

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0262.4
(180 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
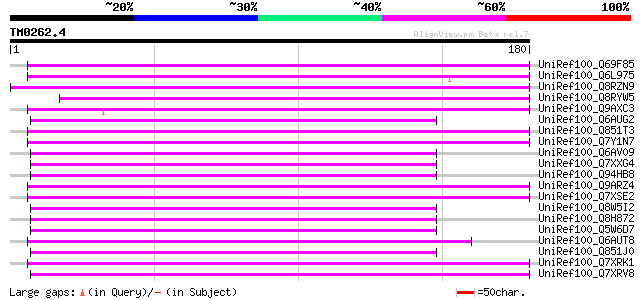
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q69F85 Gag-pol polyprotein [Phaseolus vulgaris] 99 4e-20
UniRef100_Q6L975 GAG-POL [Vitis vinifera] 99 5e-20
UniRef100_Q8RZN9 Putative polyprotein [Oryza sativa] 86 3e-16
UniRef100_Q8RYW5 OSJNBa0066C06.12 protein [Oryza sativa] 85 8e-16
UniRef100_Q9AXC3 Hypothetical protein [Antirrhinum hispanicum] 85 1e-15
UniRef100_Q6AUG2 Putative polyprotein [Oryza sativa] 82 5e-15
UniRef100_Q851T3 Putative gag-pol [Oryza sativa] 81 1e-14
UniRef100_Q7Y1N7 Putative polyprotein [Oryza sativa] 81 1e-14
UniRef100_Q6AV09 Putative polyprotein [Oryza sativa] 81 1e-14
UniRef100_Q7XXG4 OSJNBb0089K24.6 protein [Oryza sativa] 80 2e-14
UniRef100_Q94HB8 Putative retroelement [Oryza sativa] 79 5e-14
UniRef100_Q9ARZ4 Putative polyprotein [Oryza sativa] 78 9e-14
UniRef100_Q7XSE2 OSJNBb0033G08.12 protein [Oryza sativa] 78 9e-14
UniRef100_Q8W5I2 Putative gag-pol [Oryza sativa] 78 9e-14
UniRef100_Q8H872 Putative polyprotein [Oryza sativa] 78 1e-13
UniRef100_Q5W6D7 Putative polyprotein [Oryza sativa] 77 2e-13
UniRef100_Q6AUT8 Putative gag-pol [Oryza sativa] 77 2e-13
UniRef100_Q851J0 Putative GAG-POL [Oryza sativa] 77 2e-13
UniRef100_Q7XRK1 OSJNBa0042D13.9 protein [Oryza sativa] 77 3e-13
UniRef100_Q7XRV8 OSJNBb0049I21.2 protein [Oryza sativa] 77 3e-13
>UniRef100_Q69F85 Gag-pol polyprotein [Phaseolus vulgaris]
Length = 1859
Score = 99.4 bits (246), Expect = 4e-20
Identities = 50/174 (28%), Positives = 85/174 (48%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKG 66
PF+ + +PD + L + Y G TDP +HL F +M + V C++FP+ +
Sbjct: 211 PFTDDIIATPLPDKWRGLTINLYDGSTDPDEHLNIFRTQMTLYTTDRTVWCKVFPTSLRE 270
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYS 126
+ WF+ LP SI F KF Q++ S+ + + L NV+Q E+L+ ++ R+S
Sbjct: 271 GPLGWFSDLPPNSIASFDALELKFTTQYATSRPHRTSSMSLLNVKQERGESLRTFMNRFS 330
Query: 127 AASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
+ I L P +LPG+ L ++ +M E+R +A ++ EE
Sbjct: 331 KVCMNIRNLNPEIAMHHLVSAILPGRFTESLIKRPPCNMDELRTRATKFMQIEE 384
>UniRef100_Q6L975 GAG-POL [Vitis vinifera]
Length = 549
Score = 99.0 bits (245), Expect = 5e-20
Identities = 51/175 (29%), Positives = 92/175 (52%), Gaps = 1/175 (0%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKG 66
PF + + P Y G +DP DH++++ M + +DA+ C++FP+ +G
Sbjct: 5 PFCSHIINYEPPRGFLVPKFSTYDGTSDPFDHIMHYRQLMTLDIGNDALLCKVFPASLQG 64
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYS 126
