

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0262.8
(312 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
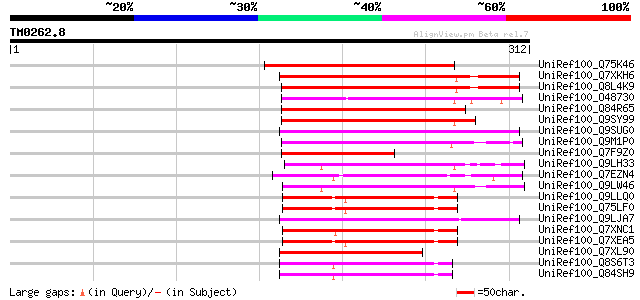
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q75K46 Hypothetical protein OSJNBb0108E08.7 [Oryza sat... 133 5e-30
UniRef100_Q7XKH6 OSJNBb0033P05.11 protein [Oryza sativa] 127 5e-28
UniRef100_Q8L4K9 Hypothetical protein OSJNBa0079H13.17 [Oryza sa... 124 2e-27
UniRef100_O48730 En/Spm-like transposon protein [Arabidopsis tha... 124 4e-27
UniRef100_Q84R65 Hypothetical protein OSJNBb0113I20.11 [Oryza sa... 121 3e-26
UniRef100_Q9SY99 T25B24.14 protein [Arabidopsis thaliana] 118 2e-25
UniRef100_Q9SUG0 Hypothetical protein AT4g08200 [Arabidopsis tha... 108 2e-22
UniRef100_Q9M1P0 Hypothetical protein T18B22.40 [Arabidopsis tha... 107 4e-22
UniRef100_Q7F9Z0 OSJNBa0079C19.27 protein [Oryza sativa] 107 4e-22
UniRef100_Q9LH33 Gb|AAF63124.1 [Arabidopsis thaliana] 106 9e-22
UniRef100_Q7EZN4 OJ1005_B10.22 protein [Oryza sativa] 102 1e-20
UniRef100_Q9LW46 Similarity to En/Spm-like transposon protein [A... 102 1e-20
UniRef100_Q9LLQ0 Hypothetical protein DUPR11.2 [Oryza sativa] 102 2e-20
UniRef100_Q75LF0 Hypothetical protein OSJNBb0079B16.19 [Oryza sa... 101 2e-20
UniRef100_Q9LJA7 Similarity to En/Spm-like transposon protein [A... 101 3e-20
UniRef100_Q7XNC1 OSJNBa0029C04.7 protein [Oryza sativa] 101 3e-20
UniRef100_Q7XEA5 Putative glycine-rich RNA-binding protein [Oryz... 100 5e-20
UniRef100_Q7XL90 OSJNBb0115I09.22 protein [Oryza sativa] 99 2e-19
UniRef100_Q8S6T3 Hypothetical protein OSJNBa0014J14.36 [Oryza sa... 94 5e-18
UniRef100_Q84SH9 Hypothetical protein OJ1058_A12.127 [Oryza sativa] 94 6e-18
>UniRef100_Q75K46 Hypothetical protein OSJNBb0108E08.7 [Oryza sativa]
Length = 767
Score = 133 bits (335), Expect = 5e-30
Identities = 63/114 (55%), Positives = 78/114 (68%)
Query: 154 Y*HAIIFYCLGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSL 213
Y H GKYYLVD+GYP G+L PYK +YHL ++R+ P G+ E FNY HSSL
Sbjct: 609 YTHVFPHSPTGKYYLVDSGYPNRPGYLAPYKGTKYHLQEYRDAPEPQGKEENFNYAHSSL 668
Query: 214 RNVIERSFGCCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFD 267
RNVIERSFG K KW++L +PSYD + Q +II AC ALHNFIR++ D+ FD
Sbjct: 669 RNVIERSFGVLKMKWRMLEKIPSYDPRKQTQIIIACCALHNFIRKSGIRDKHFD 722
>UniRef100_Q7XKH6 OSJNBb0033P05.11 protein [Oryza sativa]
Length = 877
Score = 127 bits (318), Expect = 5e-28
Identities = 72/146 (49%), Positives = 90/146 (61%), Gaps = 6/146 (4%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFG 222
