Sbjct: 2869 QIMPPKRRDKGKGIAKDTDSLKPSKESQSTPSKEKLLSSAMPIKSWIEMV--EDEEQKAL 2926
Query: 1340 SEKPKSKDQY*PPLGSSSTSKDKALAITPLKSAGSSAPATPSKSSQRLTTSQIVKTPPQN 1399
S+ S Q + S + S + LA L+ S + ++L + +I K Q+
Sbjct: 2927 SKSISSDQQVKEWMESITKSPELMLA---LQGISKSKTLSQIPEEEKLISKEITKLSSQS 2983
Query: 1400 QKAPLVPLANKYTVLTQSSSKSPVKPESTEYF 1431
Q V ++ YT SP T +F
Sbjct: 2984 QN---VVISAHYTPSDSEKRFSPFLCFCTNFF 3012
Score = 268 bits (685), Expect = 2e-69
Identities = 179/548 (32%), Positives = 302/548 (54%), Gaps = 48/548 (8%)
Query: 491 GDPSIFRERASSQLSNLYCPTMSDYRWYKDTFFSKVTLREDGNNAFWKERFIAGLPRLMQ 550
GDPS R++ + L NL C +SD++ YK TFF+++ LR+D N+ WKE+F+A LP L+
Sbjct: 1963 GDPSHLRDKNAELLHNLRCRKLSDFQLYKTTFFTRLFLRDDANHTTWKEKFLACLPTLLG 2022
Query: 551 SKVLDNLSLFNQGNPINFGSLSFGQLHNTIVHTGIQVCTDFKLQNKMQKDMQISRKEVGS 610
KV + + N I + L++G+L + + G+++C D KLQ +++++++ S++E+GS
Sbjct: 2023 EKVRNFIKAL-YDNRIPYDELTYGELVSFVNKEGLKICQDLKLQKRLKQEIRQSKRELGS 2081
Query: 611 FCEQYGVEPLQAPSSK------ARRINRSKTKPQGYKK-----RKPFQKKPYEQNPSTQQ 659
FC+Q+ +P +A +SK + R + K + ++K R+ F KKP ++
Sbjct: 2082 FCKQFNYDPFKASTSKDCNGKCSSRPYKKHYKSKSHRKLFLGHRENFYKKPTRPYKKSRF 2141
Query: 660 PTNKK-NVNPK-----KKNVCWKCGKPGHYANKCKTQQKINELDLDQKLKDSLISVLINS 713
P K PK K+ +C +CG GH A CK +K++EL LD + + ++I S
Sbjct: 2142 PRKKDFKATPKTPFNFKEAICHRCGIKGHTAKYCKMNRKLHELGLDDDILSKIAPLMIES 2201
Query: 714 EDPHYSSEYEYEGDED--QDINLVEYSSDSHSSSEEERECVKDSSGFCDCAGCLGQNINM 771
+SE GD D Q L++ + +SS+ E E + IN+
Sbjct: 2202 S----NSESSMSGDSDSLQIDELIDSDTSVSNSSDSESESYL-------------KKINV 2244
Query: 772 LTQDQTMSLISIIDKMEDSPLRDEFLQQL--NLLVKKEETLKKDISPPIN-SMNEIFNRF 828
LT+DQ + + ++ + D L+ E+L++L + K ET K I + + EI ++
Sbjct: 2245 LTKDQE-TFLELVKHISDPNLQKEYLEKLLKTMDFNKAETSKVPIVKKNSYDLTEILDKK 2303
Query: 829 RPKPTI-TLSDLQEEVKILKEEINQLKQNDLSLEFRLIELAGIKIIESQETLASASGIKQ 887
+ K ++ + DLQ+E+K +K EI LK+ S + I+L K ++ S
Sbjct: 2304 KTKKSVPNIQDLQKEIKDIKSEIKDLKEKQKS-DSETIQLLLQKHLQDDSDNESTHSENH 2362
Query: 888 IPDKKNEDNNSQEPNENYLNLLNLVITHKWHSEITLVI-NDFRINIVALIDSGADINCIQ 946
I ++N DN P++ +L +L V T K+ + TL+ NDF I+ +AL D+GAD+NCI+
Sbjct: 2363 I--EQNVDNIESVPHD-FLFVLKQVTTRKYLIKATLIFSNDFAIDAIALFDTGADLNCIR 2419
Query: 947 EGLIPTQFYEKTKEGVNSANGSKMNIQYKLSNAKICKNQICFKSSFVLVKNMTEKVILGT 1006
+ ++P +F+EKTKE +++AN SK+ + K+ A I N FK+SF+L ++ VILGT
Sbjct: 2420 QDIVPKRFHEKTKERLSAANNSKLKVDSKV-EASIHNNGFEFKTSFILTNDIHHAVILGT 2478
Query: 1007 LFICLLYP 1014
FI L+ P
Sbjct: 2479 PFINLITP 2486
Score = 243 bits (620), Expect = 5e-62
Identities = 195/689 (28%), Positives = 332/689 (47%), Gaps = 124/689 (17%)
Query: 11 SPKQETIMLEANLKRSLVQVPKKLTHEEVMSNVPEEWVFKNSLP-------QPKVHNTQV 63
S + ET++L+ +L R+ +PK + +++ N+PEEW+ + + P +P V
Sbjct: 205 SKRDETLLLQTDLSRANTVIPKPIQWKDI--NLPEEWILEGAAPPTIPKQLEPNTELQNV 262
Query: 64 RELFQEGANLTLRMNRSNSFS-------IRTPQQLYR-----VDLPRSSV---------- 101
+ L R + S+ FS I T ++ + ++LP S
Sbjct: 263 TQYSDGKVKLLFRRSMSSRFSNKESCSSIPTLERKFTKIPSVINLPYQSTKSQPRFSTSD 322
Query: 102 --SSKLKGIDTTSSPSIATPIY----------QDEESSNYSPTPSQI--NMLSRVKSSFE 147
SS + +D T++ + PIY + E S SPT S + N+++ ++ FE
Sbjct: 323 IPSSSIHSVDYTTN--VPHPIYTSSQHVQSQEEKEPSLPTSPTFSAVTENVINIIEKEFE 380
Query: 148 INKDFIRKYFMAEYNKDKRLWYFKNFSKLETENLRTLYYEEMEVSETNIYFFD*FENYCI 207
++K + F ++ NK+K++W+FK F + + ++ +YYE + + +I FFD FE Y
Sbjct: 381 LDKTLLHNDFYSDLNKEKKIWFFKQFLN-QRKEIQQIYYEFVNFHKVHILFFDWFEIYSS 439
Query: 208 KNNLNYPFWKNVNPITKIDTI--WKTPDNN-TITSEYPPQTGVKIAIKDGQ---EIEASP 261
+NN+ YPF K NPIT I WK D++ TI SE+PP + I G+ +I ASP
Sbjct: 440 ENNITYPF-KESNPITIRKKIPEWKLSDSDKTIESEHPPLR--SLTIDHGEPPIQIRASP 496
Query: 262 YKV--------EANKVHQQLNFANTMLTVMSKQLGRIEVQKPEPSTSKKSSSSF----TL 309
YK+ + + QQ NF NT L + KQL RIE Q + + + S+S
Sbjct: 497 YKIPKPNDSDSNLSSIIQQNNFCNTNLNTIGKQLTRIENQFQKSTITVSSTSPIPSKSDY 556
Query: 310 GNPLQTPIFKIPQYTK--EEFNSFNLSGSVEKLKEKLNKL-----SINHLDKDLNINKIR 362
L+ IFK Q +K ++ + S + ++E+L+++ S N + N +
Sbjct: 557 DKKLKEAIFKPFQVSKTSQKLVQESKSDFAKAIREQLDRIESASSSSNKVQIAPNTPQSS 616
Query: 363 KFG----PDNNTRAYYPRPSYSDMRFEERREFVQNTFSGTSLDEWNIDGFSEQNILDITH 418
K G D + A ++ D R + + ++ + + N +
Sbjct: 617 KIGVLEQDDQTSIASSDLEAFKDEAPTPRANKIHWELAPPTVK--SPPDLAIDNRPRVLQ 674
Query: 419 QMIMAATAYKVHNNT-DRNAALMITHGFTGQLRGWWDNIMTPEDKSTILEKKKENGEEDA 477
QM MAA AYK + T D A ++ GFTGQL+
Sbjct: 675 QMTMAANAYKTQSGTSDMAIAEILIAGFTGQLK--------------------------- 707
Query: 478 VATLIYTIILHFIGDPSIFRERASSQLSNLYCPTMSDYRWYKDTFFSKVTLREDGNNAFW 537
GDPS R++ + L NL C +SD++ YK TFF+++ LR+D N+ W
Sbjct: 708 -------------GDPSHLRDKNAELLHNLRCRKLSDFQLYKTTFFTRLFLRDDANHTTW 754
Query: 538 KERFIAGLPRLMQSKVLDNLSLFNQGNPINFGSLSFGQLHNTIVHTGIQVCTDFKLQNKM 597
KE+F+AGLP L+ KV +++ N I + L++GQL + + G+++C D KLQ ++
Sbjct: 755 KEKFLAGLPTLLGEKVRNSIKAL-YDNRIPYDELTYGQLVSFVNKEGLKICQDLKLQKRL 813
Query: 598 QKDMQISRKEVGSFCEQYGVEPLQAPSSK 626
+++++ S++E+GSFC+Q+ +P +A +SK
Sbjct: 814 KQELRQSKRELGSFCKQFNYDPFKASTSK 842
Score = 123 bits (308), Expect = 8e-26
Identities = 79/189 (41%), Positives = 108/189 (56%), Gaps = 29/189 (15%)
Query: 2290 PPDILKKTTEKLSAAEGSRLIIKYKIPSAIIKNGSLEIETPFLLVRNLSQKVIIGTPFIK 2349
P +KT E+LSAA S+L + K+ ++I NG E +T F+L ++ VI+GTPFI
Sbjct: 1060 PKRFHEKTKERLSAANNSKLKVDSKVEASIHNNG-FEFKTSFILTNDIHHAVILGTPFIN 1118
Query: 2350 KLFPYDTDVNGITVQHLGQPILFKFSEPPFDKTLNVISYKEKQINFLKEEISYKNIEVQL 2409
+ P QH+ + P++ S+ EKQI I N++++
Sbjct: 1119 LITPK---------QHM--------VDLPYEN-----SFDEKQIPTKARPIQM-NMDLER 1155
Query: 2410 QQPSVKSRIENILGNIQSSICSCKSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCW 2469
K I +++ + I +S WSCAAFYVNK +EIERGTPRLVINYKPLN+AL W
Sbjct: 1156 H---CKEEINDLVK--KGLIVKSRSPWSCAAFYVNKNSEIERGTPRLVINYKPLNKALKW 1210
Query: 2470 IRYPIPNKK 2478
IRYPIPNKK
Sbjct: 1211 IRYPIPNKK 1219
Score = 91.3 bits (225), Expect = 3e-16
Identities = 47/86 (54%), Positives = 59/86 (67%), Gaps = 1/86 (1%)
Query: 2394 NFLKEEISYKNIEVQLQQPSVKSRIENILGNIQSS-ICSCKSSWSCAAFYVNKQAEIERG 2452
+F +++I K +Q+ + E I ++ I +S WSCAAFYVNK +EIERG
Sbjct: 2599 SFDEKQIPTKARPIQMNMDLERHCKEEINDLVKKGLIVKSRSPWSCAAFYVNKNSEIERG 2658
Query: 2453 TPRLVINYKPLNQALCWIRYPIPNKK 2478
TPRLVINYKPLN+AL WIRYPIPNKK
Sbjct: 2659 TPRLVINYKPLNKALKWIRYPIPNKK 2684
Score = 84.7 bits (208), Expect = 3e-14
Identities = 54/175 (30%), Positives = 92/175 (51%), Gaps = 19/175 (10%)