A++WF LP S+ FRD S F+ Q+ S + I L N++ + E+L+++VKR+
Sbjct: 65 QALSWFHRLPPNSVGNFRDLSEAFVGQYLCSARHKQNISTLQNIKMQDNESLREFVKRFG 124
Query: 127 AASVKIEELEP*ACARAFKDGLLPG-KLNNKLSRKLARSMTEVRAQANTYILDEE 180
A +++E A + FK + PG L++K +M ++ +AN Y + E+
Sbjct: 125 QAVLQVEACSMDAVLQIFKRSICPGTPFFESLAKKPPTTMDDLFRRANKYSMLED 179
>UniRef100_Q8RZN9 Putative polyprotein [Oryza sativa]
Length = 2001
Score = 86.3 bits (212), Expect = 3e-16
Identities = 47/180 (26%), Positives = 86/180 (47%)
Query: 1 AVAEFGPFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMF 60
A+ P S ++ P N K + Y G TDP ++L + + + D K ++
Sbjct: 336 AIDTKNPLSQNLQMAPWPINFKLSNITKYKGDTDPNEYLRVYETAVEAAGGDDTTKAKIL 395
Query: 61 PSMFKGTAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQ 120
P+M +G A++W+TT+P +I + F F + + DDLY ++QL ETL+
Sbjct: 396 PTMLEGVALSWYTTIPPMTIYSWEHMRDTFRAGFIGAYEEPKEADDLYAMKQLPGETLRS 455
Query: 121 YVKRYSAASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
++ ++S +I ++ A K LLPG L L+R ++ ++ + ++ EE
Sbjct: 456 FIVKFSRVRCQIRHVDDEMLIAAAKRALLPGPLRFDLARNRPKTTKDLFERMESFARGEE 515
>UniRef100_Q8RYW5 OSJNBa0066C06.12 protein [Oryza sativa]
Length = 359
Score = 85.1 bits (209), Expect = 8e-16
Identities = 44/163 (26%), Positives = 82/163 (49%)
Query: 18 PDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGTAMAWFTTLPL 77
P N K + Y G DP ++L + + + D VK ++ P+M +G A++W+TT+P
Sbjct: 5 PTNFKLSNITKYRGDIDPNEYLRVYETIVEETGGDDTVKAKILPTMLEGVALSWYTTIPP 64
Query: 78 GSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSAASVKIEELEP 137
+I + F F + + DDLY ++QL ETL+ ++ ++S +I +++
Sbjct: 65 MTIYSWEHMMDTFRAGFVGAYEEPKETDDLYAMKQLPGETLRSFIVKFSRVRCQIRQVDD 124
Query: 138 *ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
A K LLPG L L+R + ++ ++ + ++ EE
Sbjct: 125 EMLIAAAKRALLPGPLRFDLARNIPKTAKDLFERMESFTKGEE 167
>UniRef100_Q9AXC3 Hypothetical protein [Antirrhinum hispanicum]
Length = 1455
Score = 84.7 bits (208), Expect = 1e-15
Identities = 52/177 (29%), Positives = 87/177 (48%), Gaps = 3/177 (1%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSG---GTDPKDHLLYFSMKMVISAASDAVKCRMFPSM 63
PF+ + IP +M+ Y+G DP+DH+ F M I + D C +F S
Sbjct: 156 PFAPHILAPEIPVSMRLSNPVVYTGDRNARDPRDHINQFLAVMDILSTPDHALCWIFRST 215
Query: 64 FKGTAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVK 123
G A AWF+ LP GSI F + F+ QF+ ++ TI L + + E E+L++Y++
Sbjct: 216 LTGRAQAWFSQLPRGSIRSFDELRELFVRQFACNRRYPKTIGHLMTMVEAEGESLREYMR 275
Query: 124 RYSAASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
R++ + + + P + G + N L + ARS+ E+ +A Y+L E+
Sbjct: 276 RFANTVIDVRHVGPHILSEFVAQGTRSVEFANSLVGRPARSLEELMHRAERYMLQED 332