LGK+YLVD+GYP G+L PYK YH ++ GR E FNY HSS RNVIERSFG
Sbjct: 726 LGKFYLVDSGYPNRPGYLAPYKGITYHFHEYNESTLPRGRREHFNYCHSSCRNVIERSFG 785
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFD--ANYENEDGGGENE 280
K KW+IL S+PSY + Q++II AC+ALHNFIR + +D EFD + EN D G
Sbjct: 786 VLKNKWRILFSLPSYSQEKQSRIIHACIALHNFIRDSQMADTEFDNCDHDENYDPLG--- 842
Query: 281 AGPSTVTWEEPDFQSSLQMEQIREHI 306
G S+ + E + S M Q R+ I
Sbjct: 843 -GTSSPSSEPTNELDSRVMNQFRDWI 867
>UniRef100_Q8L4K9 Hypothetical protein OSJNBa0079H13.17 [Oryza sativa]
Length = 916
Score = 124 bits (312), Expect = 2e-27
Identities = 70/145 (48%), Positives = 89/145 (61%), Gaps = 6/145 (4%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
GK+YLVD+GYP G+L PYK YH ++ G+ E FNY HSS RNVIERSFG
Sbjct: 766 GKFYLVDSGYPNRPGYLAPYKGITYHFQEYNESTLPRGKREHFNYCHSSCRNVIERSFGV 825
Query: 224 CKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFD--ANYENEDGGGENEA 281
K KW+IL S+PSY + Q++II AC+ALHNFIR + +D EFD + EN D G
Sbjct: 826 LKNKWRILFSLPSYSQEKQSRIIHACIALHNFIRDSQMADTEFDNCDHDENYDPLG---- 881
Query: 282 GPSTVTWEEPDFQSSLQMEQIREHI 306
G S+ + E + S M Q R+ I
Sbjct: 882 GTSSPSSEPTNELDSRVMNQFRDWI 906
>UniRef100_O48730 En/Spm-like transposon protein [Arabidopsis thaliana]
Length = 292
Score = 124 bits (310), Expect = 4e-27
Identities = 76/166 (45%), Positives = 96/166 (57%), Gaps = 22/166 (13%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQF-RNGPRITGRVEVFNYYHSSLRNVIERSFG 222
GKYYLVD+GYPT G+LGP+++ RYHL QF R GP +T R E+FN HS LR+VIER+FG
Sbjct: 119 GKYYLVDSGYPTRTGYLGPHRRMRYHLGQFGRGGPPVTAR-ELFNRKHSGLRSVIERTFG 177
Query: 223 CCKAKWKILG-SMPSYDLKTQNKIITACMALHNFIRRNDRSDEEF------------DAN 269
KAKW+I+ P Y L KI+TA MALHNFIR + R D +F D
Sbjct: 178 VWKAKWRIVDRKHPKYGLAKWIKIVTATMALHNFIRDSHREDHDFLQWQSIEEYHVDDEE 237
Query: 270 YENEDGG-----GENEAGPSTVTWEEPDFQ--SSLQMEQIREHIKN 308
E E+ G GE E G E ++ ME +R++I N
Sbjct: 238 EEEEEEGEEEEEGEEEEGEGEEEEEHVAYEPTGDRAMEALRDNITN 283
>UniRef100_Q84R65 Hypothetical protein OSJNBb0113I20.11 [Oryza sativa]
Length = 411
Score = 121 bits (303), Expect = 3e-26
Identities = 57/111 (51%), Positives = 71/111 (63%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
GKYYLVDAGYP G+L PYK RYH+P+FR G +G E FN+ H S+R +IER FG
Sbjct: 257 GKYYLVDAGYPNRPGYLAPYKGQRYHVPEFRRGSAPSGVKEKFNFLHCSVRTIIERCFGV 316
Query: 224 CKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFDANYENED 274
K KW++L MPS+ L Q ++ A MALHNFIR ++ D F N D
Sbjct: 317 WKMKWRVLLKMPSFPLWKQKMVVAATMALHNFIRDHNAPDRHFHRFERNPD 367