Query: 2290 PPDILKKTTEKLSAAEGSRLIIKYKIPSAIIKNGSLEIETPFLLVRNLSQKVIIGTPFIK 2349
P +KT E+LSAA S+L + K+ ++I NG E +T F+L ++ VI+GTPFI
Sbjct: 2424 PKRFHEKTKERLSAANNSKLKVDSKVEASIHNNG-FEFKTSFILTNDIHHAVILGTPFIN 2482
Query: 2350 KLFPYDTDVNGITVQHLGQPILFKFSEPPFDKTLNV--------------ISYKEKQINF 2395
+ PY + + I+ + + I+F F E P + LN+ I+ K+ + F
Sbjct: 2483 LITPYTVNYDSISFKVKNKKIIFPFIEKPKTRNLNIVKACSIYQNRINNLINSKQNDLTF 2542
Query: 2396 LKEEISYKNIEVQLQQPSVKSRIENILGNIQSSICSCKSSWSCAAFYVNKQAEIE 2450
L++++S + IE LQ+ +K +I + I+ IC+ S AF+ KQ ++
Sbjct: 2543 LQKDLSLQRIENDLQRDFIKRKIFDFKTLIEKEICADLPS----AFWNRKQHMVD 2593
>UniRef100_Q8S2M4 Hypothetical protein OSJNBa0090K04.9 [Oryza sativa]
Length = 1634
Score = 400 bits (1027), Expect = e-109
Identities = 334/1181 (28%), Positives = 557/1181 (46%), Gaps = 191/1181 (16%)
Query: 11 SPKQETIMLEANLKRSLVQVPKKLTHEEVMSNVPEEWVFKNSLPQPKVHNTQVRELFQEG 70
+PK T +L+ N + +PK L +E+ +PE+WV ++ + ++V L +
Sbjct: 12 TPKGLTTLLQTN-PNNRHTIPKTLKWDEI--TLPEKWVLSQAVEPKSMDQSEVESLIET- 67
Query: 71 ANLTLRMNRSNSFSIRTPQQLYRVDLPRSSVSSKLKGIDTTSSPSIATPIYQDEESSNYS 130
++ + + Q+ + P S+ S+ + T ++ Y+D S S
Sbjct: 68 ------LDGDVEITFASKQKAFLQSRPSVSLDSRPR----TKPQNVVYATYEDN-SDEPS 116
Query: 131 PTPSQINML--------SRVKSSFEINKDFIRKYFMAEYNKDKRLWYFKNFSKLETENLR 182
+ IN++ + + FEI+K+ +R+ N+ K Y + + +R
Sbjct: 117 ISDFDINVIELDVGFVIALEEEEFEIDKELLRREIRLPKNRAKTKRYLEEVDESFRMKIR 176
Query: 183 TLYYEEMEVSETNIYFFD*FENYCIKNNLNYPFWKNV------------NPITKIDTIWK 230
+++ EM NI+FFD +EN I + + KN+ I K+ T W+
Sbjct: 177 EVWHNEMREQRRNIFFFDWYENSQIIHFEEFFKRKNMIKKEQKSEAEDLTVIKKVSTEWE 236
Query: 231 TPDNNTITSEYPPQTGVKIAIKDGQ-----EIEASPYKVEANKVH-----QQLNFANTML 280
T N + S +PP ++++ G+ I + Y A H +Q N+AN L
Sbjct: 237 TASGNKVDSVHPPFESIQLSHNGGKACPLKSISKNTYGETAKVEHIGHLVEQQNYANISL 296
Query: 281 TVMSKQLGRIEV--------QKPE-----PSTSKKSSSSFT--LGNPLQTPIFKI----- 320
+ +Q RIE +PE PS S+ SSS + P P K
Sbjct: 297 RSLGQQTDRIETILMEGYKTGRPEVKINIPSNSQSSSSQSVSPMFVPTIDPNIKFGKQKA 356
Query: 321 --PQYTKEEFNSFNLSGSVEKLKEKLNKLSINHLDKDLNINKIRKFGP-DNNTRAYYPRP 377
P ++E N L + K+ + +N++S N +K +NKI K + TR YYPRP
Sbjct: 357 FGPAISEELVNELALKLNNLKVNKNINEISDN--EKYDIVNKIFKPSTLTSTTRNYYPRP 414
Query: 378 SYSDMRFEERREFVQNTF-SGTSLDEWNIDGFSEQNILDITHQMIMAATAYKVHNNTDRN 436
+Y+D++FEE + T+ +G + EWN+DGF+E I + HQMIM A A + N +R
Sbjct: 415 TYADLQFEEMPQIQNMTYYNGKEIVEWNLDGFTEYQIFTLCHQMIMYANACIANGNKERE 474
Query: 437 AALMITHGFTGQLRGWWDNIMTPEDKSTIL--EKKKENGE---------------EDAVA 479
AA MI GF+GQL+GWW+N + + IL K+ + G DA+A
Sbjct: 475 AANMIVIGFSGQLKGWWNNYLNETQRQEILCAVKRDDQGRPLPDRDGNGNPTGNISDALA 534
Query: 480 TLIYTIILHFIGDPSIFRERASSQLSNLYCPTMSDYRWYKDTFFSKVTLREDGNNAFWKE 539
TLIY II HF G+ E+ QL NL C TMSD+RWYKDTF SK+ + N FWKE
Sbjct: 535 TLIYNIIYHFAGNYHDIYEKNREQLINLKCKTMSDFRWYKDTFLSKLYTLPEPNQDFWKE 594
Query: 540 RFIAGLPRLMQSKVLDNLSLFNQGNPINFGSLSFGQLHNTIVHTGIQVCTDFKLQNKMQK 599
++I+GLP L KV ++L G IN+ L G++ I G ++C D K++++++K
Sbjct: 595 KYISGLPPLFAEKVRNSLRK-EGGGSINYHYLDIGKITQKIQLVGAELCNDLKIKDQLKK 653
Query: 600 DMQISRKEVGSFCEQYGVEPLQAPSSKARRINRSK--TKPQGYK-------KRKPFQKKP 650
+ ++E+G FC Q+G Q P +R SK TKP KRKP K+
Sbjct: 654 QRILGKREMGDFCYQFG---FQDPYVYRKRKTHSKPMTKPNDKSKMSFQATKRKP--KRI 708
Query: 651 YEQNPSTQQPTNKKNVNPKKKNVCWKCGKPGHYANKC---KTQQKINELDLDQKLKD--S 705
Y +N TQ K+ +C+KCG GH AN+C K +++I L LD + +D
Sbjct: 709 YNKNIRTQD-------TESKETICYKCGLKGHIANRCFKSKVKKEIQAL-LDSESEDVKE 760
Query: 706 LISVLINSEDPHYSSEYEYEGDEDQDINLVEYSSDS---HSSSEEERECVKDSSGFCDCA 762
+ ++N+ D SS+ E ++ +IN + S S +SEEE +
Sbjct: 761 KLEAILNNIDNDLSSDEE----KNAEINCCQDSGCSCCEPDNSEEESD------------ 804
Query: 763 GCLGQNINMLTQDQTMSLISIIDKMEDSPLRDEFLQQLNLLVKKE-ETLKKDI-SPPINS 820
+NI +LT + ++ + ++D + L++ VK + + LKKDI S
Sbjct: 805 ----ENILVLTSLEEF-VLDTFETIQDPEEKRRVLEKFLSRVKTDKDKLKKDIQKSKFLS 859
Query: 821 MNEIFNRF----RPKPTITLSDLQEEVKILKEEINQLKQNDLSLEFRLIELAGIKIIESQ 876
++E+F R + L L E+ KI+K+++ ++++ LE + G + E
Sbjct: 860 IDEVFKRLDEQKKKNEQPDLISLFEDQKIMKQDLEEIRKRLYMLESK----EGFHMEEKD 915
Query: 877 ETLASASGIKQIPDKKNEDNNSQEPNENYLNLLNLVITHKWHSEITL-VINDFRINIVAL 935
E + + +++ + + + KW++E+ I+ + L
Sbjct: 916 EPI--------------------QEDDHVVGTIQKYMKQKWYTEVMYRFIDGSYFQHITL 955
Query: 936 IDSGADINCIQEGLIPTQFYEKTKEGVNSANGSKMNIQYKLSNAKICKNQICFKSSFVLV 995
IDSGAD++CI+EG+IP +++ K + A+G + ++Y++ IC +++C K+SF+LV
Sbjct: 956 IDSGADVSCIREGIIPHKYFYKAAHRIRGADGRLLTVEYQIPEIYICISEVCIKTSFLLV 1015
Query: 996 KNMTEKVILGTLFICLLYPKVHT------------IALPY---IDEFNEKDIPTKARPIQ 1040
KN+ + VILGT F+ L+ P + T ++L + DE ++ I TK +
Sbjct: 1016 KNLKQDVILGTPFLSLIRPFLVTNEDIQFKIMDKQVSLRFSSNTDEILDQLIQTKREQV- 1074
Query: 1041 MNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASE 1081
+N YL YL K + K+ W Y+L A++
Sbjct: 1075 VNTIYLH--DNSFPSYLPKSMDLPGKTSWE--DRYLLKATK 1111
Score = 57.4 bits (137), Expect = 6e-06
Identities = 32/107 (29%), Positives = 54/107 (49%)
Query: 2287 GSFPPDILKKTTEKLSAAEGSRLIIKYKIPSAIIKNGSLEIETPFLLVRNLSQKVIIGTP 2346
G P K ++ A+G L ++Y+IP I + I+T FLLV+NL Q VI+GTP
Sbjct: 968 GIIPHKYFYKAAHRIRGADGRLLTVEYQIPEIYICISEVCIKTSFLLVKNLKQDVILGTP 1027
Query: 2347 FIKKLFPYDTDVNGITVQHLGQPILFKFSEPPFDKTLNVISYKEKQI 2393
F+ + P+ I + + + + +FS + +I K +Q+
Sbjct: 1028 FLSLIRPFLVTNEDIQFKIMDKQVSLRFSSNTDEILDQLIQTKREQV 1074
>UniRef100_Q6Z3S5 Hypothetical protein OSJNBa0025J22.29 [Oryza sativa]
Length = 1275
Score = 384 bits (985), Expect = e-104
Identities = 316/1159 (27%), Positives = 522/1159 (44%), Gaps = 216/1159 (18%)
Query: 11 SPKQETIMLEANLKRSLVQVPKKLTHEEVMSNVPEEWVFKNSLPQPKVHNTQVRELFQEG 70
+PK T +L+ N + +PK L +E+ +PE+WV ++ + ++V L +
Sbjct: 161 TPKGLTTLLQTN-PNNRHTIPKTLKWDEI--TLPEKWVLSQAVEPKSMDQSEVESLIETP 217
Query: 71 ANLTLRMNRSNSFSIRTPQQLYRVDLPRSSVSSKLKGIDTTSSPSIATPIYQDEESSNYS 130
+ + + Q+ + P S+ S+ + T ++ Y+D S S
Sbjct: 218 -------DGDVEITFASKQKAFLQSRPSVSLDSRPR----TKPQNVVYATYEDN-SDEPS 265
Query: 131 PTPSQINMLSR--------VKSSFEINKDFIRKYFMAEYNKDKRLWYFKNFSKLETENLR 182
+ IN++ + FEI+KD +++ + N+ K YF+ + +R
Sbjct: 266 ISDFDINVIELDVGFVIAIEEDEFEIDKDLLKRELRLQKNRPKMKRYFERVDEPFRLKIR 325
Query: 183 TLYYEEMEVSETNIYFFD*FENYCIKNNLNYPFWKNV------------NPITKIDTIWK 230
L+++EM NI+FFD +E+ +++ + KN+ I K+ T W+
Sbjct: 326 ELWHKEMREQRKNIFFFDWYESSQVRHFEEFFKRKNMIKKEQKSEAEDLTVIKKVSTEWE 385
Query: 231 TPDNNTITSEYPPQTGVKIAIKDGQ-----EIEASPYKVEANKVH-----QQLNFANTML 280