>UniRef100_Q6AUG2 Putative polyprotein [Oryza sativa]
Length = 1194
Score = 82.4 bits (202), Expect = 5e-15
Identities = 44/141 (31%), Positives = 71/141 (50%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
F +R+V P+ + A+E Y G TDPK+ L +S + + A D P+ KG+
Sbjct: 434 FGRSLRDVRWPERFRPGAIEKYDGSTDPKEFLQVYSTVLYAAGADDNALANYLPTALKGS 493
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP SI + D +F+ F + + DDL+ + Q E+L++YV+R +
Sbjct: 494 ARSWLMHLPPYSISSWADLRQQFIANFQGTYKRHAIEDDLHALTQNSGESLREYVRRLNE 553
Query: 128 ASVKIEELEP*ACARAFKDGL 148
I E+ + RAFK G+
Sbjct: 554 CRNTIPEITDASVIRAFKSGV 574
>UniRef100_Q851T3 Putative gag-pol [Oryza sativa]
Length = 971
Score = 81.3 bits (199), Expect = 1e-14
Identities = 44/174 (25%), Positives = 83/174 (47%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKG 66
P S ++ IP K L + YSG TDPK+ L + + + + K ++ G
Sbjct: 357 PLSGGIKNRPIPPQFKFLPVPRYSGETDPKEFLSIYESAIEAAHGDENTKAKVIHLALDG 416
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYS 126
A +W+ LP SI + F++ F + + T L +RQ E++++Y++R+S
Sbjct: 417 IARSWYFNLPANSIYSWEQLRDVFVLNFRGTYEEPKTQQHLLGIRQRPGESIREYMRRFS 476
Query: 127 AASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
A ++E++ + A GLL G+L K++ K +++ + + + EE
Sbjct: 477 QARCQVEDITEASVINAASAGLLEGELTRKIANKEPQTLEHLVRIIDGFAKSEE 530
>UniRef100_Q7Y1N7 Putative polyprotein [Oryza sativa]
Length = 518
Score = 81.3 bits (199), Expect = 1e-14
Identities = 45/174 (25%), Positives = 83/174 (46%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKG 66
P S ++ IP K + YSG TD K+ L + + + + K ++ + G
Sbjct: 282 PLSWGIKNHPIPPQFKYPPVPQYSGETDSKEFLSIYESAIEAAHGDENTKAKLIHLVLDG 341
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYS 126
A +W+ LP+ SI + F++ F + + T DL + Q + E+ ++Y++R+
Sbjct: 342 IARSWYFNLPVNSIYSWEQLRDVFVLNFRGTYEEPKTQQDLVGIHQRQGESTREYIRRFL 401
Query: 127 AASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
A +++++ + A GLL G+L KL RK R++ + + Y DEE
Sbjct: 402 QARCQVQDITEASVIHAASAGLLEGELTRKLVRKEPRTLEHLFRIIDGYARDEE 455
>UniRef100_Q6AV09 Putative polyprotein [Oryza sativa]
Length = 1551
Score = 80.9 bits (198), Expect = 1e-14
Identities = 43/141 (30%), Positives = 72/141 (50%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
F +R+V P+ ++ A+E Y G TDP++ L +S + + A D P+ KG+
Sbjct: 379 FGRSLRDVRWPERLRPGAIEKYDGSTDPEEFLQVYSTVLYAAGADDNALANYLPTALKGS 438
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP SI + D +F+ F + + DDL+ + Q E+L++YV+R++
Sbjct: 439 ARSWLMHLPPYSISSWIDLWQQFVANFQGTYKRHAIEDDLHTLTQNSGESLREYVRRFNE 498
Query: 128 ASVKIEELEP*ACARAFKDGL 148
I E+ RAFK G+
Sbjct: 499 CRNTIPEITDAFVIRAFKSGV 519