>UniRef100_Q9SY99 T25B24.14 protein [Arabidopsis thaliana]
Length = 404
Score = 118 bits (295), Expect = 2e-25
Identities = 61/121 (50%), Positives = 80/121 (65%), Gaps = 4/121 (3%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
GKYYLVD+GYPT G+LGP+++ RYHL F G T E+FN HSSLR+VIER+FG
Sbjct: 252 GKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTFGV 311
Query: 224 CKAKWKILG-SMPSYDLKTQNKIITACMALHNFIRRNDRSDEEF---DANYENEDGGGEN 279
KAKW+IL P Y++K KI+T+ MALHN+IR + + D +F + E G EN
Sbjct: 312 WKAKWRILDRKHPKYEVKKWIKIVTSTMALHNYIRDSQQEDSDFRHWEIVESYEQHGDEN 371
Query: 280 E 280
+
Sbjct: 372 D 372
>UniRef100_Q9SUG0 Hypothetical protein AT4g08200 [Arabidopsis thaliana]
Length = 202
Score = 108 bits (270), Expect = 2e-22
Identities = 57/144 (39%), Positives = 83/144 (57%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFG 222
+ KYYLVD+GY G+L Y++++YH F+N E FN HSSLR V ER+F
Sbjct: 50 ISKYYLVDSGYGLHRGYLISYRQSQYHPSHFQNQAPPNNYKEKFNRLHSSLRLVTERTFR 109
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFDANYENEDGGGENEAG 282
K KWKI+ + YD++T K++ MALHNF+R+++ D +F+AN+E +D +
Sbjct: 110 VWKGKWKIMHNRARYDVRTTKKLVVETMALHNFVRKSNILDPDFEANWEQDDNHQPSLKE 169
Query: 283 PSTVTWEEPDFQSSLQMEQIREHI 306
V + F S ME IR+ I
Sbjct: 170 EVEVQDDGQIFDSRQYMEGIRDDI 193
>UniRef100_Q9M1P0 Hypothetical protein T18B22.40 [Arabidopsis thaliana]
Length = 377
Score = 107 bits (267), Expect = 4e-22
Identities = 64/149 (42%), Positives = 83/149 (54%), Gaps = 12/149 (8%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRN-GPRITGRVEVFNYYHSSLRNVIERSFG 222
GK+YLVD GY FL P+++ RYHL FR G + E+FN H+SLRNVIER FG
Sbjct: 221 GKFYLVDCGYANRRKFLAPFRRTRYHLQDFRGQGRDPKTQNELFNLRHASLRNVIERIFG 280
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDE---EFDANYENEDGGGEN 279
K+++ I S P Y KTQ +++ AC LHNF+ + RSDE E N EN D EN
Sbjct: 281 IFKSRFLIFKSAPPYPFKTQTELVLACAGLHNFLHQECRSDEFPPETSINEENSDDNEEN 340
Query: 280 EAGPSTVTWEEPDFQSSLQMEQIREHIKN 308
+EE L+ +Q RE+ N
Sbjct: 341 -------NYEENGDVGILESQQ-REYANN 361
>UniRef100_Q7F9Z0 OSJNBa0079C19.27 protein [Oryza sativa]
Length = 728
Score = 107 bits (267), Expect = 4e-22
Identities = 48/68 (70%), Positives = 56/68 (81%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
GK+YLVD+GYP MGFL PYK +YHLP+FR GPR +G+ EVFN+ HSSLRNVIERSFG
Sbjct: 648 GKFYLVDSGYPNRMGFLAPYKGTKYHLPEFRAGPRPSGKKEVFNHLHSSLRNVIERSFGV 707
Query: 224 CKAKWKIL 231
K KW+IL
Sbjct: 708 LKMKWRIL 715
>UniRef100_Q9LH33 Gb|AAF63124.1 [Arabidopsis thaliana]
Length = 270