T N + S +PP V+++ G+ I + Y A H +Q N+AN L
Sbjct: 386 TASGNKVDSVHPPFESVQLSHNGGKACPLKSISKNTYGETAKVEHIGHLVEQQNYANISL 445
Query: 281 TVMSKQLGRIEVQKPEPSTSKKSSSSFTLGNPLQTPIFKIPQYTKEEFNSFNLSGSVEKL 340
+ +Q RIE E + G P
Sbjct: 446 RSLGQQTNRIETILME---------GYKTGRP---------------------------- 468
Query: 341 KEKLNKLSINHLDKDLNINKIRKFGP-DNNTRAYYPRPSYSDMRFEERREFVQNT--FSG 397
+K +NKI K + TR YYPRP+Y+D++FEE + +QN ++G
Sbjct: 469 ------------EKYDMVNKIFKPSTLTSTTRNYYPRPTYADLQFEEMPQ-IQNMIYYNG 515
Query: 398 TSLDEWNIDGFSEQNILDITHQMIMAATAYKVHNNTDRNAALMITHGFTGQLRGWWDNIM 457
+ EWN+DGF+E I + HQMIM A A + N +R AA MI GF+GQL+GWW+N +
Sbjct: 516 KEIVEWNLDGFTEYQIFTLCHQMIMYANACIANGNKEREAANMIVIGFSGQLKGWWNNYL 575
Query: 458 TPEDKSTIL--EKKKENGE---------------EDAVATLIYTIILHFIGDPSIFRERA 500
+ IL K+ + G DA+ATLIY II HF G+ E+
Sbjct: 576 NETQRQEILCAVKRDDQGRPLPDRDGNGNPTGNISDALATLIYNIIYHFAGNYHDIYEKN 635
Query: 501 SSQLSNLYCPTMSDYRWYKDTFFSKVTLREDGNNAFWKERFIAGLPRLMQSKVLDNLSLF 560
QL NL C TMSD+RWYKDTF SK+ + N FWKE++I+GLP L KV ++L
Sbjct: 636 REQLINLKCKTMSDFRWYKDTFLSKLYTLPEPNQDFWKEKYISGLPPLFAEKVRNSLRK- 694
Query: 561 NQGNPINFGSLSFGQLHNTIVHTGIQVCTDFKLQNKMQKDMQISRKEVGSFCEQYGVEPL 620
G IN+ L G++ I G ++C D K++++++K + ++E+G FC Q+G
Sbjct: 695 EGGGSINYHYLDIGKITQKIQLVGAELCNDLKIKDQLKKQRILGKREMGDFCYQFG---F 751
Query: 621 QAPSSKARRINRSK--TKPQGYK-------KRKPFQKKPYEQNPSTQQPTNKKNVNPKKK 671
Q P +R SK TKP KRKP K+ Y +N TQ K+
Sbjct: 752 QDPYVHRKRKTHSKPMTKPNDKSKMSFQATKRKP--KRIYNKNIRTQD-------TESKE 802
Query: 672 NVCWKCGKPGHYANKCKTQQKINELDLDQKLKDSLISVLINSEDPHYSSEYEYEGDEDQD 731
+C+KCG GH AN+C K+K I L++SE + E
Sbjct: 803 TICYKCGLKGHIANRC----------FKSKVKKE-IQALLDSESEDVKEKLE------AI 845
Query: 732 INLVEYSSDSHSSSEEERECVKDSSGFCDCAGCLGQNINMLTQDQTMSLISI-------I 784
+N ++ S S E C +DS C+ C N + + + L S+
Sbjct: 846 LNNIDNDSSSDEEKNAEINCCQDSG----CSCCEPDNSEEESDENILVLTSLEEFVLDTF 901
Query: 785 DKMEDSPLRDEFLQQLNLLVKKE-ETLKKDI-SPPINSMNEIFNRF----RPKPTITLSD 838
+ ++D + L++ VK + + LKKDI S++E+F R + L
Sbjct: 902 ETIQDPEEKRRVLEKFLSRVKTDKDKLKKDIQKSKFLSIDEVFKRLDEQKKKNEQPDLIS 961
Query: 839 LQEEVKILKEEINQLKQNDLSLEFRLIELAGIKIIESQETLASASGIKQIPDKKNEDNNS 898
L E+ KI+K+++ ++++ LE + G + E E +
Sbjct: 962 LFEDQKIMKQDLEEIRKRLYMLESK----EGFHMEEKDEPI------------------- 998
Query: 899 QEPNENYLNLLNLVITHKWHSEITL-VINDFRINIVALIDSGADINCIQEGLIPTQFYEK 957
+ +++ + + + KW++E+ I+ + LIDSGAD+NCI+EG+IP +++ K
Sbjct: 999 -QEDDHVVGTIQKYMKQKWYTEVMYRFIDGSYFQHITLIDSGADVNCIREGIIPHKYFYK 1057
Query: 958 TKEGVNSANGSKMNIQYKLSNAKICKNQICFKSSFVLVKNMTEKVILGTLFICLLYPKVH 1017
+ A+G + ++Y++ IC ++C K+SF+LVKN+ + VILGT F+ L+ P +
Sbjct: 1058 AAHRIRGADGGLLTVEYQIPEIYICITEVCIKTSFLLVKNLKQDVILGTPFLSLIRPFLV 1117
Query: 1018 T------------IALPY---IDEFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLI 1062
T ++L + DE ++ + TK + +N YL YL K +
Sbjct: 1118 TNEDIQFKIMGKQVSLRFSSNTDEILDQLVQTKREQV-VNTIYLH--DNSFPSYLPKSMD 1174
Query: 1063 RHSKSPWSCIGFYVLNASE 1081
K+ W Y+L A++
Sbjct: 1175 LPGKTSWE--DRYLLKATK 1191
Score = 59.3 bits (142), Expect = 1e-06
Identities = 32/107 (29%), Positives = 55/107 (50%)
Query: 2287 GSFPPDILKKTTEKLSAAEGSRLIIKYKIPSAIIKNGSLEIETPFLLVRNLSQKVIIGTP 2346
G P K ++ A+G L ++Y+IP I + I+T FLLV+NL Q VI+GTP
Sbjct: 1048 GIIPHKYFYKAAHRIRGADGGLLTVEYQIPEIYICITEVCIKTSFLLVKNLKQDVILGTP 1107
Query: 2347 FIKKLFPYDTDVNGITVQHLGQPILFKFSEPPFDKTLNVISYKEKQI 2393
F+ + P+ I + +G+ + +FS + ++ K +Q+
Sbjct: 1108 FLSLIRPFLVTNEDIQFKIMGKQVSLRFSSNTDEILDQLVQTKREQV 1154
>UniRef100_Q7XFZ6 Hypothetical protein [Oryza sativa]
Length = 1204
Score = 290 bits (741), Expect = 5e-76
Identities = 213/688 (30%), Positives = 333/688 (47%), Gaps = 137/688 (19%)
Query: 368 NNTRAYYPRPSYSDMRFEERREFVQNTF-SGTSLDEWNIDGFSEQNILDITHQMIMAATA 426
+ TR YYPRP+Y+D++FEE + T+ +G + EWN+DGF+E I + HQMIM A A
Sbjct: 13 STTRNYYPRPTYADLQFEEMPQIQNMTYYNGKEIVEWNLDGFTEYQIFTLCHQMIMYANA 72
Query: 427 YKVHNNTDRNAALMITHGFTGQLRGWWDNIMTPEDKSTIL--EKKKENGE---------- 474
+ N +R AA MI GF+GQL+GWW+N + + IL K+ + G
Sbjct: 73 CIANGNKEREAANMIVIGFSGQLKGWWNNYLNETQRQEILCAVKRDDQGRPLPDRDGNGN 132
Query: 475 -----EDAVATLIYTIILHFIGDPSIFRERASSQLSNLYCPTMSDYRWYKDTFFSKVTLR 529
DA+ATLIY II HF G+ E+ QL NL C TMSD+RWYKDTF SK+
Sbjct: 133 PTGNISDALATLIYNIIYHFAGNYHDIYEKNREQLINLKCKTMSDFRWYKDTFLSKLYTL 192
Query: 530 EDGNNAFWKERFIAGLPRLMQSKVLDNLSLFNQGNPINFGSLSFGQLHNTIVHTGIQVCT 589
+ N FWKE++I+GLP L KV ++L G IN+ L G++ I G+++C
Sbjct: 193 PEPNQDFWKEKYISGLPPLFAEKVRNSLRK-EGGGSINYHYLDIGKITQKIQLVGVELCN 251
Query: 590 DFKLQNKMQKDMQISRKEVGSFCEQYGVEPLQAPSSKARRINRSK--TKPQGYK------ 641
D K++++++K + ++E+G FC Q+G Q P +R SK TKP
Sbjct: 252 DLKIKDQLKKQRILGKREMGDFCYQFG---FQDPYVYRKRKTHSKPMTKPNDKSKMSFQA 308
Query: 642 -KRKPFQKKPYEQNPSTQQPTNKKNVNPKKKNVCWKCGKPGHYANKC---KTQQKINELD 697
KRKP K+ Y +N TQ K+ +C+KCG GH AN+C K +++I L
Sbjct: 309 TKRKP--KRIYNKNIRTQD-------TESKETICYKCGLKGHIANRCFKSKVKKEIQAL- 358
Query: 698 LDQKLKD--SLISVLINSEDPHYSSEYEYEGDED--QDINLVEYSSDSHSSSEEERECVK 753
LD + +D + ++N+ D SS+ E + + QD Y D +SEEE +
Sbjct: 359 LDSESEDVKEKLEAILNNIDNDSSSDEEKNAEINCCQDSGCSCYEPD---NSEEESD--- 412
Query: 754 DSSGFCDCAGCLGQNINMLTQDQTMSLISIIDKMEDSPLRDEFLQQLNLLVKKE-ETLKK 812
+NI +LT + ++ + ++D + L++ VK + + LKK
Sbjct: 413 -------------ENILVLTSLEEF-VLDTFETIQDPEEKRRVLEKFLSRVKTDKDKLKK 458
Query: 813 DI-SPPINSMNEIFNRF----RPKPTITLSDLQEEVKILKEEINQLKQNDLSLEFRLIEL 867
DI S++E+F R + L L E+ +I+K+++ ++++ +RL
Sbjct: 459 DIQKSKFLSIDEVFKRLDEQKKKNEKPDLISLFEDQRIMKQDLEEIRKR----LYRLESK 514
Query: 868 AGIKIIESQETLASASGIKQIPDKKNEDNNSQEPNENYLNLLNLVITHKWHSEITL-VIN 926
G + E E + + ++ + + + KW++E+ I+
Sbjct: 515 EGFHMEEKDEPI--------------------QEDDQVVGTIQKYMKQKWYTEVMYRFID 554
Query: 927 DFRINIVALIDSGADINCIQEGLIPTQFYEKTKEGVNSANGSKMNIQYKLSNAKICKNQI 986
+ LIDSGAD+NCI+E +
Sbjct: 555 GSYFQHITLIDSGADVNCIRE--------------------------------------V 576
Query: 987 CFKSSFVLVKNMTEKVILGTLFICLLYP 1014
C K+SF+LVKN+ + VILGT F+ L+ P
Sbjct: 577 CIKTSFLLVKNLKQDVILGTPFLSLIRP 604
Score = 43.9 bits (102), Expect = 0.064
Identities = 22/67 (32%), Positives = 38/67 (55%)
Query: 2327 IETPFLLVRNLSQKVIIGTPFIKKLFPYDTDVNGITVQHLGQPILFKFSEPPFDKTLNVI 2386
I+T FLLV+NL Q VI+GTPF+ + P+ I + +G+ + +FS + ++
Sbjct: 578 IKTSFLLVKNLKQDVILGTPFLSLIRPFLVTNEDIQFKIMGKQVSLRFSSNTDEILDQLV 637
Query: 2387 SYKEKQI 2393
K +Q+
Sbjct: 638 QSKREQV 644