>UniRef100_Q7XXG4 OSJNBb0089K24.6 protein [Oryza sativa]
Length = 1400
Score = 80.1 bits (196), Expect = 2e-14
Identities = 43/141 (30%), Positives = 71/141 (49%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
FS +R+V P+ + A+E Y G TDP++ L +S + + A D P+ KG
Sbjct: 429 FSRSLRDVRWPERFRPRAIEKYDGSTDPEEFLQVYSTVLYAAGADDNALANYLPTALKGY 488
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP SI + D +F+ F + + DDL+ + Q E+L++YV+R++
Sbjct: 489 ARSWLMHLPPYSISSWADLWQQFVANFQGTYKRHAIEDDLHALTQNSGESLREYVRRFNE 548
Query: 128 ASVKIEELEP*ACARAFKDGL 148
I E+ + RA K G+
Sbjct: 549 CRNTIPEITDASVIRALKSGV 569
>UniRef100_Q94HB8 Putative retroelement [Oryza sativa]
Length = 2079
Score = 79.0 bits (193), Expect = 5e-14
Identities = 42/141 (29%), Positives = 71/141 (49%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
F +R+V P+ + A+E Y G DP++ L +S + + A D P+ KG+
Sbjct: 565 FGRSLRDVRWPERFRPGAIEKYDGSIDPEEFLQVYSTVLYAAGADDNAVANYLPTALKGS 624
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP SI + D +F+ F + + DDL+ + Q E+L++YV+R++
Sbjct: 625 ARSWLMHLPPYSISSWADLWQQFVANFQGTYKRHAIEDDLHALTQNSGESLREYVRRFNE 684
Query: 128 ASVKIEELEP*ACARAFKDGL 148
I E+ + RAFK G+
Sbjct: 685 CRNTIPEITDASVIRAFKSGV 705
>UniRef100_Q9ARZ4 Putative polyprotein [Oryza sativa]
Length = 1579
Score = 78.2 bits (191), Expect = 9e-14
Identities = 42/174 (24%), Positives = 83/174 (47%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKG 66
P S ++ IP K + YSG TDPK+ L + + + + K ++ G
Sbjct: 49 PLSGGIKTRPIPPQFKFPPVPCYSGETDPKEFLSIYESAIEAAHGDENTKAKVIHLTLDG 108
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYS 126
A +W+ LP SI + F++ F + + T L +RQ E++++Y++R+S
Sbjct: 109 IARSWYFNLPANSIYSWEQLRDVFVLNFRGTYEEPKTQQHLLGIRQRPGESIREYMRRFS 168
Query: 127 AASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
A +++++ + A GLL G+L K++ K +++ ++ + + EE
Sbjct: 169 QARCQVQDITEASVINAASAGLLEGELTRKIANKEPQTLKQLLRIIDGFARGEE 222
>UniRef100_Q7XSE2 OSJNBb0033G08.12 protein [Oryza sativa]
Length = 1847
Score = 78.2 bits (191), Expect = 9e-14
Identities = 42/174 (24%), Positives = 83/174 (47%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKG 66
P S ++ IP K + YSG TDPK+ L + + + + K ++ G
Sbjct: 417 PLSGGIKTRPIPPQFKFPPVPRYSGETDPKEFLSIYESAIEAAHGDENTKAKVIHLALDG 476
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYS 126
A +W+ LP SI + F++ F + + T L +RQ + E++++Y++R+S
Sbjct: 477 IARSWYFNLPANSIYSWEQLRDVFVLNFRGTYKEPKTQQHLLGIRQKQGESIREYMRRFS 536
Query: 127 AASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
A +++++ + A GLL G+L K++ K +++ + + + EE
Sbjct: 537 QARCQVQDITKASVINAASAGLLEGELTRKIANKEPQTLEHLLRIIDGFARGEE 590