Score = 106 bits (264), Expect = 9e-22
Identities = 64/154 (41%), Positives = 84/154 (53%), Gaps = 16/154 (10%)
Query: 166 YYLVDAGYPTFMGFLGPYKKN-----RYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERS 220
YY+VD GYP GFL PY+ + RYH+ QF NGP + E+ N H+SLR+VIER+
Sbjct: 114 YYVVDYGYPNKQGFLAPYRSSQNGVVRYHMSQFYNGPPPRNKQELVNRCHASLRSVIERT 173
Query: 221 FGCCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEF-----DANYENEDG 275
FG KW+IL P YD+ + +++ A M LHNFIR + DE+F D N N +
Sbjct: 174 FGVWMKKWRILCEFPRYDIDVKKRMVMATMGLHNFIRILNYFDEDFVEVMGDTNINNVN- 232
Query: 276 GGENEAGPSTVTWEEPDFQSSLQMEQIREHIKNM 309
EN+ +PD M IRE I NM
Sbjct: 233 -SENDL-DDMERGAKPD---GNHMANIRERIANM 261
>UniRef100_Q7EZN4 OJ1005_B10.22 protein [Oryza sativa]
Length = 305
Score = 102 bits (255), Expect = 1e-20
Identities = 66/154 (42%), Positives = 86/154 (54%), Gaps = 10/154 (6%)
Query: 159 IFYCLGKYYLVDAGYPTFMGFLGPYKKNRYHLPQF--RNGPRITGRVEVFNYYHSSLRNV 216
I + G YYLVDAGY GFL PY+ RYHL +F RN PR E FN H+S RN+
Sbjct: 145 INHLAGCYYLVDAGYTNADGFLAPYRGQRYHLGRFTARNPPR--SAEEYFNMRHASARNI 202
Query: 217 IERSFGCCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFDANYENEDGG 276
+ERSFG K +W IL S + +KTQ +II AC LHN I + SD F+ E+ED
Sbjct: 203 VERSFGRLKGRWAILRSPSYFPIKTQCRIIMACALLHNLILQKMSSD-PFEDEDEDED-- 259
Query: 277 GENEAGPSTVTWE--EPDFQSSLQMEQIREHIKN 308
E+E + E EP+F S++ + +N
Sbjct: 260 -EDEIPLDILEGELAEPEFISAISTSDDWTNFRN 292
>UniRef100_Q9LW46 Similarity to En/Spm-like transposon protein [Arabidopsis thaliana]
Length = 355
Score = 102 bits (255), Expect = 1e-20
Identities = 59/155 (38%), Positives = 86/155 (55%), Gaps = 16/155 (10%)
Query: 165 KYYLVDAGYPTFMGFLGPYKKN-----RYHLPQFRNGPRITGRVEVFNYYHSSLRNVIER 219
KYYLVD+GYP GFL Y+ + RYH+ QF GP + E+FN H+SLR+VIER
Sbjct: 184 KYYLVDSGYPNKQGFLALYRSSQNRVVRYHMSQFYFGPPPRNKHELFNQCHASLRSVIER 243
Query: 220 SFGCCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEF-----DANYENED 274
+FG K KW+IL P Y++ Q +++ A + L+N IR ++ S+ +F +A N D
Sbjct: 244 TFGVWKKKWRILSDFPRYNVHVQKRVVMATVGLYNIIRISNFSNVDFVDVMTEAIINNRD 303
Query: 275 GGGENEAGPSTVTWEEPDFQSSLQMEQIREHIKNM 309
+ + E D M QI+++I NM
Sbjct: 304 FNHDLD------DIEATDIPDGEHMTQIKDNIANM 332
>UniRef100_Q9LLQ0 Hypothetical protein DUPR11.2 [Oryza sativa]
Length = 809
Score = 102 bits (253), Expect = 2e-20
Identities = 57/109 (52%), Positives = 67/109 (61%), Gaps = 7/109 (6%)
Query: 165 KYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRIT---GRVEVFNYYHSSLRNVIERSF 221
KY LVD+GYP+ MGFL PY + RYH QF+ GPR GR E FNY H+ LRN+IER F