>UniRef100_Q6XKE5 Polyprotein 1 [Petunia vein clearing virus]
Length = 1886
Score = 250 bits (638), Expect = 4e-64
Identities = 133/340 (39%), Positives = 198/340 (58%), Gaps = 11/340 (3%)
Query: 981 ICKNQICFKSSFVLVKNMTEKVILGTLFICLLYPKVHTIALPYIDEFNEKDIPTKARPIQ 1040
+ KNQ+C +S + + + L F I LP+ + NE PTKA
Sbjct: 1345 VIKNQLCAESHVDFLSKCSHPLWLNQDFF---------IQLPF--KRNENINPTKASHSG 1393
Query: 1041 MNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYKPLNKVL 1100
MN ++L+ +E E LI S S W+C FYV SE RG LRLVINY+PLN L
Sbjct: 1394 MNPEHLQLALKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFL 1453
Query: 1101 KWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMP 1160
+ ++P+PNK L L K +FSKFD+KSG++Q+ + +R KT F +P H++W VMP
Sbjct: 1454 QDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWKVMP 1513
Query: 1161 QGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVV 1220
GLK APS +Q M IF P + +VY+DD+L+FS+ ++ H++ L +FI ++KK G+++
Sbjct: 1514 FGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFGVML 1573
Query: 1221 SAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCVNYVADF 1280
SAKKM + + K +FLG + G +P +KFPD Q+Q+FLG VNY+ DF
Sbjct: 1574 SAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYIRDF 1633
Query: 1281 IPNIRIVCAPLFKRLRKNSPPWSEEMSHSVREVKKLVQSL 1320
IP + +PL L+K P W + ++V+++K+L Q +
Sbjct: 1634 IPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQV 1673
Score = 64.7 bits (156), Expect = 3e-08
Identities = 132/670 (19%), Positives = 256/670 (37%), Gaps = 125/670 (18%)
Query: 136 INMLSRVKSSFEINKDFIRKYFMAEYNKDKR-------LWYFKNFSKLETENLRT---LY 185
+ + RV N+ IR + +KR L++F + K E +L+ L
Sbjct: 562 VTKVDRVSDQSSKNQGLIRVLEQQLQDLNKRICPPGTSLFHFFDQQKSEMASLKEQIRLL 621
Query: 186 YEEMEVSETNIYFFD*FENYCIKNNLNYPFWKNVNPITKIDTIWKTPDNN-TITSEYPPQ 244
E+ + +ET+ + +Y +N + P+ + P + TP ++ S+YP Q
Sbjct: 622 KEQPQKNETDTPSYQ--SSYQPFHNFSSPYMPSNPPNSPFTNFANTPQPQPSLFSQYPIQ 679
Query: 245 TGVKIAIKDGQEIEASPYKVEANKVHQQLNFANTMLTVMSKQLGRIEVQ----------K 294
SP + K+ + A K+L + EV+ +
Sbjct: 680 P-------------KSPNTFDLAKLVWEKKDAIAEEKRAKKKLQKDEVKQKTSLPPESKR 726
Query: 295 PEPSTSKKSSSSFTLGNPLQTPIFKI--PQYTKEEFNSFNLSGSVEKLKEKLNKLSINHL 352
P+P +S F + +P ++++ P E+ +S + + E +++ + ++
Sbjct: 727 PDPQSSSHLGDQFMISDPTLPKVYELNEPSVPSEDTSSQSYISTEESVEDTDSFSVVSEE 786
Query: 353 DKDLN-INKIRKFGPDNNT----RAYYPRPSYSDMRF--------------EERREFVQN 393
L+ ++ P+NN + + RP+ ++ E R E
Sbjct: 787 STQLSQLSSSSNDSPENNENTLPQTFMVRPTEPEISEVEDEVDGMTEEPIPERRPEITPP 846
Query: 394 TFSGTSLDEWNIDGFS----EQNILDITHQMIMAATAYKVHNNTDRNAALMI---THGFT 446
GT +++D S + I D M+ +R L++ T +
Sbjct: 847 KMVGTGFHTFSLDDISITKWPERIQDFHTWMLTKQLV-------EREPFLILSEFTARLS 899
Query: 447 GQLRGWWDNIMTPEDKSTILEKKKENGEEDAVATLIYTIILHFIGDPSIFRERASSQLSN 506
G LR WW+++ P+DK+ L + I + +F GD S +E Q+
Sbjct: 900 GTLREWWNSV-GPDDKNRFLTSQDFTWN-------IRILYSYFCGDQSQNKEELRRQIFE 951
Query: 507 LYCPTMSDYRWYKDTFFSKVT--LREDGNNAFWKERFIAGLPRLMQSKVLDNLSLFNQGN 564
+ C +S R D F ++ G + K+ FI+ LP ++ ++ + + +G
Sbjct: 952 MKC--LSYDRKKIDRHFQRMIKLFYHIGGDISLKQAFISSLPPILSERI--SALIKERGT 1007
Query: 565 PINFGSLSFGQLHNTIVHTGIQVCTDFKLQNKMQK---DMQ--------ISRKEVGSFCE 613
+ + G + T + +C+ K N+M+K D++ I + + G C
Sbjct: 1008 SVT--QMHVGDIRQTAFYVLDDLCSKRKFFNQMKKMSRDLEKACTKSDLIIKGDKG--CS 1063
Query: 614 QYGVEPLQAPSSKARRINRSKTKP-QGYKKRKPFQKKPYEQNPSTQQPTNKKNVNPKKKN 672
Y P + K ++ K + + Y+KR+ F +K Q+P +
Sbjct: 1064 GY-CNPSRRRKYKRFKLPSFKERDGRQYRKRRRFFRKSKTSKAMRQKPRS---------- 1112
Query: 673 VCWKCGKPGHYANKCKTQ----------QKINELDLDQKLKDSLISVLINSEDPHYSSEY 722
C+ CGK GH++ C QK +D++ L+ SED +S E+
Sbjct: 1113 -CFTCGKIGHFSRNCPQNKKSIKLISEIQKYTGIDIEDDLESVFSIEDEPSEDTLFSLEF 1171
Query: 723 --EYEGDEDQ 730
EY G++ Q
Sbjct: 1172 YEEYAGEQYQ 1181
Score = 58.2 bits (139), Expect = 3e-06
Identities = 27/44 (61%), Positives = 33/44 (74%)
Query: 2434 SSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
S W+C AFYVNK++E RG RLVINY+PLN L ++PIPNK
Sbjct: 1420 SQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFLQDDKFPIPNK 1463
>UniRef100_Q6XKE6 Polyprotein 1 [Petunia vein clearing virus]
Length = 2180
Score = 250 bits (638), Expect = 4e-64
Identities = 133/340 (39%), Positives = 198/340 (58%), Gaps = 11/340 (3%)
Query: 981 ICKNQICFKSSFVLVKNMTEKVILGTLFICLLYPKVHTIALPYIDEFNEKDIPTKARPIQ 1040
+ KNQ+C +S + + + L F I LP+ + NE PTKA
Sbjct: 1345 VIKNQLCAESHVDFLSKCSHPLWLNQDFF---------IQLPF--KRNENINPTKASHSG 1393
Query: 1041 MNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYKPLNKVL 1100
MN ++L+ +E E LI S S W+C FYV SE RG LRLVINY+PLN L
Sbjct: 1394 MNPEHLQLALKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFL 1453
Query: 1101 KWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMP 1160
+ ++P+PNK L L K +FSKFD+KSG++Q+ + +R KT F +P H++W VMP
Sbjct: 1454 QDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWKVMP 1513
Query: 1161 QGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVV 1220
GLK APS +Q M IF P + +VY+DD+L+FS+ ++ H++ L +FI ++KK G+++
Sbjct: 1514 FGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFGVML 1573
Query: 1221 SAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCVNYVADF 1280
SAKKM + + K +FLG + G +P +KFPD Q+Q+FLG VNY+ DF
Sbjct: 1574 SAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYIRDF 1633
Query: 1281 IPNIRIVCAPLFKRLRKNSPPWSEEMSHSVREVKKLVQSL 1320
IP + +PL L+K P W + ++V+++K+L Q +
Sbjct: 1634 IPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQV 1673
Score = 62.4 bits (150), Expect = 2e-07
Identities = 130/664 (19%), Positives = 246/664 (36%), Gaps = 113/664 (17%)
Query: 136 INMLSRVKSSFEINKDFIRKYFMAEYNKDKR-------LWYFKNFSKLETENLRT---LY 185
+ + RV N+ IR + +KR L++F + K E +L+ L
Sbjct: 562 VTKVDRVSDQSSKNQGLIRVLEQQLQDLNKRICPPGTSLFHFFDQQKSEMASLKEQIRLL 621
Query: 186 YEEMEVSETNIYFFD*FENYCIKNNLNYPFWKNVNPITKIDTIWKTPDNN-TITSEYPPQ 244
E+ + +ET+ + +Y +N + P+ + P + TP ++ S+YP Q
Sbjct: 622 KEQPQKNETDTPSYQ--SSYQPFHNFSSPYMPSNPPNSPFTNFANTPQPQPSLFSQYPIQ 679
Query: 245 TGVKIAIKDGQEIEASPYKVEANKVHQQLNFANTMLTVMSKQLGRIEVQ----------K 294
SP + K+ + A K+L + EV+ +
Sbjct: 680 P-------------KSPNTFDLAKLVWEKKDAIAEEKRAKKKLQKDEVKQKTSLPPESKR 726
Query: 295 PEPSTSKKSSSSFTLGNPLQTPIFKI--PQYTKEEFNSFNLSGSVEKLKEKLNKLSINHL 352
P+P +S F + +P ++++ P E+ +S + + E +++ + ++