>UniRef100_Q8W5I2 Putative gag-pol [Oryza sativa]
Length = 698
Score = 78.2 bits (191), Expect = 9e-14
Identities = 41/141 (29%), Positives = 71/141 (50%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
F +R+V P+ + A++ Y G TDP++ L +S + + A D P+ KG+
Sbjct: 202 FGRSLRDVRWPERFRPGAIKKYDGSTDPEEFLQVYSTVLYAAGADDNALANYLPTALKGS 261
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP SI + D +F+ F + + DDL+ + Q E+L++YV+ ++
Sbjct: 262 ARSWLMHLPPYSISSWADLWQQFIANFQGTYKRHAIEDDLHALTQNSGESLREYVRHFNE 321
Query: 128 ASVKIEELEP*ACARAFKDGL 148
I E+ + RAFK G+
Sbjct: 322 CRNTIPEITDASVIRAFKSGV 342
>UniRef100_Q8H872 Putative polyprotein [Oryza sativa]
Length = 1469
Score = 77.8 bits (190), Expect = 1e-13
Identities = 41/141 (29%), Positives = 70/141 (49%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
F +R+V P+ + A++ Y G TDP++ L +S + + A D P+ KG+
Sbjct: 329 FGRSLRDVRWPERFRPRAIKKYDGSTDPEEFLQVYSTVLYAAGADDNALANYLPTALKGS 388
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP SI + D +F+ F + DDL+ + Q E+L++YV+R++
Sbjct: 389 ARSWLMHLPPYSISSWADLWQQFVANFQGTYKHHAIEDDLHALTQNSGESLREYVRRFNE 448
Query: 128 ASVKIEELEP*ACARAFKDGL 148
I E+ + R FK G+
Sbjct: 449 CRNTIPEITDSSVIRTFKSGV 469
>UniRef100_Q5W6D7 Putative polyprotein [Oryza sativa]
Length = 1185
Score = 77.4 bits (189), Expect = 2e-13
Identities = 43/141 (30%), Positives = 72/141 (50%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
F+ +R+V P+ + A+E Y G TDP++ L +S + A D P+ KG+
Sbjct: 310 FNRSLRDVRWPERFRPGAIEKYDGSTDPEEFLQVYSTVPYAAGADDNALANYLPTALKGS 369
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP SI + D +F+ F + + + DDL+ + Q E+L+ YV+R++
Sbjct: 370 ARSWLMHLPPYSISSWADLWQQFVGNFQGTYKRHMIEDDLHALTQNPGESLRDYVRRFND 429
Query: 128 ASVKIEELEP*ACARAFKDGL 148
I E+ + RAFK G+
Sbjct: 430 CRNTIPEITDASVIRAFKTGV 450
>UniRef100_Q6AUT8 Putative gag-pol [Oryza sativa]
Length = 1631
Score = 77.4 bits (189), Expect = 2e-13
Identities = 40/154 (25%), Positives = 75/154 (47%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKG 66
P S ++ IP K L + YSG TDPK+ L + + + + +K ++ G
Sbjct: 335 PLSGGIKTRPIPPQFKFLLVPRYSGETDPKEFLSIYESAIEAAHGDENIKAKVIHLALDG 394
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYS 126
A +W+ L SI + F++ F + + T L +RQ E++++Y++R+S
Sbjct: 395 IARSWYFNLLANSIYSWEQLHDSFVLNFRGTYEEPKTQQHLLGIRQRPGESIREYMRRFS 454
Query: 127 AASVKIEELEP*ACARAFKDGLLPGKLNNKLSRK 160
A +++++ + A GLL G+L K++ K
Sbjct: 455 QARCQVQDITEASVINAASAGLLEGELTRKIANK 488
>UniRef100_Q851J0 Putative GAG-POL [Oryza sativa]
Length = 831
Score = 77.0 bits (188), Expect = 2e-13
Identities = 43/141 (30%), Positives = 71/141 (49%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