Sbjct: 661 KYLLVDSGYPSRMGFLAPYPRVRYHKDQFK-GPRAPPPEGREEKFNYIHAKLRNIIERQF 719
Query: 222 GCCKAKWKILGSMPSYDLK-TQNKIITACMALHNFIRRNDRSDEEFDAN 269
G K +WKIL +P K Q+ II A LHNF R D +F AN
Sbjct: 720 GIVKKQWKILKGIPYNPYKNVQSNIILAAFCLHNF--RIDSKQNDFQAN 766
>UniRef100_Q75LF0 Hypothetical protein OSJNBb0079B16.19 [Oryza sativa]
Length = 693
Score = 101 bits (252), Expect = 2e-20
Identities = 57/109 (52%), Positives = 67/109 (61%), Gaps = 7/109 (6%)
Query: 165 KYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRIT---GRVEVFNYYHSSLRNVIERSF 221
KY LVD+GYP+ MGFL PY + RYH QF+ GPR GR E FNY H+ LRN+IER F
Sbjct: 545 KYLLVDSGYPSRMGFLAPYPRVRYHKDQFK-GPRAPPPEGREEKFNYIHAKLRNIIERQF 603
Query: 222 GCCKAKWKILGSMPSYDLK-TQNKIITACMALHNFIRRNDRSDEEFDAN 269
G K +WKIL +P K Q+ II A LHNF R D +F AN
Sbjct: 604 GIVKKQWKILKGIPYNPYKNVQSDIILAAFCLHNF--RIDSKQNDFQAN 650
>UniRef100_Q9LJA7 Similarity to En/Spm-like transposon protein [Arabidopsis thaliana]
Length = 329
Score = 101 bits (251), Expect = 3e-20
Identities = 57/144 (39%), Positives = 79/144 (54%), Gaps = 1/144 (0%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFG 222
+GKYYLVD+GY G+LGP+++ YH QF+N E FN+ HS LR VIER+FG
Sbjct: 178 IGKYYLVDSGYALRRGYLGPFRQTWYHHNQFQNQAPPNNHKEKFNWRHSLLRCVIERTFG 237
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFDANYENEDGGGENEAG 282
K KW+I+ Y++ T KI+ A MALHNF+ + D +FD ++ +D
Sbjct: 238 VWKGKWRIMQDRAWYNIVTTRKIMVATMALHNFVWKLGIPDLDFDIDW-MQDNDHHPTLD 296
Query: 283 PSTVTWEEPDFQSSLQMEQIREHI 306
T E+ S ME R+ I
Sbjct: 297 DEDETVEQDMTGSRQHMEGFRDEI 320
>UniRef100_Q7XNC1 OSJNBa0029C04.7 protein [Oryza sativa]
Length = 758
Score = 101 bits (251), Expect = 3e-20
Identities = 55/108 (50%), Positives = 66/108 (60%), Gaps = 5/108 (4%)
Query: 165 KYYLVDAGYPTFMGFLGPYKKNRYHLPQFR--NGPRITGRVEVFNYYHSSLRNVIERSFG 222
KY LVD+GYP+ MGFL PY + RYH QF+ + P GR E FNY H+ LRN+IER FG
Sbjct: 610 KYLLVDSGYPSRMGFLAPYPRVRYHKDQFKGPHAPPPEGREEKFNYIHAKLRNIIERQFG 669
Query: 223 CCKAKWKILGSMPSYDLK-TQNKIITACMALHNFIRRNDRSDEEFDAN 269
K +WKIL +P K Q+ II A LHNF R D +F AN
Sbjct: 670 IVKKQWKILKGIPYNPYKNVQSDIILAAFCLHNF--RIDSKQNDFQAN 715
>UniRef100_Q7XEA5 Putative glycine-rich RNA-binding protein [Oryza sativa]
Length = 777
Score = 100 bits (249), Expect = 5e-20
Identities = 56/109 (51%), Positives = 67/109 (61%), Gaps = 7/109 (6%)
Query: 165 KYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRIT---GRVEVFNYYHSSLRNVIERSF 221
KY LVD+GYP+ MGFL PY + RYH QF+ GPR GR E FNY H+ LRN+IER F