Sbjct: 727 PDPQSSSHLGDQFMISDPTLPKVYELNEPSVPSEDTSSQSYISTEESVEDTDSFSVVSEE 786
Query: 353 DKDLN-INKIRKFGPDNNT----RAYYPRPSYSDMRF--------------EERREFVQN 393
L+ ++ P+NN + + RP+ ++ E R E
Sbjct: 787 STQLSQLSSSSNDSPENNENTLPQTFMVRPTEPEISEVEDEVDGMTEEPIPERRPEITPP 846
Query: 394 TFSGTSLDEWNIDGFS----EQNILDITHQMIMAATAYKVHNNTDRNAALMI---THGFT 446
GT +++D S + I D M+ +R L++ T +
Sbjct: 847 KMVGTGFHTFSLDDISITKWPERIQDFHTWMLTKQLV-------EREPFLILSEFTARLS 899
Query: 447 GQLRGWWDNIMTPEDKSTILEKKKENGEEDAVATLIYTIILHFIGDPSIFRERASSQLSN 506
G LR WW+++ P+DK+ L + I + +F GD S +E Q+
Sbjct: 900 GTLREWWNSV-GPDDKNRFLTSQDFTWN-------IRILYSYFCGDQSQNKEELRRQIFE 951
Query: 507 LYCPTMSDYRWYKDTFFSKVT--LREDGNNAFWKERFIAGLPRLMQSKVLDNLSLFNQGN 564
+ C +S R D F ++ G + K+ FI+ LP ++ ++
Sbjct: 952 MKC--LSYDRKKIDRHFQRMIKLFYHIGGDISLKQAFISSLPPILSERI---------SA 1000
Query: 565 PINFGSLSFGQLH-NTIVHTGIQVCTDFKLQNKMQKDMQISRKEVGSFCEQY-----GVE 618
I S Q+H I TG V D + K M+ +++ C + G +
Sbjct: 1001 LIKERGTSGTQMHVGDIRQTGFYVLDDLCSKRKFFNQMKKMSRDLEKACTKSDLIIKGDK 1060
Query: 619 PLQAPSSKARRINRSKTKPQGYKKRKPFQKKPYEQNPSTQQPTNKKNVNPKKKNVCWKCG 678
+ +RR + K +K+R Q Y + + + +K C+ CG
Sbjct: 1061 GCSGYCNPSRRRKYKRFKLPSFKERDGRQ---YRKRRRFFRKSKTSKAMRQKPRSCFTCG 1117
Query: 679 KPGHYANKCKTQ----------QKINELDLDQKLKDSLISVLINSEDPHYSSEY--EYEG 726
K GH++ C QK +D++ L+ SED +S E+ EY G
Sbjct: 1118 KIGHFSRNCPQNKKSIKLISEIQKYTGIDIEDDLESVFSIEDEPSEDTLFSLEFYEEYAG 1177
Query: 727 DEDQ 730
++ Q
Sbjct: 1178 EQYQ 1181
Score = 58.2 bits (139), Expect = 3e-06
Identities = 27/44 (61%), Positives = 33/44 (74%)
Query: 2434 SSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
S W+C AFYVNK++E RG RLVINY+PLN L ++PIPNK
Sbjct: 1420 SQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFLQDDKFPIPNK 1463
>UniRef100_Q91DM0 ORF I polyprotein [Petunia vein clearing virus]
Length = 2179
Score = 249 bits (635), Expect = 1e-63
Identities = 133/340 (39%), Positives = 197/340 (57%), Gaps = 11/340 (3%)
Query: 981 ICKNQICFKSSFVLVKNMTEKVILGTLFICLLYPKVHTIALPYIDEFNEKDIPTKARPIQ 1040
+ KNQ+C S + + + L F I LP+ + NE PTKA
Sbjct: 1344 VIKNQLCADSHVDFLSKCSHPLWLNQDFF---------IQLPF--KKNENINPTKASHSG 1392
Query: 1041 MNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYKPLNKVL 1100
MN ++L+ +E E LI S S W+C FYV SE RG LRLVINY+PLN L
Sbjct: 1393 MNPEHLQLAIKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFL 1452
Query: 1101 KWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMP 1160
+ ++P+PNK L L K +FSKFD+KSG++Q+ + +R KT F +P H++W VMP
Sbjct: 1453 QDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWKVMP 1512
Query: 1161 QGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVV 1220
GLK APS +Q M IF P + +VY+DD+L+FS+ ++ H++ L +FI ++KK G+++
Sbjct: 1513 FGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFGVML 1572
Query: 1221 SAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCVNYVADF 1280
SAKKM + + K +FLG + G +P +KFPD Q+Q+FLG VNY+ DF
Sbjct: 1573 SAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYIRDF 1632
Query: 1281 IPNIRIVCAPLFKRLRKNSPPWSEEMSHSVREVKKLVQSL 1320
IP + +PL L+K P W + ++V+++K+L Q +
Sbjct: 1633 IPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQV 1672
Score = 62.4 bits (150), Expect = 2e-07
Identities = 94/483 (19%), Positives = 186/483 (38%), Gaps = 67/483 (13%)
Query: 291 EVQKPEPSTSKKSSSSFTLGNPLQTPIFKI--PQYTKEEFNSFNLSGSVEKLKEKLNKLS 348
E ++P+P +S F + +P ++ + P E+ +S + + E +++ +
Sbjct: 722 ESKRPDPQSSSHLGDQFMISDPALPKVYALNEPSVPSEDTSSQSYISTEESVEDTDSFSV 781
Query: 349 INHLDKDLN-INKIRKFGPDNNT----RAYYPRPSYSDMRF--------------EERRE 389
++ L+ ++ P+NN + + RP+ ++ E R E
Sbjct: 782 VSEESTQLSQLSSSSNDSPENNENTLPQTFMVRPTEPEISEVEDEVDGMTEEPIPERRPE 841
Query: 390 FVQNTFSGTSLDEWNIDGFS----EQNILDITHQMIMAATAYKVHNNTDRNAALMI---T 442
GT +++D S + I D M+ +R L++ T
Sbjct: 842 ITPPKMVGTGFHTFSLDDISITKWPERIQDFHTWMLTKQLV-------EREPFLILSEFT 894
Query: 443 HGFTGQLRGWWDNIMTPEDKSTILEKKKENGEEDAVATLIYTIILHFIGDPSIFRERASS 502
+G LR WW+++ P+DK+ L + I + +F GD S +E
Sbjct: 895 ARLSGTLREWWNSV-GPDDKNRFLTSQDFTWN-------IRILYSYFCGDQSQNKEELRR 946
Query: 503 QLSNLYCPTMSDYRWYKDTFFSKVT--LREDGNNAFWKERFIAGLPRLMQSKVLDNLSLF 560
Q+ + C +S R D F ++ G + K+ FI+ LP ++ ++ + +
Sbjct: 947 QIFEMKC--LSYDRKKIDRHFQRMIKLFYHIGGDISLKQAFISSLPPILSERI--SALIK 1002
Query: 561 NQGNPINFGSLSFGQLHNTIVHTGIQVCTDFKLQNKMQK-DMQISRKEVGSFCEQYGVEP 619
+G + + G + T + +C+ K N+M+K + + S G +
Sbjct: 1003 ERGTSVT--QMHVGDIRQTAFYVLDDLCSKRKFFNQMKKMSRDLEKACTKSDLIIKGDKG 1060
Query: 620 LQAPSSKARRINRSKTKPQGYKKRKPFQKKPYEQNPSTQQPTNKKNVNPKKKNVCWKCGK 679
+ +RR + K +K+R Q Y + + + +K C+ CGK
Sbjct: 1061 CSGYCNPSRRRKYKRFKLPSFKERDGRQ---YRKRRRFFRRSKTSKAMRQKPRSCFTCGK 1117
Query: 680 PGHYANKCKTQ----------QKINELDLDQKLKDSLISVLINSEDPHYSSEY--EYEGD 727
GH++ C QK +D++ L+ SED +S E+ EY G+
Sbjct: 1118 IGHFSRNCPQNKKSIKLISEIQKYTGIDIEDDLESVFSIEDEPSEDTLFSLEFYEEYAGE 1177
Query: 728 EDQ 730
+ Q
Sbjct: 1178 QYQ 1180
Score = 58.2 bits (139), Expect = 3e-06
Identities = 27/44 (61%), Positives = 33/44 (74%)
Query: 2434 SSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
S W+C AFYVNK++E RG RLVINY+PLN L ++PIPNK
Sbjct: 1419 SQWACEAFYVNKRSEQVRGKLRLVINYQPLNHFLQDDKFPIPNK 1462
>UniRef100_Q83169 Reverse transcriptase [Cauliflower mosaic virus]
Length = 680
Score = 223 bits (568), Expect = 6e-56
Identities = 116/288 (40%), Positives = 176/288 (60%), Gaps = 1/288 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K +P++ + + ++I E L+ +I+ SKSP F V N +E RG R+V+NYK
Sbjct: 249 KVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYK 308
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
+NK Y LPNK +L+ + IFS FD KSG++Q+ + +E R TAF P GHY
Sbjct: 309 AMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHY 368
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EWNV+P GLK APS +Q M++ F + F VY+DD+LVFS + H+ H+ ++
Sbjct: 369 EWNVVPFGLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCN 428
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
++G+++S KK ++F+ K FLG EI +G P LE KFPD L++K QLQRFLG +
Sbjct: 429 QHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGIL 488
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLP 1321
Y +D+IP + + PL +L++N P W++E + +++VKK +Q P
Sbjct: 489 TYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFP 536