FS +R+V P+ + A+E Y G T+P++ L +S + + A D+ P+ KG+
Sbjct: 337 FSRSLRDVRWPERFRPGAIEKYDGSTNPEEFLQVYSTVLYAAGADDSALVNYLPTALKGS 396
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP SI + D +F+ F + + DDL+ + Q E+L YV R++
Sbjct: 397 ARSWLMHLPPYSISLWADLWQQFVANFQGTYKRHAIEDDLHALTQNPGESLSDYVWRFNE 456
Query: 128 ASVKIEELEP*ACARAFKDGL 148
I E+ + RAFK G+
Sbjct: 457 CRNTIPEITDASVIRAFKSGV 477
>UniRef100_Q7XRK1 OSJNBa0042D13.9 protein [Oryza sativa]
Length = 303
Score = 76.6 bits (187), Expect = 3e-13
Identities = 42/174 (24%), Positives = 82/174 (46%)
Query: 7 PFSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKG 66
P S ++ IP K + YSG TDPK+ L + + + + K ++ G
Sbjct: 49 PLSGGIKTRPIPPQFKFPPVSRYSGETDPKEFLSIYESAIEAAHGDENTKAKVIHLALDG 108
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYS 126
A +W+ LP SI + F++ F + + T L +RQ E++++Y++R+S
Sbjct: 109 IARSWYFNLPANSIYSWEQLRDVFVLNFRGTYEEPKTQQHLLGIRQRPGESIREYMRRFS 168
Query: 127 AASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
A +++++ + A GLL G+L K++ K +++ + + + EE
Sbjct: 169 QARCQVQDITEASVINAALAGLLEGELTKKIANKEPQTLEHLLRIIDGFARGEE 222
>UniRef100_Q7XRV8 OSJNBb0049I21.2 protein [Oryza sativa]
Length = 1843
Score = 76.6 bits (187), Expect = 3e-13
Identities = 46/173 (26%), Positives = 79/173 (45%)
Query: 8 FSAVVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGT 67
F+ +R V P K +E Y G T+P+ L + + + + P +
Sbjct: 427 FTDDLRRVDWPAGFKPTGIEKYDGTTNPESWLTVYGLAIRAAGGDSKAMANYLPVALADS 486
Query: 68 AMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNVRQLERETLKQYVKRYSA 127
A +W LP G+I + + +F+ F + + T DLYNV Q E+L++Y++R+S
Sbjct: 487 ARSWLHGLPRGTIGSWAELRDRFIANFQGTFERPGTQYDLYNVIQKSGESLREYIRRFSE 546
Query: 128 ASVKIEELEP*ACARAFKDGLLPGKLNNKLSRKLARSMTEVRAQANTYILDEE 180
KI ++ AF G+ +L K RK R++ + +AN Y E+
Sbjct: 547 QRNKISDITDDVIIAAFTKGIRHEELVGKFGRKPPRTVKLMFEKANEYAKAED 599
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.135 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 257,101,518
Number of Sequences: 2790947
Number of extensions: 8784274
Number of successful extensions: 29413
Number of sequences better than 10.0: 487
Number of HSP's better than 10.0 without gapping: 446
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 28824
Number of HSP's gapped (non-prelim): 572
length of query: 180
length of database: 848,049,833
effective HSP length: 119
effective length of query: 61
effective length of database: 515,927,140
effective search space: 31471555540
effective search space used: 31471555540
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0262.4