Sbjct: 629 KYLLVDSGYPSRMGFLAPYPRVRYHKDQFK-GPRAPPPEGREEKFNYIHAKLRNIIERQF 687
Query: 222 GCCKAKWKILGSMPSYDLK-TQNKIITACMALHNFIRRNDRSDEEFDAN 269
G K +WKIL +P K Q+ II A LHNF R D ++ AN
Sbjct: 688 GIVKKQWKILKGIPYNPYKNVQSDIILAAFCLHNF--RIDSKQNDYQAN 734
>UniRef100_Q7XL90 OSJNBb0115I09.22 protein [Oryza sativa]
Length = 804
Score = 98.6 bits (244), Expect = 2e-19
Identities = 48/86 (55%), Positives = 58/86 (66%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFG 222
LGK+YLVD+GYP G+L PYK YH ++ GR E FNY HSS RNVIERSFG
Sbjct: 719 LGKFYLVDSGYPNRPGYLAPYKGITYHFQEYNESTLPRGRREHFNYCHSSCRNVIERSFG 778
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITA 248
K KW+IL S+PSY + Q++II A
Sbjct: 779 VLKNKWRILFSLPSYSQEKQSRIIHA 804
>UniRef100_Q8S6T3 Hypothetical protein OSJNBa0014J14.36 [Oryza sativa]
Length = 396
Score = 94.0 bits (232), Expect = 5e-18
Identities = 52/107 (48%), Positives = 62/107 (57%), Gaps = 5/107 (4%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQF--RNGPRITGRVEVFNYYHSSLRNVIERS 220
LG+Y LVD+G+PT MGFL PY RYH Q P GR E FN+ HS+LR ++ER
Sbjct: 244 LGRYLLVDSGFPTRMGFLAPYPHVRYHRDQLAVEGAPPPVGREETFNHRHSTLRGIVERQ 303
Query: 221 FGCCKAKWKILGSMPSY-DLKTQNKIITACMALHNFIRRNDRSDEEF 266
FG K WKIL +P Y D +II A ALHNF R D D +
Sbjct: 304 FGIAKKMWKILKEIPYYRDEDIPARIIHAAFALHNF--RLDSKDPTY 348
>UniRef100_Q84SH9 Hypothetical protein OJ1058_A12.127 [Oryza sativa]
Length = 396
Score = 93.6 bits (231), Expect = 6e-18
Identities = 52/107 (48%), Positives = 62/107 (57%), Gaps = 5/107 (4%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQF--RNGPRITGRVEVFNYYHSSLRNVIERS 220
LG+Y LVD+G+PT MGFL PY RYH Q P GR E FN+ HS+LR ++ER
Sbjct: 244 LGRYLLVDSGFPTRMGFLAPYPHVRYHRDQLAVEGAPPPVGREETFNHRHSTLRGIVERQ 303
Query: 221 FGCCKAKWKILGSMPSY-DLKTQNKIITACMALHNFIRRNDRSDEEF 266
FG K WKIL +P Y D +II A ALHNF R D D +
Sbjct: 304 FGIAKKMWKILKEIPYYRDDDIPARIIHAAFALHNF--RLDSKDPTY 348
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.342 0.152 0.496
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 497,144,976
Number of Sequences: 2790947
Number of extensions: 20184716
Number of successful extensions: 62625
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 62430
Number of HSP's gapped (non-prelim): 150
length of query: 312
length of database: 848,049,833
effective HSP length: 127
effective length of query: 185
effective length of database: 493,599,564
effective search space: 91315919340
effective search space used: 91315919340
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0262.8