Score = 43.5 bits (101), Expect = 0.083
Identities = 21/45 (46%), Positives = 27/45 (59%)
Query: 2433 KSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
KS AF VN +AE RG R+V+NYK +N+A Y +PNK
Sbjct: 280 KSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNLPNK 324
>UniRef100_Q7XRB3 OSJNBa0006B20.7 protein [Oryza sativa]
Length = 685
Score = 223 bits (568), Expect = 6e-56
Identities = 123/295 (41%), Positives = 178/295 (59%), Gaps = 2/295 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K PI+ + ++ + I E L G IR S+SP F V N +E RG R+VINYK
Sbjct: 182 KTSPIEATPKDIEEFKMHIEELLKLGAIRESRSPHRSAAFIVRNHAEEVRGKSRMVINYK 241
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
LN Y +PNK + I R+ FSKFD+K+G++Q+ + EE TAF P GHY
Sbjct: 242 RLNNNTIEDAYNIPNKQEWINRIQGSKYFSKFDLKAGFWQVKMAEESIEWTAFTCPQGHY 301
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EW VMP GLKNAP+ +Q M +IF F +VY+DD+LVFS+ +H+ HLE FI ++
Sbjct: 302 EWLVMPLGLKNAPAIFQRKMQNIFNENQEFILVYIDDLLVFSRTYKEHIAHLEIFIRKVE 361
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
+NGL++S KKM+I + K FLG+EI +G+I + + +FPD + +K +LQ+FLG V
Sbjct: 362 QNGLILSKKKMEICKEKINFLGHEIGEGKIHLQEHIAKKILQFPDAMNDKKKLQQFLGLV 421
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSPPW-SEEMSHSVREVKKLVQSL-PIVTPFQ 1327
NY + I N+ + PL+ +LRKN + + E VR +K+ V+ L P+ P +
Sbjct: 422 NYARNHINNLAKLAGPLYAKLRKNGQRYFNSEDIKLVRLIKERVKELKPLELPLE 476
Score = 43.1 bits (100), Expect = 0.11
Identities = 34/94 (36%), Positives = 44/94 (46%), Gaps = 9/94 (9%)
Query: 2390 EKQINFLKEEISYKNIEVQLQQPSV---KSRIENIL--GNIQSSICSCKSSWSCAAFYVN 2444
E +IN + E K ++ + K IE +L G I+ S +S AAF V
Sbjct: 169 ECKINIINPEYIIKTSPIEATPKDIEEFKMHIEELLKLGAIRES----RSPHRSAAFIVR 224
Query: 2445 KQAEIERGTPRLVINYKPLNQALCWIRYPIPNKK 2478
AE RG R+VINYK LN Y IPNK+
Sbjct: 225 NHAEEVRGKSRMVINYKRLNNNTIEDAYNIPNKQ 258
>UniRef100_Q7XEP7 Putative rice tungro bacilliform virus; reverse transcriptase [Oryza
sativa]
Length = 2877
Score = 223 bits (567), Expect = 8e-56
Identities = 122/295 (41%), Positives = 176/295 (59%), Gaps = 2/295 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K PI+ + ++ + I E L G IR S+SP F V N +E RG R+VINYK
Sbjct: 2295 KTSPIEATPKDIEEFKMHIEELLKLGAIRESRSPHRSAAFIVRNHAEEVRGKSRMVINYK 2354
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
LN Y +PNK + I R+ FSKFD+K+G++Q+ + EE TAF P GHY
Sbjct: 2355 RLNDNTIEDAYNIPNKQEWINRIQGSKYFSKFDLKAGFWQVKMAEESIEWTAFTCPQGHY 2414
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EW VMP GLKNAP+ +Q M +IF F +VY+DD+L+FSK H+ HLE F ++
Sbjct: 2415 EWLVMPLGLKNAPALFQRKMQNIFNDNQEFVLVYIDDLLIFSKTYKDHIAHLELFFRKVE 2474
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
+NGL++S KKM+I + K FLG+EI +G+I + + +FPD + +K +LQ+FLG V
Sbjct: 2475 QNGLILSKKKMEICKEKVNFLGHEIGEGKIHLQEHIAKKILQFPDAMNDKKKLQQFLGLV 2534
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSPPW-SEEMSHSVREVKKLVQSL-PIVTPFQ 1327
NY + I N+ + PL+ +LRKN + + E VR +K+ V+ L P+ P +
Sbjct: 2535 NYARNHINNLAKLAGPLYTKLRKNGQKYFNSEDIKLVRLIKEKVKELKPLELPLE 2589
Score = 43.1 bits (100), Expect = 0.11
Identities = 34/94 (36%), Positives = 44/94 (46%), Gaps = 9/94 (9%)
Query: 2390 EKQINFLKEEISYKNIEVQLQQPSV---KSRIENIL--GNIQSSICSCKSSWSCAAFYVN 2444
E +IN + E K ++ + K IE +L G I+ S +S AAF V
Sbjct: 2282 ECKINIINPEYIIKTSPIEATPKDIEEFKMHIEELLKLGAIRES----RSPHRSAAFIVR 2337
Query: 2445 KQAEIERGTPRLVINYKPLNQALCWIRYPIPNKK 2478
AE RG R+VINYK LN Y IPNK+
Sbjct: 2338 NHAEEVRGKSRMVINYKRLNDNTIEDAYNIPNKQ 2371
>UniRef100_P03554 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 679
Score = 223 bits (567), Expect = 8e-56
Identities = 116/288 (40%), Positives = 176/288 (60%), Gaps = 1/288 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K +P++ + + ++I E L+ +I+ SKSP F V N +E RG R+V+NYK
Sbjct: 248 KVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYK 307
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
+NK Y LPNK +L+ + IFS FD KSG++Q+ + +E R TAF P GHY
Sbjct: 308 AMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHY 367
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EWNV+P GLK APS +Q M++ F + F VY+DD+LVFS + H+ H+ ++
Sbjct: 368 EWNVVPFGLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCN 427
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
++G+++S KK ++F+ K FLG EI +G P LE KFPD L++K QLQRFLG +
Sbjct: 428 QHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGIL 487
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLP 1321
Y +D+IP + + PL +L++N P W++E + +++VKK +Q P
Sbjct: 488 TYASDYIPKLAQIRKPLQAKLKENVPWRWTKEDTLYMQKVKKNLQGFP 535
Score = 43.5 bits (101), Expect = 0.083
Identities = 21/45 (46%), Positives = 27/45 (59%)
Query: 2433 KSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
KS AF VN +AE RG R+V+NYK +N+A Y +PNK
Sbjct: 279 KSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNLPNK 323
>UniRef100_Q02964 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 679
Score = 223 bits (567), Expect = 8e-56
Identities = 116/288 (40%), Positives = 176/288 (60%), Gaps = 1/288 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K +P++ + + ++I E L+ +I+ SKSP F V N +E RG R+V+NYK
Sbjct: 248 KVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYK 307
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
+NK Y LPNK +L+ + IFS FD KSG++Q+ + +E R TAF P GHY
Sbjct: 308 AMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHY 367
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EWNV+P GLK APS +Q M++ F + F VY+DD+LVFS + H+ H+ ++
Sbjct: 368 EWNVVPFGLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCN 427
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
++G+++S KK ++F+ K FLG EI +G P LE KFPD L++K QLQRFLG +
Sbjct: 428 QHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGIL 487
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLP 1321
Y +D+IP + + PL +L++N P W++E + +++VKK +Q P
Sbjct: 488 TYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFP 535
Score = 43.5 bits (101), Expect = 0.083
Identities = 21/45 (46%), Positives = 27/45 (59%)
Query: 2433 KSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
KS AF VN +AE RG R+V+NYK +N+A Y +PNK
Sbjct: 279 KSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATIGDAYNLPNK 323
>UniRef100_P03555 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 679
Score = 223 bits (567), Expect = 8e-56
Identities = 116/288 (40%), Positives = 176/288 (60%), Gaps = 1/288 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K +P++ + + ++I E L+ +I+ SKSP F V N +E RG R+V+NYK
Sbjct: 248 KVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYK 307
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
+NK Y LPNK +L+ + IFS FD KSG++Q+ + +E R TAF P GHY
Sbjct: 308 AMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHY 367
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EWNV+P GLK APS +Q M++ F + F VY+DD+LVFS + H+ H+ ++
Sbjct: 368 EWNVVPFGLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCN 427
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
++G+++S KK ++F+ K FLG EI +G P LE KFPD L++K QLQRFLG +
Sbjct: 428 QHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGIL 487
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLP 1321
Y +D+IP + + PL +L++N P W++E + +++VKK +Q P
Sbjct: 488 TYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFP 535
Score = 43.5 bits (101), Expect = 0.083
Identities = 21/45 (46%), Positives = 27/45 (59%)
Query: 2433 KSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
KS AF VN +AE RG R+V+NYK +N+A Y +PNK
Sbjct: 279 KSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATIGDAYNLPNK 323
>UniRef100_Q66162 ORF V [Cauliflower mosaic virus]
Length = 680
Score = 222 bits (566), Expect = 1e-55
Identities = 116/288 (40%), Positives = 176/288 (60%), Gaps = 1/288 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K +P++ + + ++I E L+ +I+ SKSP F V N +E RG R+V+NYK
Sbjct: 249 KVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYK 308
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
+NK Y LPNK +L+ + IFS FD KSG++Q+ + +E R TAF P GHY
Sbjct: 309 AMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHY 368
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EWNV+P GLK APS +Q M++ F + F VY+DD+LVFS + H+ H+ ++
Sbjct: 369 EWNVVPFGLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCN 428
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
++G+++S KK ++F+ K FLG EI +G P LE KFPD L++K QLQRFLG +
Sbjct: 429 QHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGIL 488
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLP 1321
Y +D+IP + + PL +L++N P W++E + +++VKK +Q P
Sbjct: 489 TYASDYIPKLAQIRKPLQAKLKENVPWRWTKEDTLYMQKVKKNLQGFP 536
Score = 43.5 bits (101), Expect = 0.083
Identities = 21/45 (46%), Positives = 27/45 (59%)
Query: 2433 KSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
KS AF VN +AE RG R+V+NYK +N+A Y +PNK
Sbjct: 280 KSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATIGDAYNLPNK 324
>UniRef100_Q00962 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 680
Score = 222 bits (565), Expect = 1e-55
Identities = 116/288 (40%), Positives = 177/288 (61%), Gaps = 1/288 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K +P++ + + ++I E L+ +I+ SKSP F V N +E RG R+V+NYK
Sbjct: 249 KVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAENGRGNKRMVVNYK 308
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
+NK Y LPNK +L+ + IFS FD KSG++Q+ + +E R TAF P GHY
Sbjct: 309 AMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHY 368
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EWNV+P GLK APS +Q M++ F + F VY+DD++VFS + H+ H+ ++
Sbjct: 369 EWNVVPFGLKQAPSIFQRHMDEAFRVFRKFCCVYVDDIVVFSNNEEDHLLHVAMILQKCN 428
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
++G+++S KK ++F+ K FLG EI +G P LE KFPD L++K QLQRFLG +
Sbjct: 429 QHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGIL 488
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLP 1321
Y +D+IPN+ + PL +L++N P W++E + +++VKK +Q P
Sbjct: 489 TYASDYIPNLAQMRQPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFP 536
Score = 43.1 bits (100), Expect = 0.11
Identities = 21/45 (46%), Positives = 27/45 (59%)
Query: 2433 KSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
KS AF VN +AE RG R+V+NYK +N+A Y +PNK
Sbjct: 280 KSPHMAPAFLVNNEAENGRGNKRMVVNYKAMNKATVGDAYNLPNK 324
>UniRef100_Q9WI36 Reverse transcriptase [Cauliflower mosaic virus]
Length = 674
Score = 221 bits (563), Expect = 2e-55
Identities = 116/288 (40%), Positives = 175/288 (60%), Gaps = 1/288 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K +P++ + + ++I E L+ +I+ SKSP F V N +E RG R+V+NYK
Sbjct: 243 KVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYK 302
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
+NK Y LPNK +L+ + IFS FD KSG++Q+ +E R TAF P GHY
Sbjct: 303 AMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLPDQESRPLTAFTCPQGHY 362
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EWNV+P GLK APS +Q M++ F + F VY+DD+LVFS + H+ H+ ++
Sbjct: 363 EWNVVPFGLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNTEEDHLLHVAMILQKCN 422
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
++G+++S KK ++F+ K FLG EI +G P LE KFPD L++K QLQRFLG +
Sbjct: 423 QHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGIL 482
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLP 1321
Y +D+IP + + PL +L++N P W++E + +++VKK +Q P
Sbjct: 483 TYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFP 530
Score = 43.5 bits (101), Expect = 0.083
Identities = 21/45 (46%), Positives = 27/45 (59%)
Query: 2433 KSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
KS AF VN +AE RG R+V+NYK +N+A Y +PNK
Sbjct: 274 KSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNLPNK 318
>UniRef100_P03556 Enzymatic polyprotein [Contains: Aspartic protease (EC 3.4.23.-);
Endonuclease; Reverse transcriptase (EC 2.7.7.49)]
[Cauliflower mosaic virus]
Length = 674
Score = 220 bits (561), Expect = 4e-55
Identities = 115/288 (39%), Positives = 175/288 (59%), Gaps = 1/288 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K +P++ + + ++I E L+ +I+ SKSP F V N +E RG R+V+NYK
Sbjct: 243 KVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYK 302
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
+NK Y PNK +L+ + IFS FD KSG++Q+ + +E R TAF P GHY
Sbjct: 303 AMNKATVGDAYNPPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHY 362
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EWNV+P GLK APS +Q M++ F + F VY+DD+LVFS + H+ H+ ++
Sbjct: 363 EWNVVPFGLKQAPSIFQRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCN 422
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
++G+++S KK ++F+ K FLG EI +G P LE KFPD L++K QLQRFLG +
Sbjct: 423 QHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGIL 482
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLP 1321
Y +D+IP + + PL +L++N P W++E + +++VKK +Q P
Sbjct: 483 TYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFP 530
Score = 41.6 bits (96), Expect = 0.32
Identities = 21/45 (46%), Positives = 26/45 (57%)
Query: 2433 KSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 2477
KS AF VN +AE RG R+V+NYK +N+A Y PNK
Sbjct: 274 KSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNPPNK 318
>UniRef100_Q7XQK4 OSJNBa0017B10.4 protein [Oryza sativa]
Length = 429
Score = 219 bits (558), Expect = 8e-55
Identities = 119/290 (41%), Positives = 175/290 (60%), Gaps = 1/290 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K PI+ + ++ + I E L G IR S+SP F V N +E RG R+VINYK
Sbjct: 71 KTSPIEATPKDIEEFKMHIEELLKLGAIRESRSPHRSAAFIVRNHAEEVRGKSRMVINYK 130
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
LN Y +PNK + I R+ FSKFD+K+G++Q+ + EE TAF P GHY
Sbjct: 131 RLNDNTIEDAYNIPNKQEWINRIQGSKYFSKFDLKAGFWQVKMAEESIEWTAFTCPQGHY 190
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EW VMP GLKNAP+ +Q + +IF F +VY+DD+LVFS+ +H+ HLE F ++
Sbjct: 191 EWLVMPLGLKNAPAIFQRKIQNIFNENQEFILVYIDDLLVFSRTYKEHIAHLEIFFRKVE 250
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
+NGL++S KKM+I + K FLG+EI +G+I + + +FPD + +K +LQ+FLG V
Sbjct: 251 QNGLILSKKKMEICKEKINFLGHEIGEGKIHLQEHIAKKILQFPDAMNDKKKLQQFLGLV 310
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSPPW-SEEMSHSVREVKKLVQSLPIV 1323
NY + I N+ + PL+ +LRKN + + E VR +K+ V+ L ++
Sbjct: 311 NYARNHINNLAKLAGPLYAKLRKNGQRYFNSEDIKLVRLIKERVKELKLL 360
Score = 43.1 bits (100), Expect = 0.11
Identities = 34/94 (36%), Positives = 44/94 (46%), Gaps = 9/94 (9%)
Query: 2390 EKQINFLKEEISYKNIEVQLQQPSV---KSRIENIL--GNIQSSICSCKSSWSCAAFYVN 2444
E +IN + E K ++ + K IE +L G I+ S +S AAF V
Sbjct: 58 ECKINIINPEYIIKTSPIEATPKDIEEFKMHIEELLKLGAIRES----RSPHRSAAFIVR 113
Query: 2445 KQAEIERGTPRLVINYKPLNQALCWIRYPIPNKK 2478
AE RG R+VINYK LN Y IPNK+
Sbjct: 114 NHAEEVRGKSRMVINYKRLNDNTIEDAYNIPNKQ 147
>UniRef100_Q688G8 Putative polyprotein [Oryza sativa]
Length = 1326
Score = 216 bits (551), Expect = 5e-54
Identities = 117/295 (39%), Positives = 175/295 (58%), Gaps = 2/295 (0%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYK 1094
K PI+ + ++ + I E L G IR S+SP F V N +E RG R+V NYK
Sbjct: 862 KTSPIEATPKDIEQFKMHIEELLKLGAIRESRSPHRSAAFIVRNHAEEVRGKSRMVKNYK 921
Query: 1095 PLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHY 1154
LN Y +PNK + I R+ FSKFD+K+G++Q+ + EE TAF P GHY
Sbjct: 922 RLNDNTIEDAYNIPNKQEWINRIQGSKYFSKFDLKTGFWQVKMAEESIEWTAFTCPQGHY 981
Query: 1155 EWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIK 1214
EW VMP GLKNAP+ +Q M +IF F +VY++D+LVFS+ +H+ HLE F ++
Sbjct: 982 EWLVMPLGLKNAPAIFQRKMQNIFNENQEFILVYINDLLVFSRTYKEHIAHLEIFFRKVE 1041
Query: 1215 KNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCV 1274
+NGL++S KKM+I + K FLG+EI +G+I + + +FPD + +K QLQ+FLG V
Sbjct: 1042 QNGLILSKKKMEICKEKINFLGHEIREGKIHLQEHIAKKILQFPDAINDKKQLQQFLGLV 1101
Query: 1275 NYVADFIPNIRIVCAPLFKRLRKNSPPW--SEEMSHSVREVKKLVQSLPIVTPFQ 1327
NY + I N+ + PL+ +LRKN + SE++ + +++ + P+ P +
Sbjct: 1102 NYARNHINNLAKLAGPLYAKLRKNGQRYFNSEDIKLVILIKERVKELKPLELPLE 1156
Score = 40.4 bits (93), Expect = 0.70
Identities = 33/94 (35%), Positives = 43/94 (45%), Gaps = 9/94 (9%)
Query: 2390 EKQINFLKEEISYKNIEVQLQQPSV---KSRIENIL--GNIQSSICSCKSSWSCAAFYVN 2444
E +IN + E K ++ + K IE +L G I+ S +S AAF V
Sbjct: 849 ECKINIINPEYIIKTSPIEATPKDIEQFKMHIEELLKLGAIRES----RSPHRSAAFIVR 904
Query: 2445 KQAEIERGTPRLVINYKPLNQALCWIRYPIPNKK 2478
AE RG R+V NYK LN Y IPNK+
Sbjct: 905 NHAEEVRGKSRMVKNYKRLNDNTIEDAYNIPNKQ 938
>UniRef100_Q7XFN9 Putative retroelement [Oryza sativa]
Length = 1183
Score = 211 bits (537), Expect = 2e-52
Identities = 111/270 (41%), Positives = 164/270 (60%)
Query: 1029 EKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALR 1088
+ ++P ++P + N + ++ + I E L G IR S+SP F V N E RG R
Sbjct: 701 QNNVPYISKPNRSNFEDIEEFKMHIEELLKLGAIRESRSPHRSAAFIVRNHVEEVRGKSR 760
Query: 1089 LVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFV 1148
++INYK LN Y +PNK + I R+ FSKFD+K+G++Q+ + EE TAF
Sbjct: 761 MIINYKRLNDNTIEDAYNIPNKQEWINRIQGSKYFSKFDLKAGFWQVKMAEESIEWTAFT 820
Query: 1149 VPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTIVYLDDVLVFSKGIDQHVQHLEK 1208
P GHYEW VMP GLKNAP+ +Q M +IF F +VY+DD+LVFS+ +H+ HLE
Sbjct: 821 CPQGHYEWLVMPLGLKNAPAIFQRKMQNIFNENQEFILVYIDDLLVFSRTYKEHIAHLEI 880
Query: 1209 FIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQ 1268
F +++N L++S KKM+I + K FLG+EI +G+I + + +FPD + +K +LQ
Sbjct: 881 FFRKVEQNRLILSKKKMEICKEKINFLGHEIGEGKIHLQEHIAKKILQFPDAMNDKKKLQ 940
Query: 1269 RFLGCVNYVADFIPNIRIVCAPLFKRLRKN 1298
+FLG VNY + I N+ + PL+ +L KN
Sbjct: 941 QFLGLVNYARNHINNLAKLAGPLYAKLIKN 970
Score = 38.5 bits (88), Expect = 2.7
Identities = 27/66 (40%), Positives = 33/66 (49%), Gaps = 6/66 (9%)
Query: 2415 KSRIENIL--GNIQSSICSCKSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRY 2472
K IE +L G I+ S +S AAF V E RG R++INYK LN Y
Sbjct: 722 KMHIEELLKLGAIRES----RSPHRSAAFIVRNHVEEVRGKSRMIINYKRLNDNTIEDAY 777
Query: 2473 PIPNKK 2478
IPNK+
Sbjct: 778 NIPNKQ 783
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,057,428,377
Number of Sequences: 2790947
Number of extensions: 178355803
Number of successful extensions: 594091
Number of sequences better than 10.0: 28772
Number of HSP's better than 10.0 without gapping: 1948
Number of HSP's successfully gapped in prelim test: 26837
Number of HSP's that attempted gapping in prelim test: 569064
Number of HSP's gapped (non-prelim): 37220
length of query: 2478
length of database: 848,049,833
effective HSP length: 144
effective length of query: 2334
effective length of database: 446,153,465
effective search space: 1041322187310
effective search space used: 1041322187310
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 84 (37.0 bits)
Lotus: description of TM0283.1


